Abstract
Control of transcription and enzyme activities are two interwoven regulatory systems essential for the function of a metabolic node. Saccharomyces cerevisiae strains differing in enzyme activities at the chorismate branch point of aromatic amino acid biosynthesis were constructed by recombinant DNA technology. Expression of an allosterically unregulated, constitutively activated chorismate mutase encoded by the ARO7T226I (ARO7c) allele depleted the chorismate pool. The resulting tryptophan limitation caused growth defects, which could be counteracted only by transcriptional induction of TRP2 encoding the competing enzyme anthranilate synthase. ARO7 expression is not transcriptionally regulated by amino acids. Transcriptional activation of the ARO7c allele led to stronger growth retardation upon tryptophan limitation. The same effect was achieved by removing the competing enzyme anthranilate synthase, which is encoded by the TRP2 gene, from the transcriptional control. The allelic situation of ARO7c being under general control instead of TRP2 resulted in severe growth defects when cells were starved for tryptophan. In conclusion, the specific regulatory pattern acting on enzymatic activities at the first metabolic node of aromatic amino acid biosynthesis is necessary to maintain proper flux distribution. Therefore, the evolution of the sophisticated allosteric regulation of yeast chorismate mutase requires as prerequisite (i) that the encoding ARO7 gene is not transcriptionally regulated, whereas (ii) the transcription of the competing feedback-regulated anthranilate synthase-encoding gene is controlled by availability of amino acids.
Supply of precursor metabolites and energy for anabolic pathways to synthesize cellular components is necessary for growth and maintenance of a living cell. Metabolic pathways are numerous and extremely plastic, and different modes of regulation are possible to channel intermediates from the input reactions to the formation of end products. Branched reaction cascades are of special interest, as most metabolic networks are constituted by such pathways. To ensure proper distribution of intermediates, specific regulatory systems have evolved to trigger the enzymatic activities at a metabolic node. Two main mechanisms are possible to regulate catalytic turnover at a given enzyme. Either the amount of protein is altered by means of gene expression, protein synthesis, or protein degradation, or enzymatic activity itself is varied by the action of effectors, modifications, or conformational changes.
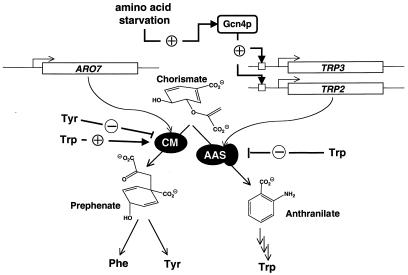
In the baker's yeast, Saccharomyces cerevisiae, specific mechanisms contribute to regulation of catalytic turnover, with transcriptional regulation being the most important feature to control protein levels. Amino acid biosynthesis is the target of a key regulatory network acting upon amino acid starvation conditions and imbalances (reviewed in ref. 1). Starvation for almost any of the 20 amino acids found in proteins can initiate the network response. The final effector of this “general control of amino acid biosynthesis” system is the transcription factor Gcn4p, which binds as a homodimer to conserved sequence elements within promoters of specified target genes (2). Biosynthesis of aromatic amino acids in S. cerevisiae is a model pathway for a strictly regulated, branched reaction cascade (reviewed in ref. 3). From the last common intermediate, chorismic acid, two main branches emerge to initiate the tyrosine/phenylalanine- and tryptophan-specific routes, respectively. The enzymatic activities constituting this metabolic node, chorismate mutase (CM; EC 5.4.99.5) and anthranilate synthase (AAS; EC 4.1.3.27), are regulated in their activities by different means (Fig. 1). While the amount of CM molecules is not varied by the general control transcriptional system, the expression of both genes encoding the competing AAS complex is transcriptionally regulated by the general control activator Gcn4p. In addition, both enzymatic activities are targets of allosteric effectors, namely the pathway end products tyrosine and tryptophan, that modulate catalytic turnover rates.
Figure 1.
Interplay between transcriptional and enzyme regulation at the first branch point of aromatic amino acid biosynthesis in yeast. The metabolic node emerging from chorismate is schematically shown with the enzymatic activities chorismate mutase (CM) and anthranilate synthase (AAS) as black ovals. The encoding genomic loci are drawn as bars with transcriptional start sites indicated by broken arrows. Positive feedbacks and inductions are indicated by ⊕, feedback inhibition by ⊝.
