Abstract
The maize Myb transcription factor C1 depends on the basic helix–loop–helix (bHLH) proteins R or B for regulatory function, but the closely related Myb protein P does not. We have used the similarity between the Myb domains of C1 and P to identify residues that specify the interaction between the Myb domain of C1 and the N-terminal region of R. Substitution of four predicted solvent-exposed residues in the first helix of the second Myb repeat of P with corresponding residues from C1 is sufficient to confer on P the ability to physically interact with R. However, two additional Myb domain amino acid changes are needed to make the P regulatory activity partially dependent on R in maize cells. Interestingly, when P is altered so that it interacts with R, it can activate the Bz1 promoter, normally regulated by C1 + R but not by P. Together, these findings demonstrate that the change of a few amino acids within highly similar Myb domains can mediate differential interactions with a transcriptional coregulator that plays a central role in the regulatory specificity of C1, and that Myb domains play important roles in combinatorial transcriptional regulation.
Combinatorial interactions between transcription factors are of central importance to regulation of gene expression in eukaryotes. These interactions can either modulate transcription factor activity or contribute to the biological specificity of factors with very similar DNA-interaction motifs. Elucidation of the mechanisms by which proteins with very similar DNA-binding domains achieve regulatory specificity remains a fundamental question in biology today.
Proteins containing the Myb-homologous DNA-binding domain are widespread in eukaryotes (reviewed in refs. 1 and 2). The vertebrate c-myb gene plays an essential regulatory role in the proliferation and differentiation of hematopoietic cells. Besides c-myb, at least two other myb-related genes (A-myb and B-myb) are present in vertebrates (3). The products of these genes have Myb domains, each consisting of three head-to-tail Myb motifs (R1, R2, and R3). Oncogenic versions of c-myb, such as v-myb, contain only R2 and R3, as do hundreds of plant Myb-domain proteins (4). Myb domains formed by the R2 and R3 Myb motifs bind DNA. Each Myb motif contains three α-helices, and the third helix of each Myb motif makes sequence-specific DNA contacts. The second and third helices of each Myb motif form a helix–turn–helix structure when bound to DNA, similar to motifs found in the λ repressor and in homeo domains (5). In addition to their well-established roles in DNA binding, Myb domains are also emerging as important protein–protein interaction motifs. These Myb domain-mediated protein–protein interactions play key roles in the biological specificity of the corresponding factors (6–13). However, the mechanisms by which protein–protein interactions contribute to the regulatory specificity of Myb domain proteins are poorly understood.
In flowering plants, several hundred genes containing the conserved Myb DNA-binding domain have been identified, which now makes the Myb proteins the largest described family of transcription factors. Most of them correspond to the R2R3 Myb gene family, characterized by the presence of Myb domains with just the R2 and R3 Myb repeats (14, 15). However, a small number of plant genes encoding proteins with three Myb repeats, closely related to those present in animals, have also been found (16).
The control of flavonoid biosynthesis by the maize R2R3 Myb domain proteins C1 and P is an excellent system to investigate how regulatory specificity by Myb domain proteins is achieved through combinatorial interactions with additional regulatory proteins. Anthocyanin accumulation in maize is controlled by two classes of regulatory proteins acting in concert: those with a Myb domain [C1 or Pl, two closely related homologs (17)], and those with a basic helix–loop–helix (bHLH) domain [R or B, members of the R/B family (18)]. Extensive studies have shown that the myb-homologous C1 or Pl genes (do not confuse Pl with P) require a member of the bHLH-containing R or B gene family to activate transcription of the anthocyanin biosynthetic genes (reviewed in ref. 19). Indeed, the R/B- and C1-encoded proteins physically interact, and this interaction is mediated by the Myb domain of C1 and the N-terminal region of B (13). The maize P gene controls the accumulation of 3-deoxy flavonoids and red phlobaphene pigments by activating a subset of the anthocyanin biosynthetic genes controlled by C1 and R/B. P and C1 activate the expression of some common genes in the flavonoid biosynthetic pathway such as A1, by interacting with different affinities to identical cis-acting regulatory elements in the A1 gene promoter (20, 21). In contrast, C1, but not P, binds and activates transcription of the Bz1 gene, specific for anthocyanin biosynthesis (20, 21). Furthermore, the ability of P to activate gene expression is independent of the R/B coactivators, despite the fact that the Myb domains of P and C1 are over 70% identical (20). Thus, the interaction of C1 or PL with R or B is very specific, providing these two Myb proteins with unique regulatory activities.
Here, we have used chimeric Myb domains of P and C1 to determine which residues specify the interaction of C1 with the bHLH coactivator R. We show that residues in the first helix of the R3 Myb repeat of C1 are sufficient for the specificity of this interaction. Replacement of four solvent-exposed residues in the Myb domain of P with the corresponding residues in the Myb domain of C1 is sufficient to transfer the interaction with R to P in yeast two-hybrid experiments. The replacement of six residues in the R3 Myb repeat of P for the corresponding residues of C1 allows R to enhance the regulatory activity of P in maize cells. In addition, when P is altered in this way, it can now activate the Bz1 promoter, normally regulated only by C1 + R. Together, these findings identify amino acid residues that allow Myb domains to act as efficient protein–protein interaction motifs that help confer unique regulatory specificity to closely related transcription factors containing Myb domains.
