Abstract
The mechanism whereby the morphology and connectivity of the dendritic tree is regulated depends on an actin dynamics that, in turn, is controlled by Rho GTPases, a family of small GTP-binding proteins encompassing Rho, Rac, and Cdc42 subfamilies. Cytotoxic necrotizing factor 1 (CNF1), a protein toxin from Escherichia coli, constitutively activates Rho GTPases, thus leading to remodeling of the actin cytoskeleton in intact cells. Here, we show that the modulation of cerebral RhoA and Rac1 activity induced by CNF1 in mice leads to (i) rearrangement of cerebral actin cytoskeleton, (ii) enhanced neurotransmission and synaptic plasticity, and (iii) improved learning and memory in various behavioral tasks. The effects persist for weeks and are not observed in mice treated with a recombinant CNF1, in which the enzymatic activity was abolished by substituting serine to cysteine at position 866. The results suggest that learning ability can be improved through pharmacological manipulation of neural connectivity.
Keywords: cytotoxic necrotizing factor 1, brain, bacterial toxins, dendritic spines, drug therapy
Memory formation is thought to involve the rearrangement of synaptic connections in neural networks. Dendritic spines, which receive the majority of excitatory synapses (1, 2), undergo highly dynamic, experience-dependent changes (3). In addition, changes in the morphology of dendritic spines have been observed during long-term potentiation (LTP), a phenomenon that models the activity-dependent changes of synaptic efficacy that are believed to represent the cellular basis of learning (4, 5). Experimental evidence indicates that dendritic spine morphology and its rearrangement are governed by the neuronal actin cytoskeleton (6, 7).
Actin assembly and polymerization and actomyosin contraction are chiefly regulated by small GTPases belonging to the Rho family, a class of hydrolases that are highly conserved during evolution (8–10). The Rho GTPases, encompassing the subfamilies Rho, Rac, and Cdc42, are molecular switches for several signaling pathways and are ubiquitously expressed in eukaryotic cells (11). They oscillate between a GTP-bound active form targeted to specific membrane locations, and a GDP-bound inactive form sequestered in the cytosol. Active Rho GTPases can bind multiple effectors playing major roles in a plethora of cell functions, including cell differentiation, organization of the actin cytoskeleton, as well as DNA transcription (11). Thus, by controlling changes of neuron morphology (12–15), Rho GTPases might also play an essential role in learning and memory.
In fact, data coming from studies in humans confirm the potential involvement of this class of proteins in conditions associated with learning impairment. It has long been reported that the shape of dendrites and dendritic spines is affected in mental retardation (MR) (16, 17). The derangement of Rho GTPase signaling, which is consistently found in MR, is believed to underlie this feature (18–21). In particular, a deficit in activated Rho GTPases or their effector proteins has been demonstrated in some forms of MR (18, 21–23). This finding is compatible with the activating effects of Rac-GTP and Cdc42-GTP on actin cytoskeleton (8, 24). Therefore, the activation of Rho GTPases may possibly represent a strategy for treatments aimed at enhancing cognitive functions, such as memory and intelligence. Molecules endowed with such an effect could represent potential cognition enhancers. Yet, to the best of our knowledge, there is no direct evidence that selective pharmacological manipulation of Rho GTPase activity affects learning and memory.
To achieve this result, we injected intracerebroventricularly (i.c.v.) in C57BL/6 mice a 114 kDa protein toxin from Escherichia coli, named cytotoxic necrotizing factor 1 (CNF1). CNF1 catalyzes by deamidation the posttranslational modification of the glutamine 63/61 of Rho family members into glutamic acid (25, 26), leading to their permanent activation. Activation of Rho GTPases by CNF1 is followed by the deactivation of these regulatory proteins via their degradation by the ubiquitin–proteasome pathway (27). This activity requires the presence of cysteine in position 866 and histidine in position 881 (28). After a recovery time of 10 days or longer after the toxin injection, behavioral and electrophysiological parameters were assessed. We performed the study in two different mouse strains, the inbred C57BL/6 and the outbred CD1, obtaining overlapping results. However, taking into account the pitfalls of using outbred mouse strains (29), and the retinal degeneration observed in CD1 mice, which might affect the results of some behavioral tests (30), we have herein reported only results achieved with the inbred stock C57BL/6; behavioral responses to CNF1 of CD1 mice are reported in supporting information (SI) Figs. 6 and 7.
Taken altogether, the findings herein reported demonstrate that a single administration of CNF1 induces sustained enhancement of cognitive performances, thus highlighting the possibility to use this molecule as a pharmacological tool in neurological disorders.
Results
Enhancement of Fear Conditioning in CNF1-Treated Mice.
Given the ability of CNF1 to modulate the Rho GTPase activity together with the putative role of these proteins in learning and memory, we analyzed the toxin effects in behavioral tasks. First, the mice were tested for fear conditioning, a paradigm in which associative emotional memories are created. The animals learn to associate the fear of an electric shock with both the context and a conditioned tone. The first component is tested by placing the animal in the test cage and scoring the freezing time that represents an index of fear. The second component is tested by placing the mice in a different environment and measuring the freezing induced by the test tone. The amygdala is essential for conditioning in both cued and context tests. However, context conditioning is also dependent on hippocampal functioning (31). The experiments were performed in mice that had received (i) 0.6 fmol/kg CNF1, (ii) saline, and (iii) 0.6 fmol/kg CNF1 C866S, a recombinant toxin in which the change of cysteine with serine at position 866 abrogates the enzymatic activity on Rho GTPases (28). CNF1 C866S was used to verify whether the behavioral responses were due to CNF1-induced Rho GTPase activation.
