Abstract
The suprachiasmatic nucleus (SCN), the brain's principal circadian pacemaker, coordinates adaptive daily cycles of behavior and physiology, including the rhythm of sleep and wakefulness. The cellular mechanism sustaining SCN circadian timing is well characterized, but the neurochemical pathways by which SCN neurons coordinate circadian behaviors remain unknown. SCN transplant studies suggest a role for (unidentified) secreted factors, and one potential candidate is the SCN neuropeptide prokineticin 2 (Prok2). Prok2 and its cognate prokineticin receptor 2 (Prokr2/Gpcr73l1) are widely expressed in both the SCN and its neural targets, and Prok2 is light-regulated. Hence, they may contribute to cellular timing within the SCN, entrainment of the clock, and/or they may mediate circadian output. We show that a targeted null mutation of Prokr2 disrupts circadian coordination of the activity cycle and thermoregulation. Specifically, mice lacking Prokr2 lost precision in timing the onset of nocturnal locomotor activity; and under both a light/dark cycle and continuous darkness, there was a pronounced temporal redistribution of activity away from early to late circadian night. Moreover, the coherence of circadian behavior was significantly reduced, and nocturnal body temperature was depressed. Entrainment by light is not, however, dependent on Prokr2, and bioluminescence real-time imaging of organotypical SCN slices showed that the mutant SCN is fully competent as a circadian oscillator. We conclude that Prokr2 is not necessary for SCN cellular timekeeping or entrainment, but it is an essential link for coordination of circadian behavior and physiology by the SCN, especially in defining the onset and maintenance of circadian night.
Keywords: hypothalamus, neuropeptide, sleep, thermoregulation, gene knockout
The suprachiasmatic nucleus (SCN) of the hypothalamus constitutes an autonomous circadian pacemaker necessary for the maintenance of daily rhythms, including the cycle of sleep and wakefulness (1). The molecular basis of the cellular oscillator of the SCN is broadly characterized, but how the SCN signals circadian time to other sites in the brain and thence to peripheral tissues is unclear (2). Neuroanatomical projections from the SCN run predominantly to local hypothalamic and thalamic structures (3), including the dorsomedial nucleus (DMN), a principal site for the regulation of sleep and wakefulness and circadian arousal by behavioral cues (4, 5). How such “hard-wired” connections influence SCN targets remains open to question. Although electrical signaling by means of synaptic contacts is likely important (6), encapsulated SCN grafts unable to form synaptic contacts with host tissues are nevertheless able to establish circadian activity/rest cycles in previously arrhythmic, SCN-lesioned recipients (7). This finding indicates that paracrine factors from the SCN mediate circadian coordination within the brain. A small number of candidate factors have been explored, including TGF-α (8) and cardiotrophin-like cytokine (9). These factors are expressed by the SCN on a circadian basis, and local infusion of exogenous peptide acutely suppresses locomotor activity. There is, however, a paucity of genetic evidence to support their proposed roles.
Prokineticin 2 (Prok2) is a secretory peptide, expressed in gut and brain, including the SCN where its transcript oscillates on a circadian basis, peaking in subjective daytime (the inactive phase of nocturnal rodents). Prok2 is acutely induced by light pulses that reset the clock (10). Central infusion of recombinant Prok2 suppresses nocturnal locomotor activity in rats, and endogenous Prok2 has therefore been proposed as an inhibitory output of the SCN, preventing excessive diurnal activity in nocturnal mammals. The Prokr2 (Gpcr73l1) gene encodes a G protein-coupled receptor for Prok2 that is expressed in both the SCN and diencephalic targets of the SCN, including the medial thalamus and DMN. To test the putative contribution(s) of Prokr2-mediated signaling to circadian timing, we have generated a null mutation of the gene, and we have examined its impact on entrained and free-running activity/rest cycles, body temperature rhythms, and on the molecular oscillator of the SCN, as reported by bioluminescent gene expression in organotypic culture. Although loss of Prokr2-mediated signaling has no effect on entrainment to the light/dark cycle, it profoundly disrupts the activity/rest cycle under both entrained and free-running conditions, with imprecise activity onsets, loss of consolidation, and a relative redistribution of activity from early to late circadian night. Moreover, the nocturnal increase in core body temperature is attenuated in mutants. Importantly, loss of Prokr2 has no effect on the molecular oscillator insofar as circadian cycles of bioluminescent gene expression in the SCN are not compromised. We conclude that the receptor is not necessary for circadian pacemaking nor entrainment, but it is a necessary link in the suprachiasmatic coordination of behavior with a specific role in defining the onset and maintenance of circadian night.
Results
The prokr2Brdm1 Mutation Is a Functional Null Allele.