Materials and Methods
Plasmids Used in Transient Expression Experiments.
All P and C1 plant expression vectors include the CaMV 35S promoter, the tobacco mosaic virus (TMV) Ω′ leader and maize first Adh1-S intron in the 5′ untranslated region (5′UTR), and potato proteinase II (pinII) termination signal. Previously described plasmids (21, 22) include pBz1Luc, pA1Luc, and PPP (35S∷P-cDNA1) corresponding to plasmid pPHP1962; CCC (35S∷C1) (pPHP665); PPC (35S∷Pmyb-C1act) (pPHP4884) and R (35S∷R) (pPHP471). 35S∷BAR (pPHP611) was used for normalizing the concentration of 35S sequences delivered in each bombardment (23). Ubi∷GUS (pPHP3953) was used to normalize the efficiency of each bombardment (24). Standard site-directed mutagenesis procedures were used to modify the amino acid sequence in the following constructs: CCP (35S∷C1myb-Pact) (pPHP4885) was obtained by introducing a translationally silent SnaBI restriction site at the 3′ end of the Myb region at Y119 in P and blunt-end ligating the non-Myb region of P to the Myb region of C1 at a PvuII site at G124. CPP (35S∷C1R2-PmybR3-Pact) (pPHP6186) was obtained by introducing a silent AatII restriction site between the R2 and R3 Myb motifs at P63 in C1 (see Fig. 2A) and ligating to the AatII site in P at D64. PCP (35S∷PR2-C1R3-Pact) (pPHP6194 or pPHP7663) was obtained by fusing PR2 to C1R3 using the AatII sites described in CPP and fusing C1R3 to the non-Myb region of P using the SnaBI/PvuII sites described in CCP. CPC:65–84P (35S∷C1R2-C1R3h1-PR3h2-Pact) (pPHP7726) was obtained by introducing a silent BstEII restriction sites at the turn between the first helix of the R3 Myb motif and the second helix of the R3 Myb motif at L85 in C1 and P. The upstream C1 regions were fused to the downstream P regions at the BstEII sites to create the R3 Myb motif chimera.
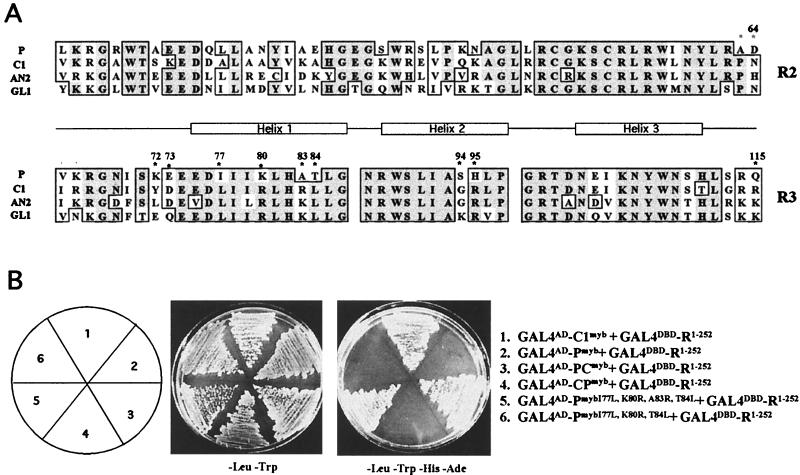
Figure 2.
Myb domain sequences that contribute to the specificity of the interaction with the bHLH cofactor R. (A) Sequence comparison between the Myb domains of P (25), and other proteins shown to interact with R, including C1 (37), AN2 (32), and GL1 (38). The position of the three α-helices that form each Myb repeat are marked, with helix 3 of each motif involved in DNA interaction. Residue numbers are based on the sequences of P and C1. Dark shading indicates identical residues, light shading indicates conservative changes. Residues focused on in this study are marked with asterisks. (B) Yeast two-hybrid interactions of the Myb domains of C1, P, or mutant versions of the Myb domain of P fused to the Gal4 activation domain (GAL4AD) with the N-terminal 252 amino acids of R fused to the GAL4 DNA-binding domain (GALDBD). The simultaneous change of the I77, K80, A83, and T84 residues in P for the corresponding residues of C1 (Gal4AD-PmybI77L,K80R,A83R,T84L) allow P to interact with R.
Plasmids Used in Yeast Two-Hybrid Experiments.