CMs are unique enzymatic activities that are found only in microorganisms and plants but never in animals (4). They accelerate the one-step conversion of chorismate to prephenate, a reaction formally resembling a Claisen rearrangement, to initiate the tyrosine/phenylalanine-specific branch at the first metabolic branch point of aromatic amino acid biosynthesis (5, 6). Whereas bacterial CM activities are often found to reside on a distinct domain within a bifunctional enzyme, all eukaryotic enzymes characterized to date are monofunctional. One prototype of bacterial CMs is represented by the CM domain (EcCM) of the Escherichia coli P-protein, which contains additional prephenate dehydratase activity (7). Both activities are subject to feedback inhibition by the end product phenylalanine, and catalytic turnover rates of the CM activity are characterized by Michaelis–Menten-like kinetics. As an exemplar of eukaryotic CMs, the enzyme of S. cerevisiae (ScCM), the ARO7 gene product, has been studied extensively (8–10). This CM activity is allosterically regulated in its activity. Substrate saturation kinetics of the unliganded enzyme display cooperativity, with the substrate chorismate acting as homotropic, positive effector. In addition, two end products of the pathway act as heterotropic effectors on ScCM. Tyrosine inhibits activity by decreasing the affinity of the enzyme toward its substrate, whereas tryptophan, the end product of the opposite branch, acts as a strong activator of catalytic turnover, resulting in Michaelis–Menten kinetics without cooperativity. Expression of the ARO7T226I mutant allele, here referred to as ARO7c, conserves the latter situation and leads to an unregulated, noncooperative yeast CM that is locked in its allosteric R state (11). In contrast to most amino acid biosynthetic genes, the CM-encoding ARO7 gene of S. cerevisiae is not transcriptionally regulated by the general control activator Gcn4p (12). The TRP2 gene of S. cerevisiae encodes the AAS activity, which is competing with the CM for the common substrate chorismate and channels it toward the tryptophan-specific branch. In vivo, an additional glutamine amidotransferase activity, encoded by part of the TRP3 gene, is necessary to fulfil catalytic activity, and both genes coding for the AAS heterodimer are regulated by Gcn4p (3, 13). Furthermore, the AAS activity is regulated in this yeast via feedback inhibition by the end product, tryptophan. A TRP2S76L (TRP2fbr) mutant allele is impaired in feedback inhibition by tryptophan and confers resistance to the structural analogue 5-methyltryptophan (5-MT) (14).
In summary, the two enzymatic activities constituting the first metabolic branch point of aromatic amino acid biosynthesis in S. cerevisiae are regulated by different means. Given the complex interwoven regulatory pattern modulating both enzymatic activities, we were interested in the impact of an unregulated, noncooperative CM enzyme upon starvation conditions. Therefore, the ARO7 gene was replaced by the ARO7c allele and growth was monitored under conditions of tryptophan starvation. Additionally, the unregulated CM was expressed in a Gcn4p-dependent manner to investigate the influence of the general control network on flux partitioning at the metabolic node. To investigate the necessity of transcriptional regulation of AAS expression, the TRP2 gene was removed from the general control system. Additionally, the impact of the feedback-unresponsive TRP2fbr allele was monitored. We found that the conserved regulatory pattern controlling CM activity is strictly necessary when AAS activity is limited and that the interplay of allosteric and transcriptional regulation is a prerequisite for proper chorismate distribution to both branches at the first metabolic node of aromatic amino acid biosynthesis in baker's yeast.
Materials and Methods
Materials.
Chorismic acid as barium salt and dl-5-MT were purchased from Sigma. 5-Fluoroorotic acid (5-FOA) was obtained from Toronto Research Chemicals (Toronto). Platinum Pfx DNA polymerase from Life Technologies (Karlsruhe, Germany) was used for PCRs. All other chemicals were supplied by Fluka or Sigma–Aldrich.
Yeast Strains and Growth Conditions.
All yeast strains in this study are isogenic to the S288C genetic background and are listed in Table 1. The aro7∷hisG and trp2∷hisG deletion mutations were introduced into the progenitor strain RH1408 [gcn4-103; ura3-52] (15) by using deletion plasmids pME1901 and pME1902, respectively, followed by counterselection on 5-FOA-supplemented medium (16). All mutant alleles of ARO7 and TRP2 were reintroduced at the homologous loci in single copy as verified by Southern hybridization analyses.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| RH1408 | MATa, ura3-52, gcn4-103 | Ref. 15 |
| RH2457 | MATa, ura3-52, gcn4-103, aro7∷hisG–URA3–hisG | This study |
| RH2458 | MATa, ura3-52, gcn4-103, aro7∷hisG | This study |
| RH2459 | MATa, ura3-52, gcn4-103, trp2∷hisG–URA3–hisG | This study |
| RH2460 | MATa, ura3-52, gcn4-103, trp2∷hisG | This study |
| RH2461 | MATa, ura3-52, gcn4-103, aro7∷hisG, trp2∷hisG–URA3–hisG | This study |
| RH2462 | MATa, ura3-52, gcn4-103, aro7∷hisG, trp2∷hisG | This study |
| RH2463 | MATa, ura3-52, gcn4-103, ARO7c | This study |
| RH2465 | MATa, ura3-52, gcn4-103, ARO7c, TRP2fbr | This study |
| RH2466 | MATa, ura3-52, gcn4-103, pTRP2∷ARO7 | This study |
| RH2467 | MATa, ura3-52, gcn4-103, pARO7∷TRP2 | This study |
| RH2468 | MATa, ura3-52, gcn4-103, pTRP2∷ARO7, pARO7∷TRP2 | This study |
| RH2469 | MATa, ura3-52, gcn4-103, pTRP2∷ARO7c | This study |
| RH2470 | MATa, ura3-52, gcn4-103, ARO7c, pARO7∷TRP2 | This study |
| RH2471 | MATa, ura3-52, gcn4-103, pTRP2∷ARO7c, TRP2fbr | This study |
| RH2472 | MATa, ura3-52, gcn4-103, ARO7c, pARO7∷TRP2fbr | This study |
| RH2473 | MATa, ura3-52, gcn4-103, pTRP2∷ARO7c, pARO7∷TRP2 | This study |
| RH2474 | MATa, ura3-52, gcn4-103, pTRP2∷ARO7c, pARO7∷TRP2fbr | This study |
Complex medium for growth of S. cerevisiae was YEPD (1% yeast extract/2% peptone/2% glucose). Minimal MV medium contained 0.14% yeast nitrogen base (without amino acids and without ammonium sulfate), 0.5% ammonium sulfate, and 2% glucose, and was buffered to acidic pH of 4.0 with succinic acid and KOH as described previously (17). Because cells harboring no functional Gcn4p starve for arginine, this amino acid was supplemented in all minimal growth media. Supplements were added according to Guthrie and Fink (18). Growth rates were determined turbidimetrically at 595 nm, and the specific growth rate is given as μ defined by (ln x2 − ln x1)/(t2 − t1), where x stands for the optical density at the corresponding time t.