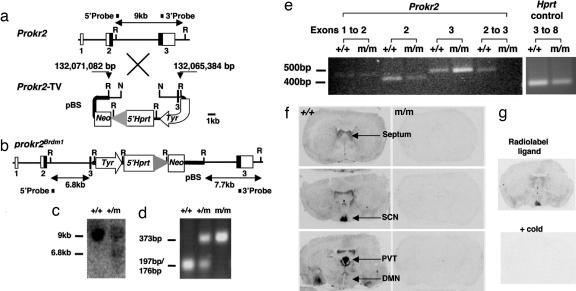
The Prokr2 insertion-targeting vector was designed to create a functional null allele for the Prokr2 gene by duplication of the exon 3 splice acceptor and 147 bp of coding sequence fused to the vector backbone of the 5′ hypoxanthine phosphoribosyltransferase (Hprt) vector (11, 12) (Fig. 1a and b). The Prokr2-targeting vector was electroporated into AB2.2 (129S7/SvEvBrdHprtb-m2-derived) ES cells, and Southern blot analysis of G418-resistant clones yielded three homologous recombinants (frequency 1.5%) (Fig. 1c), one of which yielded germ line-transmitting chimeras. The first generation backcross (N1) onto the C57BL/6-Tyrc-Brd background was genotyped by Southern blot analysis of tail DNA. The appearance of hybridizing bands in addition to the targeted and wild-type seen in heterozygous ES cells meant, however, that it was not possible to distinguish heterozygotes from homozygotes with this method. Therefore PCR-based genotyping was established by using a Prokr2-linked SNP between the mutant 129 allele and the wild-type allele of the C57BL/6 strain to which it was backcrossed (Fig. 1d).
Fig. 1.
Generation of Prokr2Brdm1 mice and validation of null mutation. (a) Genomic structure of the murine Prokr2 gene and the targeting vector (Prokr2-TV). (b) Predicted structure of the targeted allele (partial restriction maps). Chromosome 2 coordinates for the extremities of the Prokr2-TV homology arms are indicated [National Center for Biotechnology Information (NCBI) ID code m35; Ensembl version 39; www.ensembl.org]. Exon coding sequences are indicated as black bars, and untranslated sequence are shown as white bars. R, EcoRI; N, NheI; 5′Hprt, 5′ Hprt minigene (exons 1 and 2); Tyr; tyrosinase gene; Neo, neomycin resistance gene; pBS, pBluescript II vector backbone; large gray arrowhead, loxP site. (c) Southern blot analysis of ES cell DNA EcoRI digest with the 5′ external probe showing the 9-kb wild-type fragment and 6.8-kb prokr2Brdm1 fragment of a homologous recombinant ES clone. Additionally, a 3′ external probe that detects a 9-kb wild-type fragment and a 7.7-kb prokr2Brdm1 fragment was used to confirm correct targeting (data not shown). (d) PCR genotyping of C57BL/6 backcrossed mouse tail DNA by amplification of a 373-bp intron 2 fragment followed by AluI digestion. Wild-type mice have two copies of the C57BL/6 SNP with an intact AluI site, yielding 176-bp and 197-bp DNA fragments on AluI digestion of the PCR product, whereas +/m and m/m mice have one copy or two copies, respectively, of the 129S7 SNP that destroys the AluI site. (e) Prokr2 mRNA expression analysis by RT-PCR of total RNA of the brains of N1F1 prokr2Brdm1 homozygotes and wild-type littermates. Transcripts were PCR-amplified within or between Prokr2 exons as indicated, and the ethidium gel image was contrast-enhanced by using Adobe Photoshop 7.0 (Adobe, San Jose, CA). The Hprt transcript was amplified as a positive control (image not enhanced). Reactions performed in the absence of reverse transcriptase, as a control for genomic contamination, yielded no PCR products (data not shown). (f) Analysis of Prokr2 function by radioligand binding (200 μM 125I-MIT) in m/m and +/+ littermates. Note extensive expression in +/+ brain, including lateral septum, SCN, paraventricular thalamus (PVT), and DMN and complete absence of binding in m/m. (g) Displacement of specific Prokr2 binding. Autoradiographic images are shown from adjacent wild-type brain sections incubated with 200 μM 125I-MIT ligand for Prokr2 receptors, either without (Upper) or with (Lower) an excess of cold ligand.
Because the prokr2Brdm1 mutation (abbreviated here as m) duplicates but does not delete any Prokr2 exons, the mutation was characterized functionally. RT-PCR analysis of brain total RNA indicated that the Prokr2 transcript was detectable for exons 1 and 2 in both +/+ and m/m RNA (Fig. 1d), which is expected because the Prokr2 promoter remains intact, and exons 1 and 2 are 5′ to the vector insertion site. An exon 3 transcript was also detected for m/m RNA that could be the result of splicing from the 5′ Hprt cassette. However, a transcript between exon 2 and exon 3 was undetectable in m/m while being present in +/+ RNA, demonstrating that insertion of the 5′ Hprt vector backbone adjacent to the truncated exon 3 disrupts transcription from exon 2 to the intact exon 3. Functional inactivation of the Prokr2 gene was confirmed by radioligand binding of mamba intestinal toxin 1 (MIT), a peptidergic homolog of endocrine gland-derived VEGF, which binds to both Prokr2 and Prokr1 receptors (13). The latter, however, is expressed at very low levels in the rodent brain, and so the binding we observed in wild-type mouse brain likely represented Prokr2 (14, 15). In confirmation, the distribution of 125I-MIT-binding sites overlapped that of Prokr2 mRNA as described in refs. 10 and 16, with intense binding in lateral septum, SCN, midline thalamic nuclei, amygdala, anterior hippocampus, and DMN (Fig. 1f). The binding was specific, being displaced by cold ligand (Fig. 1g), and it was completely abrogated in m/m mice, confirming the targeted allele as a functional null.