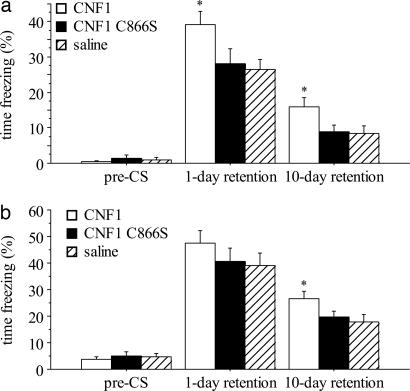
During the baseline of fear conditioning, immobility was similarly infrequent in the three groups (F2,35 = 0.334, P = 0.7182; Fig. 1a). The immediate freezing scores, i.e., those measured during conditioning for the duration of tone presentation, seemed to be similar in all groups (F2,35 = 1.666, P = 0.2036). The analysis of freezing values 24 h postconditioning demonstrated an improvement in context learning in the CNF1-treated, but not in CNF1 C866S-treated animals (F2,35 = 3.352, P = 0.0466; CNF1 significantly different from the saline-treated group, P < 0.05; Fig. 1a). Ten days postconditioning, CNF1-treated mice still performed better than the other two groups (F2,35 = 3.454, P = 0.0427; CNF1 significantly different from the saline-treated group, P < 0.05; Fig. 1a).
Fig. 1.
Enhancement of fear conditioning in CNF1-treated mice. (a) Contextual conditioning. (b) Cued fear conditioning. The value in each column represents the percentage of freezing time and is expressed as mean ± SEM. Test solutions were injected i.c.v. 10–11 days before the conditioning (0.6 fmol/kg CNF1, n = 12; saline, n = 12; 0.6 fmol/kg CNF1 C866S, n = 14 for context test and n = 11 for cued test). ∗, P < 0.05, significantly different from saline-treated group.
During the baseline time of cued test 24 h postconditioning, freezing was rarely observed (Fig. 1b), indicating that the novel context did not elicit fear in the conditioned mice. The presentation of the conditioned stimulus resulted in freezing, the degree of which was not significantly different among the three groups (F2,35 = 1.296, P = 0.2865). The animals were retested for cued conditioning 9 days later. In this test, mice treated with CNF1, but not CNF1 C866S, exhibited an increased response according to ANOVA (F2,32 = 3.311, P = 0.0493, CNF1 significantly different from saline-treated group, P < 0.05; Fig. 1b). In this experiment, we analyzed freezing dividing the time of presentation of the conditioned stimulus in 16 intervals of 20 s each. The ANOVA for repeated measurement of these data demonstrated a significant effect of time (F15,435 = 13.670, P < 0.0001), thus suggesting extinction of the conditioned fear. However, extinction was not significantly different in the three groups (time × treatment interaction: F30,435 = 1.335, P = 1.145). We then reanalyzed the results of context tests dividing the observation in five periods of 1 min each. The interaction time × treatment was again not significant (data not included).
Overall, the results indicate an increased conditioning, both cued- and context-dependent, suggesting a general improvement of associative learning that extends beyond hippocampal functioning. The enhancing effects of the toxin expand to the long-term memory of the aversive experience. Moreover, CNF1 does not seem to affect the extinction of the conditioned fear. The nonsignificant differences in cued test 24 h postconditioning might be explained by a “ceiling effect,” i.e., a conditioned response that is close to the saturation. To further verify whether the results were biased by differential sensitivity to the unconditioned stimulus, we measured the nociceptive threshold, i.e., we determined the lowest currents that elicited jumping or vocalizing in the conditioning cages. This parameter was not affected by CNF1 (304 ± 20, 287 ± 30, 314 ± 24 μA, mean ± SEM, for mice treated with saline, n = 12, 0.6 fmol/kg CNF1, n = 12, and 0.6 fmol/kg CNF1 C866S, n = 14, respectively; F2,35 = 0.298; P = 0.7445).
Improvement of Spatial Learning in CNF1-Treated Mice.
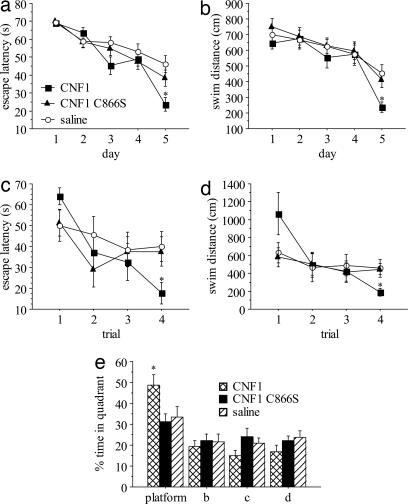
To explore the effects of 0.6 fmol/kg CNF1 or CNF1 C866S i.c.v. on spatial learning, we trained mice to find a fixed, hidden platform in a water maze paradigm. The test does not require the administration of painful aversive stimuli and it depends on hippocampal functioning (32). The performances improved across the training sessions, indicating that mice learned the platform position (effect of learning session: F4,136 = 30.963, P < 0.0001 and F4,136 = 19.020, P < 0.0001 for escape latencies and swim distance, respectively, by a three-way repeated measurement ANOVA with two ways on repetition; Fig. 2a and b). However, escape latencies decreased at different rates in the three groups (effect of the treatment × day interaction: F8,136 = 2.082, P = 0.0416). In particular, the CNF1-treated group performed better on day 5 (F2,34 = 3.337, P = 0.0475 and F2,34 = 4.699, P = 0.0158 for escape latencies and swim distance, respectively, by ANOVA for repeated measurements; P < 0.05 by post hoc comparison with saline-treated group for both; Fig. 2 a and b). Representative water maze experiments showing the performances of a saline-treated, a CNF1-treated, and a CNF1 C866S-treated mouse in the last learning trial are reported as supporting information (SI Movies 1–3, respectively).