Plasmids.
Plasmid DNAs were generally propagated in E. coli strain DH5α (19). Plasmids used in this study are listed in Table 2. Deletion cassettes for ARO7 and TRP2 were created by replacement of coding sequences by the hisG∷URA3∷hisG marker (20). Plasmid pME1905, carrying the ARO7 gene under the control of the TRP2 promoter, was constructed by separate amplification of the TRP2 promoter region from pME1903 and the ARO7 coding sequence from pME1187 by means of PCR using oligonucleotide combinations OLSK24 (5′-GGCAAAAAATGGATTTCACAAAACCAGAAAC-3′)/OLSK15 (5′-TCCTATAGAATTTATGAGCCATCG-3′) and T7 (5′-GTAATACGACTCACTATAGGGC-3′)/OLSK25 (5′-GTGAAATCCATTTTTTGCCTTTTTTCCAATC-3′), respectively, and a second PCR using both products as template in combination with T7/OLSK15. The amplified DNA was ligated as XbaI fragment to an AatII/XbaI DNA fragment comprising part of the 5′ region of ARO7 (position −1482 to −652 relative to the translational start codon) and cloned in the plasmid pGEM7(+) (Promega). Plasmid pME1907 was constructed as described for pME1905 by using oligonucleotide combinations T7/OLSK27 (5′-GCGGTCATATCTTATACCAATTTTATGCAG-3′) and OLSK26 (5′-GGTATAAGATATGACCGCTTCCATCAAAATTC-3′)/OLSK17 (5′-ACAGAGAATGCCCTTTTTAAGC-3′). The resulting pARO7∷trp2 EcoRI/Eco72I fragment carrying a chimeric construct with the ARO7 promoter, and part of the TRP ORF was ligated in pME1903 together with an AatII/EcoRI fragment comprising part of the TRP2 5′ region (position −1505 to −735). Plasmid pME1906 with the ARO7c allele driven by the TRP2 promoter (pTRP2∷ARO7c) was constructed by combination of an AatII/HindIII fragment from pME1905 and a HindIII/EcoRI fragment from pME606. For construction of pME1908 with the TRP2fbr allele under the control of the ARO7 (pARO7∷TRP2fbr) promoter, an AatII/Eco72I fragment from pME1907 was fused to an Eco72I/BamHI fragment of pME1904. Gcn4p was expressed either at low levels from plasmid p164, which carries the wild-type GCN4 gene on the low-copy vector YCp50 (21), or at high levels from p238, which carries a mutant allele of GCN4 in YCp50 with mutated upstream ORFs (22). YCp50 was used as empty vector control for strains expressing no functional Gcn4p. Plasmid pME1909 containing a 470-bp SspI/EcoRV fragment of ARO7 as well as a 465-bp Eco72I/EcoRV fragment of TRP2 in pBluescript II KS was used for probe preparation in Southern analyses and Northern experiments.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pME1901 | aro7∷hisG–URA3–hisG cassette for deletion of ARO7 ORF | This study |
| pME1902 | trp2∷hisG–URA3–hisG cassette for deletion of TRP2 ORF | This study |
| pME1187 | 2-kb EcoRI fragment containing ARO7 in pGEM7(+) | This study |
| pME606 | 2-kb EcoRI fragment containing ARO7c in pJDB207 | Ref. 8 |
| pME1903 | 2.3-kb XbaI/BamHI fragment containing TRP2 in pGEM7(+) | This study |
| pME1904 | 2.3-kb XbaI/BamHI fragment containing TRP2fbr in pGEM7(+) | This study |
| pME1905 | 3-kb AatII/EcoRI fragment containing 5′-ARO7∷pTRP2∷ARO7 replacement cassette in pGEM7(+) | This study |
| pME1906 | 3-kb AatII/EcoRI fragment containing 5′-ARO7∷pTRP2∷ARO7c replacement cassette in pUC19 | This study |
| pME1907 | 3-kb AatII/BamHI fragment containing 5′-TRP2∷pARO7∷TRP2 replacement cassette in pGEM7(+) | This study |
| pME1908 | 3-kb AatII/BamHI fragment containing 5′-TRP2∷pARO7∷TRP2fbr replacement cassette in pUC19 | This study |
| p164 | 2.8-kb SalI/EcoRI fragment containing GCN4 in YCp50 | Ref. 22 |
| p238 | 2.8-kb SalI/EcoRI fragment containing GCN4 with all four upstream ORFs mutated in YCp50 | Ref. 22 |
| pME1909 | 470-bp SspI/EcoRV fragment of ARO7 ORF and 465-bp Eco72I/EcoRV fragment of TRP2 ORF in pBluescript II KS | This study |
Transformation Procedures.
Transformation of E. coli was performed as described by Inoue et al. (23), and S. cerevisiae strains were transformed by following a modified protocol of Elble (24).
Nucleic Acid Preparation and Analyses.