Myb domains were generated by PCR and then cloned into the pAD-GAL4 vector (Stratagene) as EcoRI/SalI fragments. The long P cDNA (25) was used to obtain the wild-type and mutant P Myb domain (Pmyb). Wild-type Pmyb was generated by using primers p5pAD, which includes the first 7 amino acids of P with EcoRI and NcoI sites in the 5′ end, and p3pET, which includes amino acids 114–116 of P (25) with SnaBI, SalI, and BamHI sites in the 5′ end. Mutant Pmyb were generated by ligating two independent PCR fragments at an AgeI site, which was engineered as a silent mutation at amino acids 87–88 from P, changing these codons from AAC AGG to AAC CGG (25), and then amplifying the ligation product with p5pAD and p3pET. All of the resulting PCR products were subsequently cloned into pAD-GAL4 and sequenced. C1 Myb domains (C1myb) were generated from plasmid pPHP687 (20). Wild-type C1 Myb domain was generated by using primers C1N1, which corresponds to amino acids 2–8 with an XhoI and a EcoRI at the 5′ end, and C13pET, which corresponds to amino acids 113–118 with SnaBI, SalI, and BamHI sites at the 5′ end. R1–252 was generated by PCR using plasmid DNA pPHP687 as template and primers LcN1, which corresponds to amino acids 1–7 with a XhoI site at the 5′ end and an EcoRI and a GCG codon inserted between amino acids 1 and 2, and LcC1, which corresponds to amino acids 142–151 with a BamHI site at the 5′ end and a SalI site resulting from a mutation that changes amino acid 148 from V to D. After sequencing, the insert was cloned as an EcoRI/SalI fragment into pBD-GAL4 (Stratagene).
Microprojectile Bombardment and Gene Expression Experiments.
Bombardment conditions of suspension cells and transient expression assays for luciferase and β-glucuronidase (GUS) were performed as previously described (20). For each microprojectile preparation, the mass of DNA was adjusted to 10 μg with 35∷BAR (23) to equalize the amount of 35S promoter in each bombardment. One microgram of each of the regulators and 3 μg of reporter plasmid (pA1Luc or pBz1Luc) were used in each bombardment. To normalize luciferase activity to GUS activity, 3 μg of UBI∷GUS was included in every bombardment. Each treatment was done in triplicate, and entire experiments were repeated at least twice. The assays for luciferase and GUS and the normalization of the data were done as described (20). Data are expressed as the ratio of arbitrary light units (luciferase) to arbitrary units of fluorescence (GUS).
Yeast Two-Hybrid Experiments.
The plasmid containing R1–252 in the pBDGal4 (TRP+) vector and each of the constructs containing the mutant Myb domains of P or C1 cloned into pAD-GAL4 (LEU+) vector were cotransformed into yeast strain PJ69-4A with the genotype Mat a trp1–901 leu2–3, 112 ura3–52 his3–200 gal 4Δ gal 80Δ LYS2∷GAL1-HIS3 GAL2-ADE2 met∷GAL7-lacz (26) and plated on SC-LEU-TRP medium. Colonies were screened for growth on SC-LEU-TRP, SC-LEU-TRP-HIS, and SC-LEU-TRP-HIS-ADE.
Results
Regulatory Activity of Chimeric Myb Domains.
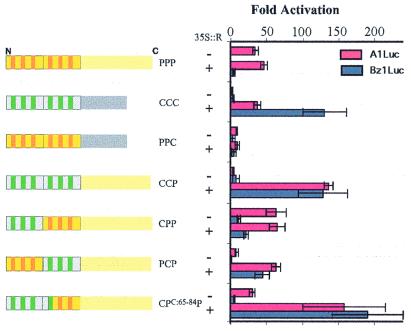
To identify regions in the Myb domain of C1 required for the specificity of the interaction with R, chimeras between P and C1 were generated. Transcriptional activation by P/C1 chimeras was assayed by transient expression in maize callus cells in the presence and absence of R. Two luciferase reporter constructs containing promoters from different flavonoid biosynthetic promoters were used: A1 (activated by C1 + R and P) and Bz1 (activated by C1 + R, but not by P). In Fig. 1, chimeric proteins are named according to the origin of the R2, R3, and C-terminal regions. For example, PCP corresponds to a protein containing the R2 Myb motif of P, the R3 Myb motif from C1, and the C-terminal region of P. As previously shown (20), P (PPP, Fig. 1) activates transcription of the A1 promoter independently of R, but fails to activate Bz1, either with or without R. C1 (CCC, Fig. 1) does not activate A1 or Bz1 expression alone, but the presence of R makes C1 a potent activator of these two promoters (20). A chimeric protein containing the Myb domain of P and the C-terminal region of C1 (PPC, Fig. 1) activates the A1 promoter very poorly, albeit in an R-independent fashion, and fails to activate Bz1, in the presence or absence of R. The chimera consisting of the Myb domain of C1 fused to the C-terminal non-Myb region of P (CCP, Fig. 1) is inactive on either A1 or Bz1 in the absence of R, but with R activates both of these promoters. These results demonstrate that, as previously shown with B (13), the Myb domain of C1 is the region that mediates the interaction with R.
Figure 1.
Activation of the A1 and Bz1 promoters. Results of transient expression after cobombardment of cultured maize cells with different chimeras of P and C1 together with A1Luc (red) or Bz1Luc (blue) reporter constructs, in the absence (−) or presence (+) of a vector that expresses R from the constitutive CaMV 35S promoter. Sequences derived from P are shown in yellow, with the helices characteristic of each of the two Myb repeats (R2 and R3) in orange. Sequences derived from C1 are shown in gray, with the helices in green. The first two letters indicate the constitution of the Myb domain, the first indicating the constitution of R2, the second of R3; and the last indicates the origin of the C-terminal region containing the transcriptional activation motif. A UBI∷GUS construct was included in every bombardment as a normalization control. Each treatment was done in triplicate, and the data were normalized for GUS activity as described (20). The fold activation was calculated as the ratio between each particular treatment and the treatment with pA1Luc or pBz1Luc constructs without activator. The average values are shown, and the error bars indicate the standard deviation of the samples.