Fig. 2.
Improved water maze performances in CNF1-treated mice. Shown is summary of escape latencies (a) and swim distances (b) in place learning. Shown is summary of escape latencies (c) and swim distances (d) in reversal learning. (e) Percentage of time spent in the pool quadrant in spatial probe. Data are expressed as mean ± SEM. Mice were injected i.c.v. with saline (n = 12), 0.6 fmol/kg CNF1 (n = 12), or 0.6 fmol/kg CNF1 C866S (n = 13) 10 days before the training. ∗, P < 0.05, significantly different from saline-treated group.
Even though the mice learned to locate the goal position, they could have been relying on the use of nonspatial strategies, such as learning that the hidden platform is at a certain distance from the edge of the pool. To determine whether the mice had formed spatial memories during the task, 4 days after place learning, the platform was removed and we measured the time spent in the four quadrants in which the pool had been arbitrarily divided. CNF1-treated mice showed a more marked preference for the platform quadrant, indicating an enhanced spatial learning (F2,34 = 4.145, P = 0.0245; P < 0.05 by post hoc test; Fig. 2e). Representative water maze experiments showing the performances of a saline-treated, a CNF1-treated, and a CNF1 C866S-treated mouse in the spatial probe are reported as supporting information (SI Movies 4–6, respectively).
On the subsequent day, we placed the hidden platform in a quadrant different from the one in which it was during place learning, and studied the ability of trained mice to locate the novel position (reversal learning). The analysis of escape latencies (Fig. 2c) shows a significant interaction between the trial number and the treatment (F6,102 = 2.191, P = 0.0497; effects of treatment: F2,34 = 0.359, ns; effects of trial: F3,102 = 8.295, P < 0.0001), as did the analysis of swim distances (F6,102 = 3.038, P = 0.0089; effects of treatment: F2,34 = 0.095, ns; effects of trial: F3,102 = 8.801, P < 0.0001; Fig. 2d). In the last trial of learning, CNF1-treated mice, but not CNF1 C866S, performed significantly better than saline-treated (F2,34 = 3.391, P = 0.0454 and F2,34 = 4.128, P = 0.0248 for escape latencies and swim distances, respectively; CNF1 significantly different from saline by post hoc comparison, P < 0.05 for both). This finding suggests a positive effect of CNF1 on the ability to locate the novel platform position, which could be at least in part explained by better spatial learning during prior training. Finally, because sensorimotor and/or motivational differences might have biased the results, we trained the mice in a modified water maze task in which the platform had been made visible. The analysis of this cued learning failed to demonstrate any significant effect of the treatment and its interactions (data not shown). In conclusion, CNF1 improves performances in water maze experiments and enhanced spatial learning seems to have specifically contributed to these effects.
CNF1-Mediated Activation of Rho GTPases and Actin Remodeling in Brain Tissue of Mice.
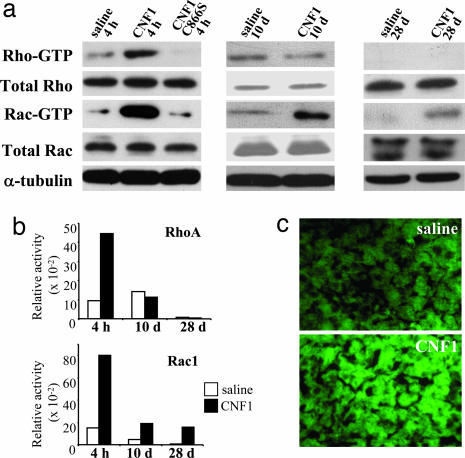
To evaluate the activity of CNF1 on cerebral Rho GTPases in vivo, we asked which was the activation state of these regulatory proteins in brain samples collected at different time points (4 h, 10 days, 28 days) after i.c.v. injection. Rho-GTP and Rac-GTP were revealed by a pull-down assay, a test based on the use of effectors capable of binding activated Rho GTPases (27). In brains collected from animals 4 h after the injection of CNF1, we observed a sharp activation of both Rac and Rho GTPases (Fig. 3a and b), demonstrating that CNF1 is able to activate Rho proteins in vivo. It is worth noting that the activation state of Rho GTPases in brain tissues from CNF1 C866S-treated mice was comparable to that observed in samples derived from saline-treated mice (Fig. 3 a and b), thus proving that the activation of Rho proteins is actually due to the enzymatic activity of the toxin. We then monitored the activation state of Rho and Rac at the beginning (10 days) and at the end (28 days) of the time frame when the behavioral tests were performed. In both cases the activation state of Rho in CNF1-treated mice was nearly identical to that observed in controls, whereas Rac resulted to be still activated, although at a reduced extent as compared with the 4 h experiment (Fig. 3 a and b).
Fig. 3.