For isolation of plasmid DNA from bacterial strains the plasmid purification system from Qiagen (Hilden, Germany) was used. Genomic DNA from yeast was isolated according to ref. 25 and analyzed by Southern hybridization (26) or diagnostic PCR (27). Total RNAs from S. cerevisiae cultures were prepared according to Cross and Tinkelenberg (28), and transcript levels were quantified by Northern hybridization (29) using a Bio-Imaging Analyzer from Fuji Photo Film (Tokyo). Sequencing reactions were carried out by using a BigDye sequencing kit (30) and analyzed on an ABI Prism 310 Genetic Analyzer (PE Biosystems, Foster City, CA).
Enzyme Assays.
Enzymatic assays were performed at 37°C with Triton X-100-treated cell suspensions prepared by the method of Miozzari et al. (17). Glutamine-dependent AAS activities were determined according to Egan and Gibson (31) at 0.5 mM substrate concentration. CM activity was measured spectrophotometrically as described (8) with the modification that permeabilized cells were spun down and resuspended in cold buffer containing 125 mM potassium phosphate (pH 7.6), 25 mM dl-dithiothreitol, 2.5 mM EDTA, and 0.125 mM phenylmethylsulfonyl fluoride before chorismate was added to 1 mM final concentration. The concentration of phenylpyruvate was determined after cells had been removed from the assay mixture by brief centrifugation. Specific activities are quantified in units (U), 1 U equaling 1 nmol of product formed in 1 min of turnover by 1 mg of total protein. Protein content of the detergent-treated cell suspensions was measured by the method of Herbert et al. (32), using the Bradford assay (33).
Results
An Unregulated CM Allele and a gcn4 Deletion Are Synthetically Lethal in Yeast After Tryptophan Starvation.
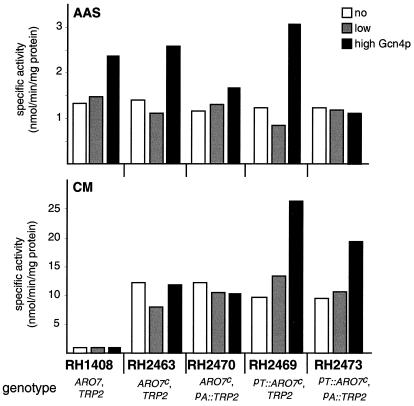
The yeast ARO7c allele encoding an unregulated CM displays high catalytic activity that preferentially channels chorismate toward the tyrosine/phenylalanine branch when expressed in high amounts (11). To modulate distribution of chorismate, we replaced the wild-type ARO7 gene in strain RH1408 [gcn4-103, ura3-52] at its original locus with the ARO7c allele, resulting in strain RH2463. Because of the gcn4 mutation these strains are unable to adapt their transcription to the availability of amino acids. Enzymatic activities of the branch point enzymes, CM and AAS, altered the flux at the metabolic node (Fig. 2). In the progenitor strain RH1408 used as wild-type control, a specific CM activity of 1.0 U was determined, and specific AAS activity was at a basal level of 1.3 U. RH2463, which expresses ARO7c, displayed a specific CM activity of 12.2 U, whereas AAS activity remained at 1.4 U. Determination of growth rate constants μ revealed no nutritional requirements of the two strains when cultured in minimal medium (Table 3). However, when both strains were starved for tryptophan by the action of the structural analogue 5-MT, significant differences in viability were present. 5-MT acts as a false feedback inhibitor on AAS and therefore decreases the input into the tryptophan-specific branch of the pathway. In minimal medium supplemented with 0.1 mM 5-MT, wild-type strain RH1408 grew at a reduced rate of 0.18 h−1. Strain RH2463 expressing the unregulated, constitutively active, CM enzyme was not viable under these conditions.
Figure 2.
Specific AAS (Upper) and CM (Lower) activities of S. cerevisiae strains with altered regulatory properties of the branch point enzymes. Activities were determined from cells cultivated in minimal medium expressing either no functional Gcn4p (−), low levels (+), or high levels (+++) of Gcn4p. The genotypes of all strains are indicated with pA∷ and pT∷ representing ARO7 and TRP2 promoter fusions, respectively. All values are the means of two independent measurements with a standard deviation not exceeding 20%.
Table 3.
Growth behavior of S. cerevisiae strains with altered regulatory properties at the branch point of aromatic amino acid biosynthesis emerging from chorismate
| Strain | Genotype | Growth rate, h−1
or % reduction
|
|||||
|---|---|---|---|---|---|---|---|
| No limitation
|
Trp
starvation
|
||||||
| − | + | +++ | − | + | +++ | ||
| RH1408 | ARO7, TRP2 | 0.21 h−1 | 0.21 h−1 | 0.17 h−1 | 0.18 h−1 | 0.20 h−1 | 0.16 h−1 |
| RH2463 | ARO7c, TRP2 | 0% | 0% | 0% | 100% | 53% | 22% |
| RH2470 | ARO7c, pARO7∷TRP2 | 0% | 0% | 0% | 100% | 53% | 41% |
| RH2469 | pTRP2∷ARO7c, TRP2 | 0% | 0% | 0% | 100% | 60% | 43% |
| RH2473 | pTRP2∷ARO7c, pARO7∷TRP2 | 0% | 0% | 0% | 100% | 78% | 76% |
Growth rates were determined in minimal medium (No limitation) and in minimal medium supplemented with 10−4 M 5-MT (Trp starvation). Strains expressed no functional Gcn4p (−), low wild-type levels (+), or high levels (+++) of the transcriptional activator as indicated. For the wild-type strain RH1408, growth rate constants μ are indicated, whereas for all other strains the growth reduction as determined from growth rates with respect to RH1408 is given. Values are the mean of three independent measurements with a standard deviation not exceeding 20%.