To further identify the region within the Myb domain of C1 that makes transcription by C1 R dependent, chimeras between the Myb domains of C1 and P were analyzed. A chimeric protein in which the R2 Myb repeat of P was replaced by the corresponding region of C1 (CPP, Fig. 1) activates the A1 promoter in the absence of R. Unlike P, CPP also weakly activates the Bz1 promoter (compare PPP and CPP, Fig. 1), although the activity of CPP was not significantly increased by the presence of R. In contrast, a chimeric factor containing the R2 Myb repeat of P fused to the R3 Myb repeat of C1 (PCP) was unable to effectively activate A1 or Bz1 in the absence of R. However, the presence of R increased transcriptional activation over 40-fold on both the A1 and the Bz1 promoters (Fig. 1).
The above results suggest that either the R3 repeat, the linker between the R2-R3 repeats (residues 63–66, Fig. 2A), or both were mediating the interaction with R. The linker region plays an important role in the DNA-binding activity of Myb domains (27, 28). Thus, it was possible that the different linker regions in P and C1 influenced their independence or dependence on R for transcriptional activation, respectively. To test this hypothesis, the PCP construct used in the experiment shown in Fig. 1, which contains the amino acid sequence linker characteristic of C1 (PNIR, see Materials and Methods), was compared with a construct identical to PCP but with the amino acid sequence of the linker region corresponding to P (ADVK). The PCP protein with the ADVK linker region activated A1 10.3 ± 1-fold without R, and 50.3 ± 12.4-fold with R, and Bz1 1.5 ± 1-fold without R, and 73.8 ± 12.2-fold with R. These results were very similar to those shown for PCP in Fig. 1, suggesting that the linker region between R2 and R3 does not contribute to R-dependent or R-independent transcription. These findings indicate that only the R3 Myb repeat of C1 (residues 67–115, Fig. 2A) is responsible for the inability of C1 to activate transcription on its own, and that the same R3 Myb repeat mediates the specificity of the interaction with R, which is required as a cofactor for activation.
To determine which residues in the R3 Myb repeat of C1 are responsible for the functional dependence of C1 on R, residues 65 to 84 in the CPP chimera were replaced by the corresponding region of C1 (CPC:65–84P, Fig. 1). In contrast with CPP, activation by CPC:65–84P was increased by R, on both the A1 as well as the Bz1 promoters. Thus, the specificity of the interaction between C1 and R is provided by a region of the R3 Myb repeat of C1 between residues 67 and 84 (Fig. 2A). Interestingly, PPP, CPP, and CPC1:65–84P (in contrast to CCC, CCP, or PCP) can each activate the A1 promoter without R. These results indicate that a region of the Myb domain of P between amino acid 85 and the end of the Myb domain allows transcription activation in an R-independent fashion. Together, these data show that the R3 Myb repeat of C1 (residues 67–115) contains a sequence that allows C1 to interact with R (within residues 67–84), as well as a region that makes C1 transcription R dependent (within residues 84–115), suggesting that these two activities map to separate regions in R3.
A comparison of the Myb domain of C1 with the Myb domains of two other proteins that interact with R and that show R-dependent activity reveals a high level of conservation in the R3 Myb repeat, further limiting the potential functionally relevant residues (Fig. 2A). The Arabidopsis GL1 protein interacts with R in coprecipitation (29) and yeast two-hybrid experiments (30), and a model has been proposed in which the interactions between GL1 (and similar proteins) with bHLH cofactors is essential for trichome and root hair formation (31). The Petunia AN2 protein is the C1 ortholog that also requires a bHLH protein for flower pigmentation (19, 32) and physically interacts with R in yeast two-hybrid experiments (Kroon, Koes, E.G., and Mol, unpublished results).
Four Residues Are Sufficient to Transfer the Interaction with R from C1 to P.
To determine whether R-dependent activation of transcription by the P and C1 chimeras tested in Fig. 1 reflect the ability of these proteins to physically interact, we conducted yeast two-hybrid experiments with the P/C1 chimeric Myb domains fused to the GAL4 activation domain (GAL4AD). If these proteins were able to interact with a protein containing the first 252 amino acids of R fused to the GAL4 DNA-binding domain (R1–252-GAL4DBD), this would result in the activation of two selectable markers (HIS3 and ADE3) driven by GAL4 binding sites, thereby conferring growth in synthetic media lacking adenine and histidine (see Materials and Methods). As previously shown with B (13), the N-terminal region of R (GAL4DBD-R1–252 physically interacts with the Myb domain of C1 (GAL4AD-C1myb, Fig. 2B, 1). Under similar conditions, however, the Myb domain of P does not interact with R (GAL4AD-Pmyb, Fig. 2B, 2), thus providing direct evidence of the specificity of the interaction of related Myb domains with R. As deduced from the transient expression experiments, the PC, but not the CP, chimeric Myb domain mediates interaction with R (compare GAL4AD-PCmyb and GAL4AD-CPmyb in Fig. 2B, 3 and 4). These results show that the region of C1 that specifically interacts with R is the same as that required for R-dependent activity.