CNF1 causes long-lasting activation of Rac and actin polymerization in brains from C57BL/6 mice. (a) Immunoblots showing the amount of both total and activated Rho and Rac (Rho-GTP and Rac-GTP) in the left hemisphere at 4 h, 10 days, and 28 days after CNF1 injection in the controlateral hemisphere. α-Tubulin was used to verify the equal amount of proteins subjected to pull-down assays. (b) Densitometric analysis of the immunoblots showed in a. The histograms represent the GTPase activity normalized for the amount of total protein loaded. (c) Fluorescence micrographs of representative sections of mouse left parietal cortex stained with FITC-phalloidin for F-actin detection. (Magnification: ×40.)
Because the ability of Rho GTPases to control the organization of the actin cytoskeleton is well known, we asked whether the modulation of these regulatory proteins by CNF1 could somehow affect the actin network in brains derived from toxin-treated mice. To answer this question, we stained cerebral sections with phalloidin, a compound that selectively binds filamentous (F)-actin (33), linked to a fluorescent probe. The observations performed 15 days after 0.6 fmol/kg CNF1 i.c.v. evidenced both an increase in staining intensity and an enrichment in stained areas throughout the whole brain (Fig. 3b; percent stained area in the parietal cortex: 65.44 ± 2.36 and 57.37 ± 2.68, for CNF1 and saline treated mice, respectively, mean ± SEM, n = 7 for both groups; significantly different by ANOVA after angular transformation of data, F1,12 = 5.036, P = 0.0453), thus indicating the ability of CNF1 to remodel actin in brain tissue in vivo.
CNF1 Reshapes the Dendritic Actin Cytoskeleton in Primary Cultures of Neural Cells.
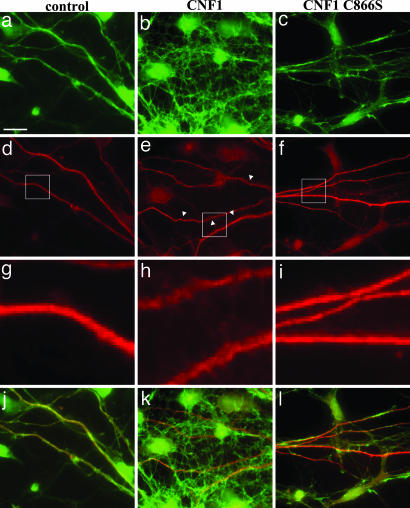
The effect on the mouse brain actin prompted us to analyze the cytoskeleton changes caused by CNF1 in cultured neural cells from the cortex of C57BL/6 mice. After 24 h of exposure to either CNF1 or CNF1 C866S, cells were double-stained with fluorescein-phalloidin for F-actin detection and with an anti-MAP2 antibody that labels dendrites. By fluorescence microscopy, we observed that challenge with CNF1 induced actin cytoskeleton enrichment (Fig. 4b) as well as changes in dendritic processes (Fig. 4 e and h), which appeared “wrinkled” (arrowheads). The above changes were absent when cells were treated with CNF1 C866S (Fig. 4 c, f, and i).
Fig. 4.
Effect of CNF1 on primary neural cultured cells from C57BL/6 mice. Fluorescence microscopy analysis of neural cells, either untreated (a, d, g, and j) or treated with CNF1 (b, e, h, and k) or with CNF1 C866S (c, f, i, and l) for 24 h and then stained for F-actin (green, a–c) and MAP2 (red, d–f) detection. g–i show details of selected areas (squares) of images in d–f. j–l represent the merge of actin and MAP2 staining. [Scale bar: 10 μm (a–f; j-l) and 2 μm (g–i).]
These results highlight the ability of CNF1 to induce, in primary neuronal cells, an increase in dendritic surface, particularly by spine-like neo-formations, accompanied by actin cytoskeleton reshape. Importantly, this effect was critically dependent on Rho GTPases' activation.
Enhancement of Hippocampal Neurotransmission in CNF1-Treated Mice.