To investigate the impact of transcriptional regulation by amino acids that acts in the wild-type situation on TRP2 expression but not on ARO7, both strains were transformed with plasmids expressing the activator protein Gcn4p at wild-type levels and in high amounts, respectively. Specific CM activities of both strains were unaffected by Gcn4p, whereas AAS activities were induced to a similar degree by high levels of the transcriptional activator to 2.4 U (RH1408) and 2.6 U (RH2463), respectively (Fig. 2). Growth rates determined in minimal medium displayed no differences between the wild-type control RH1408 and RH2463 when Gcn4p was expressed (Table 3). When cultured in the presence of 5-MT, RH1408 grew at rates comparable to rates determined in unsupplemented medium. Strain RH2463, however, was viable in the presence of 5-MT when Gcn4p was expressed at low levels but displayed a growth reduction of 53% (μ of 0.094 vs. 0.20 h−1). This reduction was diminished by high levels of Gcn4p, which resulted in a growth rate of 0.13 h−1, corresponding to a growth reduction of 22% in comparison to the wild-type ARO7 strain RH1408. Expression of the allosterically unregulated, feedback resistant, TRP2fbr allele counteracted the growth defects caused by the unregulated CM activity and resulted in a strain (RH2465) that was fully viable in the presence of 5-MT without the transcriptional activator Gcn4p (not shown).
These results suggest that depletion of the intracellular chorismate pool caused by an unregulated constitutively active CM is lethal when the competing enzyme AAS is reduced in its enzymatic activity. The general control “backup” system is able to counteract this starvation situation by increasing the amount of AAS molecules by transcriptional derepression, suggesting that the enzyme and transcriptional regulation represent strongly interwoven systems.
Removing TRP2 from the General Control Transcription System in the Presence of ARO7c Results in Strong Growth Reduction Under Tryptophan Starvation Conditions.
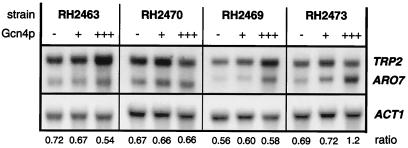
To monitor the impact of the transcriptional regulation-mediated changes in AAS activities, strains were constructed that carry the encoding TRP2 gene under the control of a Gcn4p-independent promoter. For that purpose, the ARO7 promoter seemed most appropriate. Strain RH2470 expresses the unregulated CM activity encoded by the ARO7c allele in combination with an ARO7 promoter∷TRP2 fusion integrated at the trp2∷hisG locus of strain RH2462 (Table 1). Again, this strain was transformed with plasmids to express functional Gcn4p at low and high levels. Steady-state transcript levels in RH2470 showed a constant ratio of ARO7c and TRP2 mRNAs independent of the general control network (Fig. 3). Specific activities of both branch point enzymes were not significantly changed in RH2470 at different levels of Gcn4p, with AAS activity ranging from 1.2 U to 1.7 U and CM activity from 12.2 U to 10.4 U (Fig. 2). Whereas no difference in growth of RH2470 compared with the wild-type strain RH1408 was observed under minimal conditions, starvation for tryptophan had a strong influence on viability of that strain (Table 3). Without any functional Gcn4p present, RH2470 showed no growth upon tryptophan starvation induced by 5-MT. Expression of wild-type levels of Gcn4p restored growth to a rate of 0.094 h−1, corresponding to 53% growth reduction in comparison with RH1408. High amounts of the transcriptional activator increased the growth rate slightly to 0.096 h−1, which corresponds to a decreased reduction of 41% compared with the wild-type level. Growth reductions of RH2470 were suppressed either by the TRP2fbr allele (RH2472) or by restoring allosteric regulation of Aro7p (RH2467), as both strains grew well at wild-type levels in the presence of 5-MT (not shown).
Figure 3.
Northern hybridization analyses of S. cerevisiae strains expressing ARO7c and TRP2 with altered dependence on the general control activator Gcn4p. Cells were cultivated in minimal medium and expressed no functional Gcn4p (−) or low levels (+) or high levels (+++) of Gcn4p, as indicated. In each lane, 25 μg of total RNAs was hybridized successively with probes specific for ARO7/TRP2 and ACT1 (encoding actin). For quantification, steady-state transcript levels were standardized with respect to ACT1 levels and the ratio between ARO7c and TRP2 transcript levels is indicated at the bottom. All values are the means of two independent measurements with a standard deviation not exceeding 20%.
In conclusion, these data demonstrate that transcriptional derepression of AAS activity is crucial under tryptophan starvation conditions when allosteric regulation of CM activity is not present.
Placing ARO7c Under General Control Impairs Proper Flux Partitioning at the Branch Point.