Variable Partial Penetrance of Homozygous prokr2Brdm1 Lethality.
The m/m mice exhibited partially penetrant postnatal lethality dependent on the contribution of the C57BL/6 genetic background. Survival to weaning of N2F1 generation genotypes was not significantly different from the expected Mendelian ratio (χ2 test, P = 0.23) (+/+, 15; +/m, 25; m/m, 7). At the N6F1 generation, however, a significant difference from the expected Mendelian ratio of genotypes was observed (χ2 test, P < 0.01) with very few m/m surviving to adulthood (+/+, 109; +/m, 191; m/m, 3). Surviving m/m mice weighed significantly less at 6 and 10 weeks of age than +/+ littermates (P ≤ 0.02, two-tailed t test) [see supporting information (SI) Table 1]. Postweaning, surviving m/m mutant mice were independently viable, and they exhibited normal patterns of feeding, grooming, and nesting behavior. Consistent with the reported sterility of a different Prokr2 null mutation (15), male and female m/m mice failed to breed.
Loss of Prokr2 Compromises Circadian Behavior and Thermoregulation.
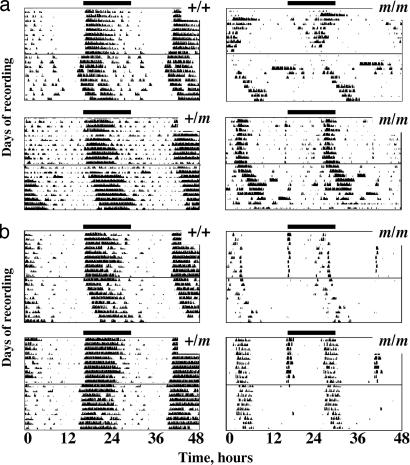
Because of severe penetrance of postnatal lethality of m/m at advanced stages of backcross into the C57BL/6 genetic background, cohorts of male and female N2, N3, and N4 C57BL/6 backcrossed homozygous mutant mice, with sex- and litter-matched wild-type and heterozygote controls, were used for behavioral analyses. Under a full photo schedule of 12 h of white light and 12 h of dim red light (LD), +/+ and +/m mice exhibited typical patterns of wheel-running behavior and general locomotor activity. Activity onset coincided with lights off, and activity continued through most of the dark phase (Fig. 2). The entrained behavior of homozygous mutants was dramatically different, often being biphasic in nature with a brief burst of activity coincident with lights off and a second more pronounced bout of activity toward the end of the dark phase. There was, however, marked interindividual variation in the relative intensity of early and late bouts. The consolidated locomotor activity in early night typical of +/+ and +/m mice was not evident in the m/m mice, and interday stability of activity was significantly lower in mutant mice (mean ± SEM: m/m, 0.55 ± 0.06; +/m, 0.74 ± 0.06; +/+, 0.79 ± 0.03; P < 0.05), whereas intradaily variability was higher (m/m, 1.25 ± 0.16; +/m, 0.84 ± 0.17; +/+, 0.81 ± 0.12; P < 0.05). The effects of the mutation on behavior were evident at all stages of backcross onto a C57BL/6 genetic background.
Fig. 2.
Disrupted entrained and circadian behavior in prokr2Brdm1 mutant mice. Shown are representative actograms of wheel-running behavior from individual +/+, +/m, and m/m mice, of C57BL/6 backcross generations N2F2 (a) or N4F1 (b). The initial 10 days of recording were under a 12:12 LD schedule (indicated by bars). Mice were then transferred to continuous dim red light on the day indicated by the horizontal line. Data are double-plotted over 48 h for clarity. Note coherent and sustained nocturnal activity rhythms under both schedules in +/+ and +/m mice and profound disorganization of behavior in m/m mice.