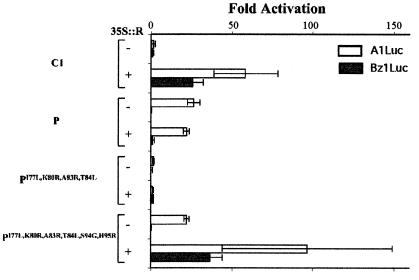
Figure 3.
Transfer of the interaction with R from C1 to P in vivo. Results of transient expression after cobombardment of cultured maize cells with P, C1, and mutants of P together with A1Luc (open bars) or Bz1Luc (filled bars) reporter constructs, in the absence (−) or presence (+) of 35S∷R. All other experimental details are described in the legend of Fig. 1.
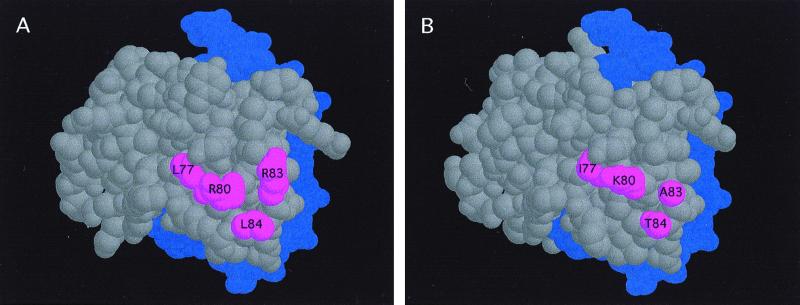
Figure 4.
Solvent-exposed surfaces of the C1 and P Myb domains. Modeling of the structure of the Myb domains of C1 (A) or P (B), based on the deduced structure of the R2R3 region of c-Myb (5). The DNA is shown in purple, and the four amino acids in C1 (L77, R80, R83, and L84) sufficient to transfer the interaction with R from C1 to P are shown in red. The position of G94 and R95 could not be precisely determined, although the polar nature of R95 makes it a candidate for a surface-exposed residue.
Based on the transient expression experiments (Fig. 1), the 67–84 region of the R3 Myb repeat of C1 is essential for the specificity of the interaction with R. To further determine which of the six residue differences between P and C1 in this region (Fig. 2A) are important for the interaction with R, two residues in C1 (Y72 and D73) were simultaneously changed to the corresponding residues in P (K and E). The resulting C1 mutant Myb domain was capable of interacting with R in yeast two-hybrid experiments (data not shown), indicating that those two residues are unnecessary for interaction with R.
To determine which residues in C1 confer specificity in the interaction with R, residues I77, K80, A83, and T84 in P (Fig. 2A) were replaced with the corresponding L, R, R, and L residues present in C1. The P Myb domain with these four changes, when fused to the GAL4AD (GAL4AD-PmybI77L,K80R,A83R,T84L), was capable of interacting with R like C1 (Fig. 2B). However, when each one of these residues in P was mutated independently or in combinations of two or three, no interaction with R was observed (see for example GAL4AD-PmybI77L,K80R,T84L in Fig. 2B). These results demonstrate that four amino acids in the Myb R3 motif of C1 are sufficient to confer on the P Myb domain the ability to specifically interact with the cofactor R. Consistent with this, when R83 in C1 was changed to the P residue A, or L84 in C1 to T, interaction between C1 and R was lost (data not shown), suggesting that R83 and L84 are necessary for C1 to interact with R.
P Mutants Interact with R in Plant Cells.
To investigate the regulatory activity in plant cells of P mutants able to interact with R in yeast, the I77L, K80R, A83R, and T84L changes were introduced into the full-length P protein. Driven from the constitutive CaMV 35S promoter, PI77L,K80R,A83R,T84L was assayed in transient expression experiments for activation of the A1 and Bz1 promoters. Surprisingly, no activation of A1 or Bz1 was observed, regardless of the presence or absence of R (Fig. 3). Because some of these factors show a dose-dependent response (E.G. and B.B., unpublished results), different concentrations of 35S∷PI77L,K80R,A83R,T84L were tested, with identical negative results (not shown). A negative result like this could be indicative of a loss of DNA-binding activity caused by the four residue changes. To test for this possibility, we expressed the PI77L,K80R,A83R,T84L protein in a yeast strain containing a reporter gene controlled by the high-affinity P-binding sites (33). In this assay, PI77L,K80R,A83R,T84L activates transcription as efficiently as P does (not shown), suggesting that, at least in yeast cells, the mutant PI77L,K80R,A83R,T84L binds DNA.