To investigate the effects when an unregulated CM activity of S. cerevisiae was expressed in a Gcn4p-dependent manner, strains were constructed in which the encoding ARO7c allele was fused to the TRP2 promoter. Strain RH2469 carries a TRP2 promoter∷ARO7c cassette integrated at the aro7∷hisG locus of RH2460 (Table 1). As a result, the synthesis of the two competing enzymatic activities at the metabolic node is increased in a general control-dependent manner driven by the TRP2 promoter after starvation for almost any amino acid. To modulate the general control in RH2469, Gcn4p was expressed from plasmids in low and high amounts. The changes in transcript levels resulted in a constant ratio between ARO7c and TRP2 mRNAs that was not altered by the level of Gcn4p (Fig. 3). Specific enzymatic activities of CM and AAS reflected the impact of the transcriptional activator (Fig. 2). In the absence of Gcn4p, AAS activity was at a basal level of 1.2 U that was elevated to 3.1 U when Gcn4p was present in high amounts. Accordingly, CM activity was induced from 9.5 U to 26.4 U by high levels of the transcriptional activator. Growth rates were determined from RH2469 expressing different levels of Gcn4p. Whereas no effect on growth fitness was observed in minimal medium, severe growth reductions were present upon starvation for the aromatic amino acid tryptophan as induced by 5-MT (Table 3). No growth in the presence of the drug was observed for RH2469 when no functional Gcn4p was expressed. In the presence of Gcn4p, growth rates were reduced in comparison with strain RH1408. Whereas a low level of Gcn4p restored growth with 60% growth reduction, high levels of the transcriptional activator counteracted the starvation situation, resulting in 43% growth reduction with respect to the wild-type situation of RH1408. Again, this reduction in viability of RH2469 was suppressed by the TRP2fbr allele (RH2471) or alternatively by an allosterically regulated CM encoded by the ARO7 wild-type gene (RH2466). Both control strains RH2471 and RH2466 displayed no growth reduction upon starvation for tryptophan (not shown).
Furthermore, inversion of chorismate flux was achieved in strain RH2473, which expresses the ARO7c allele driven by the TRP2 promoter as well as the TRP2 gene from the ARO7 promoter. Northern analyses after transformation of this strain with Gcn4p-expressing plasmids demonstrated an inversion in the ratio between ARO7c and TRP2 mRNAs when the transcriptional activator was present in high amounts (Fig. 3). This altered expression pattern was reflected by specific enzymatic activities of CM and AAS (Fig. 2). AAS activity of RH2473 was unaffected by the level of Gcn4p at a basal level of 1.2 U, in contrast to CM activities, which ranged from 9.4 U to 19.6 U in the absence and presence of the transcriptional activator, respectively. The in vivo effect of this allelic situation was monitored by determination of growth rate constants and compared with the wild-type situation of strain RH1408 (Table 3). No significant reduction in growth rates was detected when cells were cultured in minimal medium. In contrast, tryptophan limitation induced severe reductions in growth. In the absence of Gcn4p, RH2473 displayed no growth when starved for tryptophan. Reintroduction of the transcriptional activator resulted in strongly retarded growth that was not elevated by expression of Gcn4p in high amounts. These two growth reductions in comparison with RH1408 were 78% and 76%, respectively. In contrast, strain RH2474, which expresses the feedback-resistant TRP2fbr allele from the ARO7 promoter, showed no significant growth retardation under tryptophan starvation conditions induced by 5-MT. Accordingly, strain RH2468, in which an allosterically regulated CM is present, grew well in minimal medium supplemented with the false feedback inhibitor (not shown).
In summary, we demonstrate here that transcriptional regulation of an allosterically unregulated CM enzyme in S. cerevisiae leads to severe growth defects when the competing enzyme is down-modulated in its activity, even when the general control system is present. Removing AAS expression from the transcriptional control amplifies this effect, as no other mechanism is present to increase enzyme activities for catalytic turnover feeding the tryptophan trunk of the branch point.
Discussion
Biosynthesis of aromatic amino acids in the yeast S. cerevisiae is a model for a branched, strictly regulated, metabolic reaction cascade. Different modes of regulation acting on enzyme synthesis as well as catalytic turnover are present in yeast to modulate the enzymatic activities constituting the pathway. For the amount of a given enzyme, different mechanisms of modulation are possible, involving synthesis and degradation of the encoding transcript, translational regulation, or stability of the gene product. Additionally, enzymatic activity itself can be modulated by a variety of mechanisms such as specific localization, (covalent) modification, or the action of effector molecules.
Here, we present data illustrating the necessity of the interplay of allosteric and transcriptional regulation acting on the enzymatic activities at the first metabolic node of aromatic amino acid biosynthesis in S. cerevisiae. Two enzymatic activities constitute the branch point emerging from chorismate. The amount of CM enzyme is not regulated, but catalytic activity is carefully modulated and fine-tuned by a positive as well as a negative effector. In contrast to this, expression of AAS activity is transcriptionally induced upon amino acid starvation, and catalytic turnover can only be reduced by negative feedback.
We have expressed an allosterically unregulated, constitutively active, CM to favor the tyrosine/phenylalanine-specific branch of this pathway. When no starvation situation was present, no reduction in growth was determined for all strains expressing the ARO7c allele. The ARO7c allele used in this study reflects a CM activity that is not regulated allosterically. This is reminiscent of bacterial CMs, which are found to display Michaelis–Menten-like kinetics in substrate saturation assays. Accordingly, catalytic efficiency (kcat/Km) of this unregulated yeast CM activity of approximately 13 min−1⋅μM−1 is in the same range as the value reported for the E. coli P-protein CM activity: 10 min−1⋅μM−1 (7, 11). The ARO7c gene product is locked in its active R state because of an amino acid exchange in a flexible loop (loop 220s). Several mutant enzymes with substitutions in this loop have been characterized, but the precise role of this structural element in the allosteric transitions remains unclear (34).