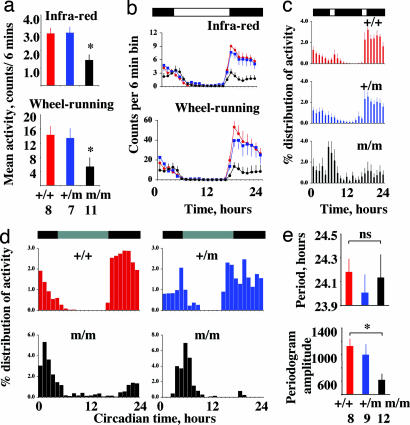
As a group, the m/m mice were significantly (P < 0.05) hypokinetic relative to other genotypes as assessed both by wheel-running and general activity (Fig. 3a). This behavior was not, however, expressed equally across all times of day and night: activity levels in the late night were comparable between genotypes whereas activity in the early night was significantly reduced in null mutants. Two-way repeated-measures ANOVA of both wheel-running and general activity therefore revealed highly significant (P < 0.01) interactions between time and genotype main effects (Fig. 3b) as well as significant (wheel-running P < 0.01, general activity P < 0.05) differences between genotypes. These effects were equally evident at all stages of backcross into a C57BL/6 genetic background, and there were no significant interactions between the number of backcrosses and behavior (wheel-running, P = 0.57; general activity, P = 0.77). The relative redistribution of wheel-running activity into the later night and loss of a clear activity onset at lights-off were also evident in m/m mice (regardless of background) entrained to a skeleton photoperiod with two 1-h light pulses at dusk and dawn (two-way repeated-measures ANOVA genotype effect, P < 0.01; genotype*time, P < 0.01) (Fig. 3c).
Fig. 3.
Temporal redistribution of entrained and circadian behavior in prokr2Brdm1 mutant mice. (a) Mean ± SEM daily levels of total activity monitored by infrared movement detectors and wheel-running. Note significantly reduced levels (P < 0.05) in m/m mice. (b) Temporal distribution of general activity and wheel-running in the final 10 days under LD before transfer to DD. Note attenuation of “dusk” behavior in m/m mice after lights-off but normal “dawn” activity levels around lights-on (mean ± SEM). (c) Temporal distribution of wheel-running under 10 days of skeleton photoperiod (after entrainment to LD) reveals selective loss of activity during early night and relative redistribution of activity to late night in the m/m compared with the +/m and +/+ mice (mean ± SEM). (d) Representative individual activity profiles plotted in circadian time from +/+, +/m, and m/m mice. Data were collected over 10 days starting on the second day of exposure to DD. Note loss of dusk activity at the onset of circadian night in m/m mice but maintenance of activity at the end of circadian night coincident with subjective dawn. (e) Free-running circadian period over the 10 days was not affected by prokr2Brdm1 allele (Upper), but the strength of the rhythm, assessed by the periodogram amplitude, was significantly (P < 0.05) attenuated in m/m mice (mean ± SEM).
On transfer to free-running conditions (DD), +/+ and m/+ mice exhibited very precise circadian profiles with marked activity onsets predicted from their entrained behavior under LD and consolidated activity during subjective night (Fig. 2). The m/m mice continued to exhibit bouts of wheel-running and general locomotion, but these activities free-ran predominantly from the later activity bouts observed under LD and skeleton schedules. The brief activity associated with lights-off was usually absent under DD, suggesting that it was a direct response to the lighting schedule and not under circadian control (Figs. 2 and 3d). Overall, the circadian period was not significantly different between groups (Fig. 3e), but the free-running activity of m/m mice in DD was less coherent and precise than that observed in +/+ and +/m mice, regardless of the C57BL/6 backcross generation. Consequently, although the behavior of null mutants was significantly rhythmic, the amplitude of the periodogram, a measure of how coherent and robust the rhythms were, was significantly reduced relative to +/+ mice, both for wheel-running (Fig. 3e) and general activity (data not shown), and interdaily stability was reduced (mean ± SEM: m/m, 0.53 ± 0.04; +/m, 0.63 ± 0.07; +/+, 0.73 ± 0.05; P < 0.05). In contrast to the effects of mutation to several core clock genes, the quality of the rhythm did not deteriorate further with prolonged exposure to DD, nor was there evidence of a multiplicity of periods, in contrast to mice lacking the Vpac2 receptor for vasoactive intestinal peptide (VIP) (17, 18).
Loss of Prokr2 was also associated with a disruption of circadian thermoregulation recorded by telemetry in an independent group of four m/m and four control (three +/m and one +/+) mice. Whereas wild-type and heterozygous mice displayed well organized nocturnal elevations of core body temperature, the nocturnal elevations in m/m mice were intermittent (SI Fig. 5a). The mean profiles under both entrained and free-running conditions showed a marked bimodality in the m/m mice, with pronounced dusk and dawn peaks but no sustained elevation between them (SI Fig. 5b). Overall, the mean body temperature of m/m mice was significantly lower than controls, and although the mean period of the body temperature cycle was not different, the amplitude of the periodogram was significantly lower in the m/m mice, consistent with a loss of temporal coherence (SI Fig. 5c).
Competent Molecular Timekeeping in Prokr2-Null SCN in Vitro.