Although we cannot rule out from our findings the possibility that the PI77L,K80R,A83R,T84L protein is unstable in maize cells, or that inhibitory interactions enable the activity of PI77L,K80R,A83R,T84L, another possibility is that other residues from C1 are required for transcription activation. There are five differences between P and C1 in the 84–115 region (Fig. 2A). The G94 and R95 residues in C1 were particularly interesting because they are conserved in other R-dependent Myb domains (Fig. 2A). In addition, the simultaneous change of G94S and R95H allowed a C1mybG94S,R95H-GAL4AD chimeric protein to activate transcription in yeast from a promoter containing the previously described high-affinity P-binding sites, in contrast to C1myb-GAL4AD, which is inactive in this assay (T. Matulnik, J.M.H., and E.G., unpublished data). Thus, we investigated the effect of changing S94 to G and H95 to R in the context of PI77L,K80R,A83R,T84L on the activation of A1 and Bz1 in transient expression experiments in plant cells. 35S∷PI77L,K80R,A83R,T84L,S94G,H95R activated transcription of A1 independently of R, similar to P (Fig. 3). However, when R is cobombarded, a significant enhancement of this activity was observed (Fig. 3), providing strong evidence that this mutant of P interacts with R in maize cells. The requirement for the six residue changes in plant cells relative to four in yeast could reflect that additional residues are necessary for interaction with R in plant cells, or that the two additional residues are required for R to mediate transcriptional activation. The first hypothesis is unlikely for several reasons. First, a C1 derivative with the G94S and R95H residue changes interacts with R in yeast two-hybrid experiments (not shown). Second, the C1mybG94S,R95H-GAL4AD fusion has R-dependent transcriptional activity in maize cells like C1Myb-GAL4AD (T. Matulnik, J.M.H., and E.G., unpublished data). Finally, a PmybS94G,H95R-GAL4AD construct does not interact with R in yeast two-hybrid experiments (not shown).
Together, these findings demonstrate that four residues in R3 are responsible for the specificity of the interaction of C1 with R and that, in the context of P, two additional residues need to be altered to enable R to enhance P transcriptional activation in plant cells.
Interaction with the Cofactor R Is Necessary for Trancriptional Activation of Bz1.
In contrast to A1, which is activated by both P and C1 + R, Bz1 is activated by C1 + R, but not by P (20). The PI77L,K80R,A83R,T84L,S94G,H95R factor, which can activate A1 without R but interacts with R in vivo, provides a unique opportunity to investigate the contribution of R to the activation of Bz1. In the absence of R, PI77L,K80R,A83R,T84L,S94G,H95R does not activate Bz1, similar to P and to C1 (Fig. 3). However, in the presence of R, a dramatic activation of Bz1 is observed. This result demonstrates that R is still required for Bz1 activation, even with an Myb-domain protein that can activate A1 without R.
Discussion
In this study, we have used the independent regulation of two branches of maize flavonoid biosynthesis by the related Myb-domain transcription factors C1 and P to elucidate the participation of the Myb domain in coactivator-dependent transcription. We identified the residues in the Myb domains of C1 that specify the interaction with the bHLH coactivator R. By replacing residues in P with the corresponding amino acids present in C1, we transferred the interaction with R to P, resulting in an activator with novel regulatory functions. Finally, we demonstrated a central role of R in the regulatory specificity of the Myb-domain proteins C1 and P.
Specificity of the Interaction Between Myb Domains and Coactivators.
Despite the higher than 70% identity between the Myb domains of C1 and P (25), our findings demonstrate that only the Myb domain of C1 interacts with the N-terminal region of the bHLH cofactor R. Transient expression and yeast two-hybrid experiments revealed that the R3 Myb repeat of C1 is responsible for the specificity of the interaction with R. Whereas R3 is necessary for the interaction of C1 with R, it is probably not sufficient. Truncation analyses indicate that in yeast two-hybrid experiments, the last 13 amino acids of R2 are also required (not shown). This construct includes most of the DNA recognition helix of R2, opening the possibility that the need for these R2 sequences involves correct folding of R3, exposing the right surface in R3 for R interaction. A Perilla frutescens Myb-domain with only an R3 interacts with Myc-rp, an R-like bHLH factor (34). In addition, it has been recently proposed that the competitive effect of the WER (an R2R3 Myb-domain protein) and CPC [which has only an R3 Myb repeat (35)] proteins in Arabidopsis root epidermal cell patterning is mediated by the ability of these two proteins to interact with an as yet unidentified bHLH transcription factor (31). These findings suggest that the R3 Myb repeat of R2R3 Myb domain proteins may provide a general surface for protein–protein interactions.
Four Amino Acid Changes Are Sufficient to Transfer the Interaction with R from C1 to P.
Our findings demonstrate that the L77, R80, R83, and L84 residues in C1 specify the interaction with R. Replacement of the corresponding residues in P for the residues present in C1 is sufficient to transfer the interaction with R from C1 to P in yeast two-hybrid experiments. A model of the Myb domain of C1 (Fig. 4A), based on the NMR structure of the R2R3 Myb domain of c-Myb (5, 36), indicates that these four residues are solvent exposed, providing a surface for the interaction with R. The smaller residues found in the Myb domain of P (Fig. 4B) make the corresponding surface of P significantly different. All four residues appear to be necessary for the specificity of the interaction, because single, double, or triple changes did not transfer the interaction with R to P (not shown). Consistent with our findings, the L77, R80, R83, and L84 residues are also conserved in the AN2 and GL1 proteins that physically interact with R and that require R for regulatory activity.