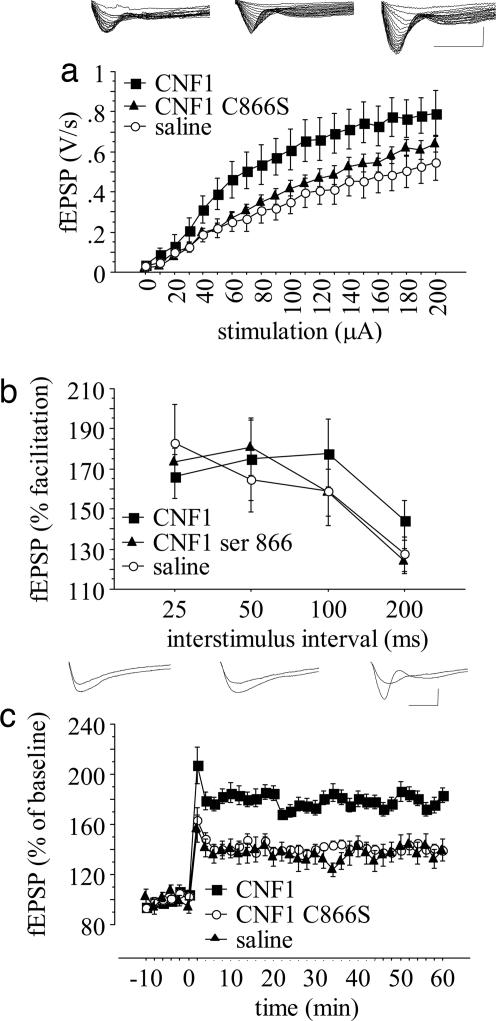
Rho GTPase activation (34), increase in spine F-actin content (6), and morphologic changes of spines (4, 5), are associated with LTP. In addition, the pharmacological manipulation of Rho GTPases affects LTP (35). Hence, to determine whether changes in synaptic plasticity might account for the cognitive enhancing properties of CNF1, we examined LTP induced by high-frequency stimulation of the Schaffer collateral/CA1 synapses, a pathway the plasticity of which is essential for spatial learning (36). The experiments were performed in slices from mice treated with 0.6 fmol/kg CNF1, 0.6 fmol/kg CNF1 C866S, or saline. The size of the potentiation observed at 60 min posttetanus was different in the three groups [F2,27 = 5.6178, P = 0.0091 by analysis of covariance (ANCOVA), Fig. 5c]: CNF1 determined a significant increase in potentiation as compared with saline (P < 0.05 by individual ANCOVA with Bonferroni's correction), whereas mice treated with the recombinant toxin matched the potentiation observed in the control group (F1,18 = 0.0305, P = 0.9565 by individual ANCOVA with Bonferroni's correction). The changes do not seem to be caused by abnormal presynaptic function. In fact, a phenomenon sensitive to presynaptic changes such as paired-pulse facilitation (PPF), is not affected by either CNF1 or CNF1 C866S (treatment: F2,23 = 0.2009, P = 0.8194; treatment × interpulse interval interaction F6,72 = 0.8665, P = 0.5238 by ANCOVA for repeated measurements; Fig. 5b). Basal neurotransmission, as assessed by the study of input-output curves, was also significantly increased by CNF1 (effects of treatment: F2,28 = 3.356, P = 0.0494, CNF1 significantly different from saline, P < 0.05; stimulation intensity: F20,560 = 126.507, P < 0.0001; treatment × intensity interaction: F40,560 = 2.314, P < 0.0001, CNF1 significantly different from saline by post hoc comparisons; Fig. 5a). A subset of slices from CNF1 treated mice were perfused with 10 μM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), which completely abolished basal responses, thus suggesting that AMPA currents account for the increased neurotransmission. Overall, the results indicate that the enzymatic activity of CNF1 enhances hippocampal excitatory neurotransmission and its long-term plasticity.
Fig. 5.
CNF1 enhances hippocampal CA1 neurotransmission. Mice were injected i.c.v. 21–28 days before the experiments. (a) Input–output function at CA3–CA1 synapse (saline, n = 11; CNF1, n = 10; CNF1 C866S, n = 10) with representative traces (Left, saline; Center, CNF1 C866S; Right, CNF1). (b) PPF (saline, n = 10; CNF1, n = 8; CNF1 C866S, n = 9) at different interstimulus intervals. (c) LTP (saline, n = 11; CNF1, n = 10; CNF1 C866S, n = 10) with representative traces (Left, saline; Center, CNF1 C866S; Right, CNF1). Means ± SEM are shown. (Horizontal scale bar: 5 ms; vertical scale bar: 0.5 mV.)
As anticipated in the Introduction, all behavioral tests have been performed also in the outbred mouse strain CD1 (SI Fig. 6, fear conditioning; SI Fig. 7, water maze; SI Fig. 8, hippocampal slice electrophysiology). Importantly, the results obtained with CD1 mice were consistent with those achieved with C57BL/6 mice (Figs. 1, 2, and 5), thus strengthening the significance of CNF1 activity in learning and memory.
Discussion
In this study, we have shown that CNF1 improves learning and memory in laboratory animals and activates brain Rho GTPases. Rho signaling is intimately linked to actin filaments and cellular morphology and, consistently, we found that polymerized actin was overexpressed throughout the brain. The Rho GTPases also play an essential role in the development and structural plasticity of dendrites and dendritic spines (37) and, accordingly, we observed actin changes and spines formation in primary neuronal cells from the mouse cortex. The activation of Rho GTPases by CNF1 seems to be mandatory for the observed behavioral changes because the recombinant CNF1 C866S, devoid of enzymatic activity (28), was ineffective in the behavioral experiments and failed in inducing Rho GTPase activation and actin changes. Furthermore, pull down experiments, showing a long-lasting activation of Rac and data on behavioral experiments are consistent with a previous report indicating that the loss of WAVE-1, a target of Rac protein, reduces learning and memory in mice (38).
Actin dynamics and neuronal morphology are differently regulated by Rho and Rac. It has been shown that transfection of neurons from rat cerebral cortex, so that they encoded for constitutively activated Rac GTPase, leads to an increase in the number of dendrites per neuron (12, 14), whereas dominant negative or inhibited forms of the proteins leads to the opposite effect (14). This increased neuronal branching might generate increased connectivity and enhanced learning ability. Accordingly, human hemizygosity of LIM-domain-containing protein kinase (LIMK), which is activated by p21-activated kinase (PAK), an effector of Rac GTPase, has been associated with MR (23). On the other hand, Rho activation has been shown to cause neurite retraction (39, 40) by means of formation of stress fibers in the growth cone (41) and to reduce growth cone mobility and dendrite branching (42, 43). The meaning of these latter actions in the physiology of learning is unclear. Possibly, the demolitive changes after Rho activation are essential for the structural plasticity associated with experience and memory formation. In support of this idea, a negative effect on learning has been reported after the inhibition of one of the main effector proteins of Rho, i.e., Rho-associated kinase (ROCK) (44, 45).