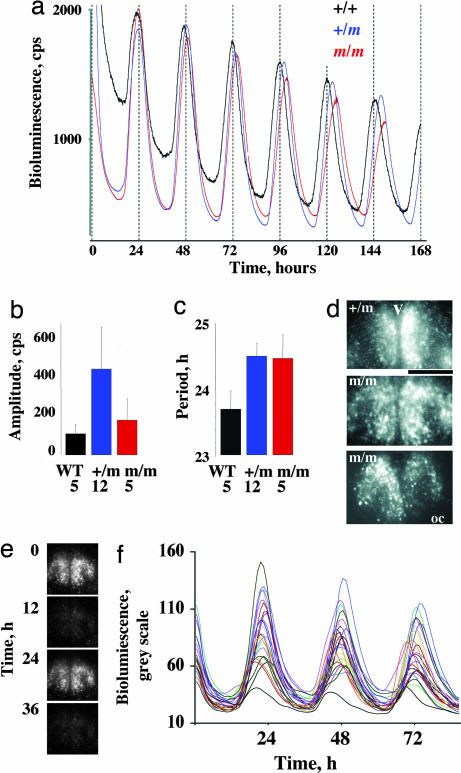
The loss of behavioral precision and coherence, the redistribution of nocturnal activity into the later night, and the disturbed thermoregulation seen in m/m mice could reflect either a disorganization of the SCN pacemaker (which normally expresses the Prokr2 receptor) and/or compromised signaling between the SCN and centers regulating sleep and body temperature that also express Prokr2. To examine the circadian competence of the SCN in m/m mice, the prokr2Brdm1 allele was crossed into a mPer1::luciferase transgenic reporter mouse line, and circadian gene expression was recorded from organotypic SCN slices cultured from neonatal pups (postnatal day 0–6, N3F3 C57BL/6 backcross) by using photomultiplier tubes (19). In +/+ and +/m slices (Fig. 4a) mPer1-driven luminescence from the slice was robustly circadian, with high-amplitude cycles and tightly regulated period. Equally competent circadian gene expression was also evident from m/m slices. Strict circadian bioluminescence was maintained over the course of recording; and not only were the period and amplitude of circadian gene expression rhythms from m/m mice not significantly different (ANOVA P = 0.09 and P = 0.49, respectively) from those of +/+ and +/m mice (Fig. 4 b and c), there was no indication of any alteration to circadian waveform in vitro that might correlate with the disrupted activity/rest profiles observed in vivo. At the tissue level, therefore, circadian timekeeping in the SCN was not altered by loss of Prokr2 signaling. Nevertheless, the compromise to circadian behavior may have arisen from a selective deficiency of cellular or regional timekeeping in the SCN not evident in ensemble records. SCN slices were therefore imaged by using a CCD to monitor cellular cycles across the subdivisions of the SCN. The regional distribution of circadian gene expression in m/m SCN slices was directly comparable with that observed in littermate control tissue (Fig. 4d and data not shown), both in the current study and as reported in refs. 19 and 20. Moreover, the amplitude and waveform of individual SCN neurons recorded from m/m slices were comparable with those of +/+ (Fig. 4 e and f), and these oscillatory neurons maintained tight synchrony over the interval of recording. Finally, genotype had no significant effect (P = 0.70 ANOVA) on individual cellular periods in either ventrolateral (mean ± SEM: +/+, 25.4 ± 0.4 h; m/m, 25.4 ± 0.3 h) or dorsomedial (+/+, 24.8 ± 0.3 h; m/m, 25.1 ± 0.3 h) SCN subdivisions.
Fig. 4.
Competent molecular circadian timekeeping in SCN of prokr2Brdm1 mutant mice revealed by bioluminescence. (a) Representative real-time recordings of bioluminescence driven by mPer1::luciferase reporter gene from organotypical SCN slices of +/+, +/m, and m/m mice. Note comparable expression levels and circadian waveform in all three genotypes. (b and c) Neither amplitude nor period of circadian gene expression from SCN slices was significantly affected by genotype (P > 0.05) (mean ± SEM). (d) Representative CCD images from one +/m and two m/m SCN slices confirm that the distribution of cellular circadian gene expression was comparable between both genotypes. v, third ventricle; oc, optic chiasm. (Scale bar, 500 μm.) (e) Serial CCD images from a m/m slice reveal circadian variation in cellular bioluminescence. (f) Recordings of mPer1-driven bioluminescence from 25 individual cells from a second m/m SCN slice reveal robust cellular timekeeping and tight synchrony between oscillatory neurons.