This surface may play an important role in other protein–protein interactions. The Myb domains of c-Myb and A-Myb, but not of B-Myb, were shown to interact with nucleolin in animal cells, and the solvent-exposed R161 residue present in c-Myb and A-Myb, but not B-Myb, is crucial for this interaction (10). Strikingly, the R161 in c-Myb coincides in position with L84 in C1 (Fig. 2A), one of the key residues in the interaction of C1 with R. Thus, a variety of coactivators may recognize similar regions with distinct surface-exposed residues in Myb domains to modulate Myb protein activity.
R Contributes to the Regulatory Specificity of Myb Transcription Factors.
R is absolutely essential for C1 to activate transcription of all of the genes in the anthocyanin pathway, including A1 and Bz1, whereas P activates transcription of a subset of the C1-regulated genes (including A1 but not Bz1) independently of R (20). Is the function of R to make C1 active, or does R contribute to the regulatory specificity of C1? The PI77L,K80R,A83R,T84L,S94G,H95R protein provided us with unique tools to address these issues. Similar to P, PI77L,K80R,A83R,T84L,S94G,H95R activates transcription of A1 independently of R (Table 1). However, R can interact with PI77L,K80R,A83R,T84L,S94G,H95R, enhancing its activity on the A1 promoter. When tested on the Bz1 promoter, PI77L,K80R,A83R,T84L,S94G,H95R does not activate transcription, like P. However, very robust activation of Bz1 is observed by PI77L,K80R,A83R,T84L,S94G,H95R in the presence of R. Thus, PI77L,K80R,A83R,T84L,S94G,H95R has a regulatory specificity that is different from either P or C1 (Table 1). Because PI77L,K80R,A83R,T84L,S94G,H95R does not activate Bz1 in the absence of R, we can conclude that the six residue changes in P do not allow PI77L,K80R,A83R,T84L,S94G,H95R to interact with the Bz1 promoter in a productive manner. Rather, the ability of PI77L,K80R,A83R,T84L,S94G,H95R to activate Bz1 is completely dependent on its interaction with R. These results suggest that R does not simply activate C1, but rather that it plays a key role in the regulatory specificity of C1. The mechanisms by which R or B control transcription are unknown. The presence of a conserved HLH motif suggests that they may interact with other HLH partners, and possibly be recruited to DNA. However, HLH partners have not yet been identified, nor has a DNA-binding activity been described for R or B.
Table 1.
Dependence on R of C1, P, and a P mutant for the activation of the A1 and Bz1 genes
| R interaction | A1 activation | Bz1 activation | |
|---|---|---|---|
| C1 | Yes | R-dependent | R-dependent |
| P | No | R-independent | None |
| PI77L,K80R,A83R,T84L,S94G,H95R | Yes | R-enhanced | R-dependent |
Together, our results demonstrate that, although C1 and P have very similar DNA-binding specificity (21), their ability to control the accumulation of different pigments by activating distinct sets of target genes is given by the specific interaction of the Myb-domain of C1 with R. These findings are of particular significance given the very large number of R2R3 Myb transcription factors expressed in the higher plants, which have very similar DNA-binding domains (14, 15). The regulatory specificity of these Myb factors might be largely provided by combinatorial interactions with other cellular factors, rather than by different DNA-binding preferences.
Acknowledgments
We thank Xiaoyun Dong, Kevin Johnson, Jilian Riley, Bill Ferguson, Yuling Sun, and Marcie Vaughn for excellent technical assistance. We thank Ed Braun, Todd Matulnik, and Dave Bisaro for comments on the manuscript. E.G. acknowledges Winship Herr for ideas and continuous support. We appreciate the help of Daan van Aalten with the modeling of Myb domains. M.B.S. was a Howard Hughes predoctoral fellow. This work was funded by Grants MCB-9896111 and MCB-9974474 from the National Science Foundation to E.G., and MCB-9304687 from the National Science Foundation to V.L.C.