Besides regulating the actin cytoskeleton, Rho GTPases also play a pivotal role in guiding a variety of other cellular processes, such as proliferation, transformation, and gene transcription. Moreover, they participate in the activation of nuclear factor-κB (NF-κB), a group of structurally related and evolutionarily conserved transcription factors involved in regulating the expression of several genes. As recently reviewed by Meffert and Baltimore (46), NF-κB family members are activated within the CNS and play a crucial role in learning and memory. Inhibition of NF-κB has been reported to produce deficits in a variety of learning paradigms in mice. It is worth noting that NF-κB activation has been demonstrated in CNF1-treated cells as a result of Rho GTPase stimulation (47).
Despite all evidence about the potential role of Rho GTPase in the structural plasticity of the CNS, to the best of our knowledge, there is no proof so far that their selective activation leads to increased learning abilities. Posttraining activation of Rho has already been associated with morphological changes in neuron and enhanced long-term spatial memory (45). However, in the above-mentioned study, Rho was activated by intra-hippocampal administration of oleoyl-l-α-lysophosphatidic acid (LPA). LPA effect is not selective, because it also activates extracellular-signal regulated kinase, phosphoinositide-3-kinase and protein-kinase C. All these molecules can modulate memory formation (see discussion in 45 for references).
Increased hippocampal excitatory neurotransmission might have played a role in the improved cognition induced by CNF1. We have also found that CNF1 enhances LTP. The occurrence of actin polymerization has been reported during this phenomenon (6). The polymerization begins early after potentiation and persists for at least 5 weeks. Apparently, actin polymerization is necessary but not sufficient for potentiation (6, 48, 49). Therefore, this mechanism might not occlude with the cascade of LTP. Indeed, as our results suggest, besides facilitating basal excitatory neurotransmission, increased content in synaptic F-actin might synergistically contribute to activity-dependent synaptic plasticity.
Overall, our data indicate that Rho GTPases might represent a target for pharmacological treatments aimed at improving cognition. Several cellular mechanisms are involved in learning, the majority of which do not seem to be modified in conditions associated with reduced cognitive ability. Conversely, Rho GTPase signaling and spine morphology are consistently affected in MR and it has even been hypothesized that Rho GTPases polymorphism might contribute to individual differences in intellectual functions (50). Therefore, CNF1 acts on a mechanism that might represent a rate-limiting step for cognition in humans. This consideration implies the potential pharmacological importance of the molecule. Because all experiments were performed between 10 and 28 days after a single injection, it seems conceivable a prolonged effect of the toxin on behavioral and electrophysiology parameters, which would be in accordance with the prolonged effect of CNF1 in peripheral tissues (51) and with its known mechanism of action. CNF1 can thus be regarded as endowed with long-lasting cognition enhancing properties.
Methods
CNF1 and CNF1 C866S Preparation.
CNF1 was obtained from the 392 ISS strain (kindly provided by V. Falbo, Istituto Superiore di Sanità, Rome, Italy) and purified as described (52). The plasmid coding for CNF1 C866S was kindly provided by E. Lemichez (U627 and Institut National de la Santé et de la Recherche Médicale, Nice, France), and this recombinant toxin was prepared as described (25). The toxin was purified to homogeneity and sterilized before injection.
Animals and Toxin Administration.
The experiments, which were approved by the ad hoc committee of the Italian Ministry of Health, were performed in male 2-month-old C57BL/6 mice, weighing 24–26 g (Harlan Italy, S. Pietro al Natisone, UD, Italy). Animal use and care conformed to the directives of the European Communities Council (1986). The surgical procedure is described in SI Methods.
Pull-Down Assay.
This assay was conducted immediately after collection (4 h and 10 and 28 days after injection) of fresh brain left hemispheres (controlateral to the injection site). Homogenized samples were processed for pull-down assay as described (51). Details are provided in SI Methods.
Fear Conditioning.
Mice were individually placed in the text chamber. After a baseline time (192 s), they received three tone-shock pairings. Twenty-four hours after the conditioning, the mice were placed back in the test chamber for 5 min and scored for freezing (contextual conditioning). Subsequently, they were moved to a novel chamber in a different environment, in which they were scored for freezing during a 192-s baseline time followed by a 320-s tone identical to the conditioned stimulus (cued conditioning). Both contextual and cued conditioning were reassessed 10 days postconditioning. Details are provided in SI Methods.
Water Maze.
Mice were trained to learn the position of a hidden platform during 5 consecutive days. Details of the experimental procedure, settings, and materials used are reported in SI Methods.
Hippocampal Slice Electrophysiology.
The experiments were performed 21–28 days postsurgery. Animals that had not been used for behavioral experiments were killed by decapitation under urethane anesthesia (1.5 g/kg i.p.). The head was rapidly put on ice, and the brain was removed and dissected. Electrophysiological recordings of hippocampal slices were obtained as detailed in SI Methods.
Cultured Cell Preparation and Treatments.
Primary cultures were obtained from the cortex of mice embryos on day 15 of gestation. Details are provided in SI Methods.
Fluorescence Microscopy.
Brain sections.
Brains were frozen, and cryosections of control and treated samples were obtained. After fixation with formaldehyde, samples were processed for F-actin detection. Details are available in SI Methods.
Cell cultures.