By making the ARO7c allele a target of the general control, a regulation of the amount of CM molecules was achieved that is not present in yeast wild-type cells. Because growth rates were not reduced even when CM activity was elevated, the cellular chorismate pool has to be sufficiently high to maintain the flux into the tryptophan-specific branch, even when the competing sink is strongly favored. This accounts for a large reserve capacity of the tryptophan-specific branch as initiated by the AAS activity.
Starvation for tryptophan induced by the structural analogue 5-MT, which reduces AAS activity, resulted in severe growth defects when the unregulated CM activity was expressed. In the absence of the general control effector Gcn4p, starvation for tryptophan because of depletion of the chorismate pool was so severe that the cells were no longer viable. The general control system counteracts this starvation situation but shows dependence on the transcriptional pattern at the branch point up to different levels. When both encoding genes, ARO7c and TRP2, were not under general control, growth was restored to 60% of wild-type levels. This reflects a replenished chorismate pool, as all enzymatic activities leading to chorismate are elevated by the general control system under starvation conditions. Making both branch point enzymes a target of the general control resulted in growth rates half as high as wild type. Here, the elevated chorismate pool is channeled symmetrically into both main branches as expression of both enzymatic activities is induced by high levels of Gcn4p. Inverting the regulatory pattern at the node with concern to general control dependency leads to severe growth retardation even in the presence of Gcn4p. In conclusion, the asymmetrical regulation of the two branch point enzymes by the general control as observed in the wild-type situation is necessary to maintain proper chorismate distribution under tryptophan starvation conditions. When allosteric regulation of CM activity was restored by the wild-type ARO7 allele, growth was maintained at wild-type levels even under starvation conditions.
In a first view it seemed surprising that the CM-encoding gene of S. cerevisiae is not regulated by the general control system. However, our detailed analysis in the study presented here showed that the ARO7 gene must not be regulated by the general control because of the careful interplay and fine tuning between activation and feedback regulation of a constant number of CM molecules vs. the feedback regulation of an increasable number of AAS molecules. This situation is likely to be conserved for other eukaryotic CMs. Two additional fungal CM enzymes have been characterized in detail, the aroC gene product of the filamentous fungus Aspergillus nidulans and the HARO7-encoded enzyme of the methylotrophic yeast Hansenula polymorpha (35, 36). For both fungal enzymes no induction of gene expression was monitored under amino acid starvation conditions, but strict allosteric regulation by homotropic and heterotropic effectors was present.
The allelic situation with an allosterically unregulated CM expressed in a Gcn4p-dependent manner might reflect an early evolutionary situation; the existence of a reversed Gcn4p recognition element in the ARO7 promoter region that is able to bind the transcriptional activator in vitro implies that this gene formerly was subjected to the general control system (12). From structural studies it has been deduced that the dimeric allosterically regulated CM might have evolved from a monomeric unregulated ancestor by a gene duplication/gene fusion event (37). Given the drastic effects observed for the ARO7c allele being subject to the general control, we speculate that either CM expression was removed from the general control system before this gene duplication and fusion or, alternatively, it was never subjected to it and TRP2 acquired the transcriptional regulation after this evolutionary event. By dimerization and remodeling of distinct domains, allosteric behavior was achieved for the yeast CM. Because the balance between transcriptional and enzymatic regulation is so crucial, subtle changes on one regulatory level immediately required the adaptation on the other level to guarantee that the subtle fine tuning permanently worked. This is an interesting result of coevolution of transcriptional and enzymatic regulation. In S. cerevisiae, the different regulation of the two branch point enzymes with respect to the general control provides that flux imbalances can be counteracted in an asymmetric manner. Furthermore, our results demonstrate that allosteric and transcriptional regulation at the first branch point of aromatic amino acid biosynthesis in S. cerevisiae are interconnected, implying a continuous coevolution of both regulatory mechanisms controlling CM activity as well as AAS activity.
Acknowledgments
We thank Georg Schnappauf for providing plasmids pME1903 and pME1904, Roney Graf for providing pME1187, Kerstin Helmstaedt for critical proofreading of the manuscript, and all other members of the laboratory for helpful discussions. This work was supported by grants from the Deutsche Forschungsgemeinschaft, the Fonds der Chemischen Industrie, the Volkswagen-Stiftung, and the Niedersächsischen Vorab der Volkswagen-Stiftung, Forschungsstelle für Nachwachsende Rohstoffe. Support by National Institutes of Health Grant GM06920 is acknowledged by W.N.L.