Discussion
By using a null mutation of the Prokr2 gene, we show that loss of Prok2-mediated signaling has no effect on molecular timekeeping within the SCN nor entrainment of the clock, but it severely disturbs circadian coordination of the activity/rest cycle and thermoregulation. The competence of the SCN as a circadian pacemaker in prokr2Brdm1 mutants contrasts markedly with its disorganization in mice lacking the Vpac2 receptor for VIP (18), a neuropeptide coexpressed with Prok2 in SCN cells (21). In Vpac2-null mice, the amplitude of circadian gene expression is reduced by >90%; half of SCN cells are arrhythmic, and the remainder express poorly defined, asynchronous cycles (19). Comparison of these two null mutants demonstrates that circadian synchronization and maintenance are properties of VIP signaling, independent of other products (specifically Prok2) of the synchronizing cells. Furthermore, even though Prok2 expression is induced within the SCN by light (21, 22), Prokr2-mediated signals are not necessary for photic entrainment because m/m mice were clearly nocturnal under LD and skeleton photoperiods, and on release to DD they free-ran from phases predicted by their entrained behavior.
It is clear, however, that Prok2/Prokr2 signaling is an essential coordinator of circadian behavior and physiology, even though the relative contributions of synaptic and paracrine communication to circadian outflow remain to be determined (6). Mice deficient in Prokr2 exhibited poorly consolidated nocturnal activity bouts and core body temperature rhythms, two daily cycles that are interrelated but generated independently. The prokr2Brdm1 phenotype is therefore very different from that observed after immunoneutralization of Clc signaling (another putative output pathway), where activity onset retains its precision, but it is advanced by 2 h, and overall activity levels are intensified (9). It also contrasts with EGF receptor mutants with compromised TGF-α signaling, where activity onsets are again very precise, and daytime activity is elevated (8). Of these putative SCN output pathways, therefore, deficiency of Prokr2 signaling shows the most distinctive and pronounced phenotype, and appropriate synthesis of Prok2 is probably necessary for competent circadian behavior. In R6/2 mice (a model for Huntington's disease) the progressive deterioration of the activity/rest cycle, which mimics that of patients, is accompanied by marked reduction in expression Prok2 mRNA in the SCN (23), and mutation of the human gene encoding Prok2 in Kallmann's syndrome is associated with sleep disturbance (24).
The effects of the prokr2Brdm1 mutation signal a role for Prok2/Prokr2 different from that predicted from pharmacological studies on rats, where exogenous Prok2 inhibited nocturnal locomotor activity followed by a “rebound” hyperactivity during circadian day (10). This observation suggested that endogenous Prok2 is an inhibitor of spontaneous daytime activity; hence, it would be expected that Prokr2-null mice would exhibit increased diurnal activity. In actuality, there is no increase in the daytime activity levels of m/m mice, and overall activity (most notably during early/middle night) is reduced. Clearly, the inferred pharmacological desensitization of the receptor (10) is not equivalent to its permanent loss, and these results are inconsistent with a simple model of Prok2/Prokr2 inhibiting locomotor activity. Moreover, Prok2/Prokr2 signaling is unlikely to be a generic inhibitor of activity across species because the SCN of diurnally active rodents express peak Prok2 mRNA expression during circadian daytime, i.e., the molecular cycles of nocturnal and diurnal species are in phase, as are the cycles of clock genes (25) that drive Prok2 expression. Release of Prok2 and subsequent activation of Prokr2 likely provide a paracrine signal of late circadian day, and/or falling levels of Prokr2 activation mark the onset of circadian night. The central pathways that regulate behavioral cycles and thermoregulation will utilize this time cue appropriately. In mice, these time cues consolidate activity, and body temperature rises; whereas in diurnal species, activity is suppressed.
Loss of Prok2/Prokr2 disrupts olfactory bulb development and leads to infertility (15, 24, 26). Disturbed circadian behavior may, therefore, be related indirectly to altered olfactory cues and/or infertility. Treatment of m/m mice with exogenous testosterone did not, however, alter their nocturnal activity profiles (E.S.M., unpublished data), and the effects of central infusion of Prok2 on activity are too rapid to be mediated by altered gonadal status. Moreover, olfactory bulbectomy increases rather than decreases general activity levels in mice (27). A more direct explanation is that loss of Prokr2 expression in SCN target sites in hypothalamus and/or thalamus compromises circadian behavior and thermoregulation. The DMN and the paraventricular thalamic nuclei express high levels of Prokr2 in wild-type brains, and they are known to regulate circadian activity/arousal levels (4). Our genetic data therefore enforce a reappraisal of conclusions based on gene expression and pharmacological studies. Signaling by means of Prokr2 is not necessary for pacemaker functions of the SCN nor its entrainment by retinal input. Prokr2 is necessary for circadian control of behavior and thermoregulation. Whether these two outputs are regulated in a coordinated or independent manner awaits clarification and, more specifically, Prokr2 appears necessary for precise regulation of particular elements of the activity/rest cycle associated the onset of circadian night. Because Prokr2 mediates circadian control of behavior rather than circadian timing per se, targeted intervention of Prok2/Prokr2 signaling may provide therapeutic strategies for sleep disorders (28) without compromising the central molecular clockwork.
Experimental Procedures
Gene Targeting.