Neural cells were seeded on 13-mm glass coverslips and treated with 10−10 M CNF1 or with 10−10 M CNF1 C866S for 24 h. Cells were processed for F-actin and MAP-2 staining. Details are available in SI Methods.
Statistical Analysis.
Data from fear conditioning were analyzed by ANOVA. Repeated measurement ANOVA was used to analyze water maze learning data (one way on between-group comparison; two ways on repetition for place learning, one way on repetition for reversal and cued learning). In electrophysiological experiments, input-output curves were analyzed by repeated measurement ANOVA. LTP and PPF data were analyzed by ANCOVA, by using baseline responses as covariate. A t test with Bonferroni's correction was used for post hoc individual comparisons. All calculations were performed with Statistica 5.0 for Windows.
Supplementary Material
Acknowledgments
We thank F. Malchiodi Albedi for criticisms. This work was supported in part by Istituto Superiore di Sanità Grants C3OQ-2004 (to G.D.) and C3GD-2004 (to C.F.). Financial support for G.V. was provided by the “Enrico ed Enrica Sovena” Foundation of Rome.
Abbreviations
- LTP
long-term potentiation
- MR
mental retardation
- CNF1
cytotoxic necrotizing factor 1.
Footnotes
Conflict of interest statement: A patent application has been filed under PCT/EP2006/003811.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610059104/DC1.
References
- 1.Harris KM. Curr Opin Neurobiol. 1999;9:343–348. doi: 10.1016/s0959-4388(99)80050-6. [DOI] [PubMed] [Google Scholar]
- 2.Nimchinsky EA, Sabatini BL, Svoboda K. Annu Rev Physiol. 2002;64:313–353. doi: 10.1146/annurev.physiol.64.081501.160008. [DOI] [PubMed] [Google Scholar]
- 3.Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- 4.Lang C, Barco A, Zablow L, Kandel ER, Siegelbaum SA, Zakharenko SS. Proc Natl Acad Sci USA. 2004;101:16665–16670. doi: 10.1073/pnas.0407581101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller D, Toni N, Buchs PA. Hippocampus. 2000;10:596–604. doi: 10.1002/1098-1063(2000)10:5<596::AID-HIPO10>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 6.Fukazawa Y, Saitoh Y, Ozawa F, Ohta Y, Mizuno K, Inokuchi K. Neuron. 2003;38:447–460. doi: 10.1016/s0896-6273(03)00206-x. [DOI] [PubMed] [Google Scholar]
- 7.Zito K, Knott G, Shepherd GM, Shenolikar S, Svoboda K. Neuron. 2004;44:321–334. doi: 10.1016/j.neuron.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 8.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Nat Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 9.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, et al. Science. 1996;273:245–248. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 10.Maekawa M, Ishizaki T, Boku S, Watanabe N, Fujita A, Iwamatsu A, Obinata T, Ohashi K, Mizuno K, Narumiya S. Science. 1999;285:895–898. doi: 10.1126/science.285.5429.895. [DOI] [PubMed] [Google Scholar]
- 11.Etienne-Manneville S, Hall A. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 12.Luo L, Hensch TK, Ackerman L, Barbel S, Jan LY, Jan YN. Nature. 1996;379:837–840. doi: 10.1038/379837a0. [DOI] [PubMed] [Google Scholar]
- 13.Luo L. Nat Rev Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 14.Tashiro A, Minden A, Yuste R. Cereb Cortex. 2000;10:927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- 15.Threadgill R, Bobb K, Ghosh A. Neuron. 1997;19:625–634. doi: 10.1016/s0896-6273(00)80376-1. [DOI] [PubMed] [Google Scholar]
- 16.Purpura DP. Science. 1974;186:1126–1128. doi: 10.1126/science.186.4169.1126. [DOI] [PubMed] [Google Scholar]
- 17.Kaufmann WE, Moser HW. Cereb Cortex. 2000;10:981–991. doi: 10.1093/cercor/10.10.981. [DOI] [PubMed] [Google Scholar]
- 18.Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA. Nat Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- 19.Billuart P, Bienvenu T, Ronce N, des Portes V, Vinet MC, Zemni R, Roest Crollius H, Carrie A, Fauchereau F, Cherry M, et al. Nature. 1998;392:923–926. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- 20.Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, et al. Nat Genet. 2000;26:247–250. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
- 21.Pasteris NG, Cadle A, Logie LJ, Porteous ME, Schwartz CE, Stevenson RE, Glover TW, Wilroy RS, Gorski JL. Cell. 1994;79:669–678. doi: 10.1016/0092-8674(94)90552-5. [DOI] [PubMed] [Google Scholar]
- 22.Bienvenu T, des Portes V, McDonell N, Carrie A, Zemni R, Couvert P, Ropers HH, Moraine C, van Bokhoven H, Fryns JP, et al. Am J Med Genet. 2000;93:294–298. doi: 10.1002/1096-8628(20000814)93:4<294::aid-ajmg8>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 23.Frangiskakis JM, Ewart AK, Morris CA, Mervis CB, Bertrand J, Robinson BF, Klein BP, Ensing GJ, Everett LA, Green ED, et al. Cell. 1996;86:59–69. doi: 10.1016/s0092-8674(00)80077-x. [DOI] [PubMed] [Google Scholar]