Abbreviations
- CM
chorismate mutase
- AAS
anthranilate synthase
- 5-MT
5-methyltryptophan
- U
unit(s)
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240469697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240469697
References
- 1.Hinnebusch A. In: The Molecular and Cellular Biology of the Yeast Saccharomyces. Jones E W, Pringle J R, Broach J R, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 319–414. [Google Scholar]
- 2.Arndt K, Fink G R. Proc Natl Acad Sci USA. 1986;83:8516–8520. doi: 10.1073/pnas.83.22.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braus G H. Microbiol Rev. 1991;55:349–370. doi: 10.1128/mr.55.3.349-370.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero R M, Roberts M F, Phillipson J D. Phytochemistry. 1995;40:1015–1025. doi: 10.1016/0031-9422(95)00010-5. [DOI] [PubMed] [Google Scholar]
- 5.Andrews P R, Smith G D, Young I G. Biochemistry. 1973;12:3492–3498. doi: 10.1021/bi00742a022. [DOI] [PubMed] [Google Scholar]
- 6.Weiss U, Edwards J M. The Biosynthesis of Aromatic Amino Acids. New York: Wiley; 1980. [Google Scholar]
- 7.Zhang S, Pohnert G, Kongsaeree P, Wilson D B, Clardy J, Ganem B. J Biol Chem. 1998;273:6248–6253. doi: 10.1074/jbc.273.11.6248. [DOI] [PubMed] [Google Scholar]
- 8.Schmidheini T, Sperisen P, Paravicini G, Hütter R, Braus G. J Bacteriol. 1989;171:1245–1253. doi: 10.1128/jb.171.3.1245-1253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schnappauf G, Sträter N, Lipscomb W N, Braus G H. Proc Natl Acad Sci USA. 1997;94:8491–8496. doi: 10.1073/pnas.94.16.8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schnappauf G, Krappmann S, Braus G H. J Biol Chem. 1998;273:17012–17017. doi: 10.1074/jbc.273.27.17012. [DOI] [PubMed] [Google Scholar]
- 11.Schmidheini T, Mösch H-U, Evans J N S, Braus G. Biochemistry. 1990;29:3660–3668. doi: 10.1021/bi00467a011. [DOI] [PubMed] [Google Scholar]
- 12.Schmidheini T, Mösch H-U, Graf R, Braus G H. Mol Gen Genet. 1990;224:57–64. doi: 10.1007/BF00259451. [DOI] [PubMed] [Google Scholar]
- 13.Zalkin H, Paluh J L, van Cleemput M, Moye W S, Yanofsky C. J Biol Chem. 1984;259:3985–3992. [PubMed] [Google Scholar]
- 14.Graf R, Mehmann B, Braus G H. J Bacteriol. 1993;175:1061–1068. doi: 10.1128/jb.175.4.1061-1068.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hinnebusch A G. Mol Cell Biol. 1985;5:2349–2360. doi: 10.1128/mcb.5.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boeke J D, Trueheart J, Natsoulis G, Fink G R. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- 17.Miozzari G, Niederberger P, Hütter R. Arch Microbiol. 1977;115:307–316. doi: 10.1007/BF00446457. [DOI] [PubMed] [Google Scholar]
- 18.Guthrie C, Fink G R. Methods Enzymol. 1991;194:15. doi: 10.1016/0076-6879(91)94058-k. [DOI] [PubMed] [Google Scholar]
- 19.Woodcock D M, Crowther P J, Doherty J, Jefferson S, DeCruz E, Noyer-Weidner M, Smith S S, Michael M Z, Graham M W. Nucleic Acids Res. 1989;17:3469–3478. doi: 10.1093/nar/17.9.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider B L, Steiner B, Seufert W, Futcher A B. Yeast. 1996;12:129–134. doi: 10.1002/(sici)1097-0061(199602)12:2<129::aid-yea891>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 21.Rose M D, Novick P, Thomas J H, Botstein D, Fink G R. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- 22.Müller P P, Hinnebusch A G. Cell. 1986;45:201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- 23.Inoue H, Nojima H, Okayama H. Gene. 1990;96:23–28. doi: 10.1016/0378-1119(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 24.Elble R. BioTechniques. 1992;13:18–20. [PubMed] [Google Scholar]
- 25.Hoffman C S, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 26.Southern E M. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 27.Saiki R K, Scharf S, Faloona F, Mullis K B, Horn G T, Erlich H A, Arnheim N. Bio/Technology. 1992;24:476–480. [PubMed] [Google Scholar]
- 28.Cross F R, Tinkelenberg A H. Cell. 1991;65:875–883. doi: 10.1016/0092-8674(91)90394-e. [DOI] [PubMed] [Google Scholar]
- 29.Rave N, Crkvenjakov R, Boedtker H. Nucleic Acids Res. 1979;6:3559–3567. doi: 10.1093/nar/6.11.3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heiner C R, Hunkapiller K L, Chen S M, Glass J I, Chen E Y. Genome Res. 1998;8:557–561. doi: 10.1101/gr.8.5.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egan A F, Gibson F. Biochem J. 1972;130:847–859. doi: 10.1042/bj1300847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herbert D, Phipps P J, Strange R E. In: Methods of Microbiology. Norris J R, Ribbons D W, editors. 5B. New York: Academic; 1971. pp. 209–344. [Google Scholar]
- 33.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Graf R, Dubaquie Y, Braus G H. J Bacteriol. 1995;177:1645–1648. doi: 10.1128/jb.177.6.1645-1648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krappmann S, Helmstaedt K, Gerstberger T, Eckert S, Hoffmann B, Hoppert M, Schnappauf G, Braus G H. J Biol Chem. 1999;274:22275–22282. doi: 10.1074/jbc.274.32.22275. [DOI] [PubMed] [Google Scholar]
- 36.Krappmann S, Pries R, Gellissen G, Hiller M, Braus G H. J Bacteriol. 2000;182:4188–4197. doi: 10.1128/jb.182.15.4188-4197.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sträter N, Schnappauf G, Braus G, Lipscomb W N. Structure. 1997;5:1437–1452. doi: 10.1016/s0969-2126(97)00294-3. [DOI] [PubMed] [Google Scholar]