Studies were licensed by the U.K. Home Office under the Animals (Scientific Procedures) Act 1986. The Prok2 targeting construct was generated as an insertional targeting vector incorporating the 5′ Hprt chromosomal engineering cassette (11). A 2,362-bp 5′ arm and 1,229-bp 3′ arm were PCR-amplified from a 129S7-derived BAC clone RPCI 21-497L12 by using Hi-Fidelity PCR mix (Invitrogen, Carlsbad, CA), and they were cloned into the 5′ Hprt vector as a three-piece ligation. The targeting vector was linearized for transfection by using a NheI digest. AB2.2 (129S7/SvEvBrd-Hprtb-m2) ES cells were propagated, transfected by electroporation, and selected with G418 by using standard protocols (29). Homologous recombination was confirmed by using Southern blotting of EcoRI digested with 5′ and 3′ external probes amplified from the genomic PAC clone. The 5′ probe was 858 bp, amplified with primers 5′-GCTTCATGGGGAACAGTTGGCTGGGGTG-3′ and 5′-AGAATCACAGAGCAAGGAGCATCCTTC-3′. The 3′ probe was 424 bp, amplified with primers 5′-ATCCACCTCTTCAGTGGCAGGCACCCCGC-3′ and 5′-TATGACCCTGTGCTATGCCAGGATCTCCC-3′. Chimeric mice were generated by microinjection of targeted ES cells into C57BL/6-Tyrc-Brd blastocysts. Mice were maintained by backcross onto the C57BL/6-Tyrc-Brd genetic background described by N (numbers of backcross generations) and F (numbers of familial brother sister matings as described for congenic strains) (http://informatics.jax.org). Mice were genotyped by using the following intron 2-specific primers that amplify a 373-bp PCR product containing an SNP (NCBI SNP ID code rs29958198) at position 132,076,478 bp of chromosome 2 (NCBI ID code m35): 5′-CCTCTGGGGCGTCTATTGGTCTCC-3′ and 5′-ACTTGGGGGCACTCCGCGTCTGTTC-3′. The SNP creates a restriction fragment length polymorphism on AluI digestion, the recognition site being present in C57BL/6 and absent from 129S7.
RT-PCR Analysis.
Total RNA was prepared from brains by using an RNeasy midi-prep kit (Qiagen, Valencia, CA). Reverse transcription was performed on 1 μg of total RNA by using SuperScript II (Invitrogen) and random hexamers. PCR for 30 cycles annealing at 58°C was performed by using Platinum Supermix (Invitrogen) on 5% of the cDNA product and using the following exon-specific primers: between Prokr2 exons 1 and 2 and yielding a 484-bp product, 5′-CACACGCCCACCAAGTAGG and TAGCGGGCGAGGGCAGCAATGAA-3′; within Prokr2 exon 2 and yielding a 439-bp product, 5′-GGGACCCCAGAACAGAAACACTA and AGCGATGGCCAGCAGAGC-3′; within Prokr2 exon 3 and yielding a 496-bp product, 5′-TCCACTGGCGGCCCTCTCACTACG-3′ and 5′-CCTGGATGGATGGATGGATGGATGGATA-3′; between Prokr2 exons 2 and 3 yielding a 488-bp product, 5′-CTGCCCTCGCCCGCTACAAG-3′ and 5′-ACCACGGGACCCACGAACTCAAG-3′. The Hprt-specific primers (exons 3- and 8-specific) were used as a control for mRNA quality yielding a 352-bp product: 5′-CCTGCTGGATTACATTAAAGCACTG-3′ and 5′-GTCAAGGGCATATCCAACAACAACAAAC-3′.
Behavioral Analyses and Telemetry for Body Temperature.
N2F2, N3F1, or N4F1 generation mice backcrossed into C57BL/6-Tyrc-Brd strain were housed in ventilated, light-tight cabinets in individual cages with a running wheel and a passive infrared movement detector connected to a PC running Clocklab (Actimetrics, Evanston, IL) acquisition software. Food and water were available ad libitum. The initial photoschedule was 12 h of white light between 0600 and 1800 followed by 12 h of dim red light. A skeleton photoperiod presented two 1-h pluses starting at 0600 and 1700, and the free-running circadian period was monitored after the mice were transferred to continuous dim red light. Data were analyzed by χ2 periodogram, and nonparametric circadian rhythm analysis of interdaily stability and intradaily variability (30) was conducted with Actiwatch Activity Analysis (Cambridge Neurotechnology, Cambridge, U.K.). In a separate group of four m/m mice and four littermate controls (three +/m, one +/+), body temperature was measured by using TA10TA-F20 i.p. radiotelemetry implants (DSI, St. Paul, MN). After postsurgical recovery, mice were housed individually in a temperature-controlled room (21 ± 1°C), first on a LD for 3 weeks, and then they were transferred to continuous dim red light. Temperature parameters were sampled for 10 s every 2 min by RPC-1 receivers and ART version 2.1 software.
Radioligand Autoradiography.