- 24.Takenawa T, Miki H. J Cell Sci. 2001;114:1801–1809. doi: 10.1242/jcs.114.10.1801. [DOI] [PubMed] [Google Scholar]
- 25.Flatau G, Lemichez E, Gauthier M, Chardin P, Paris S, Fiorentini C, Boquet P. Nature. 1997;387:729–733. doi: 10.1038/42743. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt G, Sehr P, Wilm M, Selzer J, Mann M, Aktories K. Nature. 1997;387:725–729. doi: 10.1038/42735. [DOI] [PubMed] [Google Scholar]
- 27.Doye A, Mettouchi A, Bossis G, Clement R, Buisson-Touati C, Flatau G, Gagnoux L, Piechaczyk M, Boquet P, Lemichez E. Cell. 2002;111:553–564. doi: 10.1016/s0092-8674(02)01132-7. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt G, Selzer J, Lerm M, Aktories K. J Biol Chem. 1998;273:13669–13674. doi: 10.1074/jbc.273.22.13669. [DOI] [PubMed] [Google Scholar]
- 29.Chia R, Achilli F, Festing MF, Fisher EM. Nat Genet. 2005;37:1181–1186. doi: 10.1038/ng1665. [DOI] [PubMed] [Google Scholar]
- 30.Serfilippi LM, Pallman DR, Gruebbel MM, Kern TJ, Spainhour CB, Chia R, Achilli F, Festing MF, Fisher EM. Comp Med. 2004;54:69–76. [PubMed] [Google Scholar]
- 31.Phillips RG, LeDoux JE. Behav Neurosci. 1992;106:274–285. doi: 10.1037//0735-7044.106.2.274. [DOI] [PubMed] [Google Scholar]
- 32.Morris RG, Garrud P, Rawlins JN, O'Keefe J. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 33.Capani F, Martone ME, Deerinck TJ, Ellisman MH. J Comp Neurol. 2001;435:156–170. doi: 10.1002/cne.1199. [DOI] [PubMed] [Google Scholar]
- 34.O'Kane EM, Stone TW, Morris BJ. J Neurochem. 2003;87:1309–1312. doi: 10.1046/j.1471-4159.2003.02102.x. [DOI] [PubMed] [Google Scholar]
- 35.O'Kane EM, Stone TW, Morris BJ. Neuropharmacology. 2004;46:879–887. doi: 10.1016/j.neuropharm.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 36.Tsien JZ, Huerta PT, Tonegawa S. Cell. 1996;87:1327–1338. doi: 10.1016/s0092-8674(00)81827-9. [DOI] [PubMed] [Google Scholar]
- 37.Li Z, Aizenman CD, Cline HT. Neuron. 2002;33:741–750. doi: 10.1016/s0896-6273(02)00621-9. [DOI] [PubMed] [Google Scholar]
- 38.Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J, Scott JD. Proc Natl Acad Sci USA. 2003;100:1723–1728. doi: 10.1073/pnas.0438033100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borisoff JF, Chan CC, Hiebert GW, Oschipok L, Robertson GS, Zamboni R, Steeves JD, Tetzlaff W. Mol Cell Neurosci. 2003;22:405–416. doi: 10.1016/s1044-7431(02)00032-5. [DOI] [PubMed] [Google Scholar]
- 40.Yuan XB, Jin M, Xu X, Song YQ, Wu CP, Poo MM, Duan S. Nat Cell Biol. 2003;5:38–45. doi: 10.1038/ncb895. [DOI] [PubMed] [Google Scholar]
- 41.Nobes CD, Hall A. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- 42.Li Z, Van Aelst L, Cline HT. Nat Neurosci. 2000;3:217–225. doi: 10.1038/72920. [DOI] [PubMed] [Google Scholar]
- 43.Wong WT, Faulkner-Jones BE, Sanes JR, Wong RO. J Neurosci. 2000;20:5024–5036. doi: 10.1523/JNEUROSCI.20-13-05024.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamprecht R, Farb CR, LeDoux JE. Neuron. 2002;36:727–738. doi: 10.1016/s0896-6273(02)01047-4. [DOI] [PubMed] [Google Scholar]
- 45.Dash PK, Orsi SA, Moody M, Moore AN. Biochem Biophys Res Commun. 2004;322:893–898. doi: 10.1016/j.bbrc.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 46.Meffert MK, Baltimore D. Trends Neurosci. 2005;28:37–43. doi: 10.1016/j.tins.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 47.Boyer L, Travaglione S, Falzano L, Gauthier NC, Popoff MR, Lemichez E, Fiorentini C, Fabbri A. Mol Biol Cell. 2004;15:1124–1133. doi: 10.1091/mbc.E03-05-0301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim CH, Lisman JE. J Neurosci. 1999;19:4314–4324. doi: 10.1523/JNEUROSCI.19-11-04314.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krucker T, Siggins GR, Halpain S. Proc Natl Acad Sci USA. 2000;97:6856–6861. doi: 10.1073/pnas.100139797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramakers GJ. Trends Neurosci. 2002;25:191–199. doi: 10.1016/s0166-2236(00)02118-4. [DOI] [PubMed] [Google Scholar]
- 51.Travaglione S, Messina G, Fabbri A, Falzano L, Giammarioli AM, Grossi M, Rufini S, Fiorentini C. Cell Death Differ. 2005;12:78–86. doi: 10.1038/sj.cdd.4401522. [DOI] [PubMed] [Google Scholar]
- 52.Falzano L, Fiorentini C, Donelli G, Michel E, Kocks C, Cossart P, Cabanie L, Oswald E, Boquet P. Mol Microbiol. 1993;9:1247–1254. doi: 10.1111/j.1365-2958.1993.tb01254.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.