Abbreviations
- bHLH
basic helix–loop–helix
- GUS
β-glucuronidase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250379897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250379897
References
- 1.Martin C, Paz-Ares J. Trends Genet. 1997;13:67–73. doi: 10.1016/s0168-9525(96)10049-4. [DOI] [PubMed] [Google Scholar]
- 2.Lipsick J S. Oncogene. 1996;13:223–235. [PubMed] [Google Scholar]
- 3.Nomura N, Takahashi M, Matsui M, Ishii S, Date T, Sasamoto S, Ishizaki R. Nucleic Acids Res. 1988;16:11075–11089. doi: 10.1093/nar/16.23.11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jin H, Martin C. Plant Mol Biol. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- 5.Ogata K, Morikawa S, Nakamura H, Sekikawa A, Inoue T, Kanai H, Sarai A, Ishii S, Nishimura Y. Cell. 1994;79:639–648. doi: 10.1016/0092-8674(94)90549-5. [DOI] [PubMed] [Google Scholar]
- 6.Ganter B, Fu S-l, Lipsick J S. EMBO J. 1998;17:255–268. doi: 10.1093/emboj/17.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mink S, Kerber U, Klempnauer K-H. Mol Cell Biol. 1996;16:1316–1325. doi: 10.1128/mcb.16.4.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hedge S P, Kumar A, Kurschner C, Shapiro L H. Mol Cell Biol. 1998;18:2729–2737. doi: 10.1128/mcb.18.5.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verbeek W, Gombart A F, Chumakov A M, Muller C, Friedman A D, Koeffler H P. Blood. 1999;93:3327–3337. [PubMed] [Google Scholar]
- 10.Ying G-G, Proost P, van Damme J, Bruschi M, Introna M, Golay J. J Biol Chem. 2000;275:4152–4158. doi: 10.1074/jbc.275.6.4152. [DOI] [PubMed] [Google Scholar]
- 11.Ness S A, Marknell A, Graf T. Cell. 1989;59:1115–1125. doi: 10.1016/0092-8674(89)90767-8. [DOI] [PubMed] [Google Scholar]
- 12.Dash A B, Orrico F C, Ness S A. Genes Dev. 1996;10:1858–1869. doi: 10.1101/gad.10.15.1858. [DOI] [PubMed] [Google Scholar]
- 13.Goff S A, Cone K C, Chandler V L. Genes Dev. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- 14.Romero I, Fuertes A, Benito M J, Malpica J M, Leyva A, Paz-Ares J. Plant J. 1998;14:273–284. doi: 10.1046/j.1365-313x.1998.00113.x. [DOI] [PubMed] [Google Scholar]
- 15.Rabinowicz P D, Braun E L, Wolfe A D, Bowen B, Grotewold E. Genetics. 1999;153:427–444. doi: 10.1093/genetics/153.1.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun E L, Grotewold E. Plant Physiol. 1999;121:21–24. doi: 10.1104/pp.121.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cone K C, Cocciolone S M, Burr F A, Burr B. Plant Cell. 1993;5:1795–1805. doi: 10.1105/tpc.5.12.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ludwig S E, Wessler S R. Cell. 1990;62:849–851. doi: 10.1016/0092-8674(90)90259-h. [DOI] [PubMed] [Google Scholar]
- 19.Mol. J, Grotewold E, Koes R. Trends Plant Sci. 1998;3:212–217. [Google Scholar]
- 20.Grotewold E, Drummond B, Bowen B, Peterson T. Cell. 1994;76:543–553. doi: 10.1016/0092-8674(94)90117-1. [DOI] [PubMed] [Google Scholar]
- 21.Sainz M B, Grotewold E, Chandler V L. Plant Cell. 1997;9:611–625. doi: 10.1105/tpc.9.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grotewold E, Peterson T. Mol Gen Genet. 1994;242:1–8. doi: 10.1007/BF00277341. [DOI] [PubMed] [Google Scholar]
- 23.Grotewold E, Chamberlain M, St. Claire G, Swenson J, Siame B A, Butler L G, Snook M, Bowen B. Plant Cell. 1998;10:721–740. [PMC free article] [PubMed] [Google Scholar]
- 24.Gordon-Kamm W J, Baszczynski C L, Bruce W B, Tomes D T. Transgenic Cereals: Zea mays (maize) London: Kluwer; 1999. [Google Scholar]
- 25.Grotewold E, Athma P, Peterson T. Proc Natl Acad Sci USA. 1991;88:4587–4591. doi: 10.1073/pnas.88.11.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.James P, Halladay J, Craig E A. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hegvold A B, Gabrielsen O S. Nucleic Acids Res. 1996;24:3990–3995. doi: 10.1093/nar/24.20.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Aalten D M F, Grotewold E, Joshua-Tor L. Methods Companion Methods Enzymol. 1998;14:318–328. doi: 10.1006/meth.1998.0587. [DOI] [PubMed] [Google Scholar]
- 29.Szymanski D B, Jilk R A, Pollock S M, Marks D. Development. 1998;125:1161–1171. doi: 10.1242/dev.125.7.1161. [DOI] [PubMed] [Google Scholar]
- 30.Larkin J C, Marks M D, Nadeau J, Sack F. Plant Cell. 1997;9:1109–1120. doi: 10.1105/tpc.9.7.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee M M, Schiefelbein J. Cell. 1999;99:473–483. doi: 10.1016/s0092-8674(00)81536-6. [DOI] [PubMed] [Google Scholar]
- 32.Quattrocchio F, Wing J, van der Woude K, Souer E, de Vetten N, Mol. J, Koes R. Plant Cell. 1999;11:1433–1444. doi: 10.1105/tpc.11.8.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grotewold E. Maize Genet Coop News. 1995;69:32. [Google Scholar]
- 34.Gong Z-Z, Yamazaki M, Saito K. Mol Gen Genet. 1999;262:65–72. doi: 10.1007/pl00008639. [DOI] [PubMed] [Google Scholar]
- 35.Wada T, Tachibana T, Shimura Y, Okada K. Science. 1997;277:1113–1116. doi: 10.1126/science.277.5329.1113. [DOI] [PubMed] [Google Scholar]
- 36.Ogata K, Morikawa S, Nakamura H, Hojo H, Yoshimura S, Zhang R, Aimoto S, Ametani Y, Hirata Z, Sarai A, Ishii S, Nishiura Y. Struct Biol. 1995;2:309–319. doi: 10.1038/nsb0495-309. [DOI] [PubMed] [Google Scholar]
- 37.Paz-Ares J, Ghosal D, Weinland U, Peterson P A, Saedler H. EMBO J. 1987;6:3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oppenheimer D G, Herman P L, Sivakumaran S, Esch J, Marks D M. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]