Brains were rapidly removed 4 h after projected lights on to coincide with peak Prok2 expression; they were then snap frozen on dry ice and stored at −70°C until sectioning (20 μm) on a cryostat. Slides were thawed at room temperature, air-dried, and preincubated in Tris acetate buffer (pH 7.4) containing 0.1% BSA for 30 min at room temperature to remove any endogenous ligand. Slides were then incubated for 1 h in 200 μM 125I-MIT (2,000 Ci/mmol; Perkin-Elmer, Wellesley, MA) in Tris acetate buffer, with alternate slides being incubated in 200 μM 125I-MIT plus 1 μM MIT (PentaBiotech, Inc., Union City, CA). After incubation, slides were rinsed 10 times for 5 s each in ice-cold Tris acetate buffer, and then they were then air-dried. Sections were apposed to film (Kodak Biomax XMR-1; GE Healthcare, Piscataway, NJ) for 4 days before being processed.
Bioluminescence Imaging of Circadian Gene Expression.
The prokr2Brdm1 heterozygous mice of the N2F2 (C57BL/6 backcrossed) generation were bred into the mPer1::luciferase line (20) (C57BL/6-N Charles River genetic background), and the N3F2 offspring were intercrossed. Organotypic slices containing SCN were made from 0- to 6-day-old N3F3 pups for recording circadian gene expression (19) by using either photomultiplier assemblies for whole-tissue emission or a CCD camera for single-cell imaging (both items from Hamamatsu Photonics U.K., Ltd., Welwyn Garden City, Hartfordshire, U.K.).
Supplementary Material
Acknowledgments
We thank E. Grau and T. Hamilton for blastocyst injection; F. Law, A. Beasley, and T. Butcher for technical support; Helen I'Anson for conducting the telemetry; and Prof. H. Okamura (Kobe, Japan) for supplying luciferase reporter mice. This work was supported by the Medical Research Council, Wellcome Trust, and Biotechnology and Biological Sciences Research Council Grants BBS/B/10765 and BB/C/006941/1.
Abbreviations
- DMN
dorsomedial nucleus
- Hprt
hypoxanthine phosphoribosyltransferase
- m
prokr2Brdm1 mutation
- MIT
mamba intestinal toxin 1
- Prok2
prokineticin 2
- SCN
suprachiasmatic nucleus
- VIP
vasoactive intestinal peptide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606884104/DC1.
References
- 1.Reppert SM, Weaver DR. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 2.Hastings MH, Reddy AB, Maywood ES. Nat Rev Neurosci. 2003;4:649–661. doi: 10.1038/nrn1177. [DOI] [PubMed] [Google Scholar]
- 3.Buijs RM, Kalsbeek A. Nat Rev Neurosci. 2001;2:521–526. doi: 10.1038/35081582. [DOI] [PubMed] [Google Scholar]
- 4.Saper CB, Scammell TE, Lu J. Nature. 2005;437:1257–1263. doi: 10.1038/nature04284. [DOI] [PubMed] [Google Scholar]
- 5.Gooley JJ, Schomer A, Saper CB. Nat Neurosci. 2006;9:398–407. doi: 10.1038/nn1651. [DOI] [PubMed] [Google Scholar]
- 6.Hastings MH, Herzog ED. J Biol Rhythms. 2004;19:400–413. doi: 10.1177/0748730404268786. [DOI] [PubMed] [Google Scholar]
- 7.Silver R, LeSauter J, Tresco P, Lehman MN. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- 8.Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, Weitz CJ. Science. 2001;294:2511–2515. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 9.Kraves S, Weitz CJ. Nat Neurosci. 2006;9:212–219. doi: 10.1038/nn1633. [DOI] [PubMed] [Google Scholar]
- 10.Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou QY. Nature. 2002;417:405–410. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- 11.Zheng B, Sage M, Cai WW, Thompson DM, Tavsanli BC, Cheah YC, Bradley A. Nat Genet. 1999;22:375–378. doi: 10.1038/11949. [DOI] [PubMed] [Google Scholar]
- 12.Zheng B, Mills AA, Bradley A. Methods. 2001;24:81–94. doi: 10.1006/meth.2001.1160. [DOI] [PubMed] [Google Scholar]
- 13.Masuda Y, Takatsu Y, Terao Y, Kumano S, Ishibashi Y, Suenaga M, Abe M, Fukusumi S, Watanabe T, Shintani Y, et al. Biochem Biophys Res Commun. 2002;293:396–402. doi: 10.1016/S0006-291X(02)00239-5. [DOI] [PubMed] [Google Scholar]
- 14.Soga T, Matsumoto S, Oda T, Saito T, Hiyama H, Takasaki J, Kamohara M, Ohishi T, Matsushime H, Furuichi K. Biochim Biophys Acta. 2002;1579:173–179. doi: 10.1016/s0167-4781(02)00546-8. [DOI] [PubMed] [Google Scholar]
- 15.Matsumoto S, Yamazaki C, Masumoto KH, Nagano M, Naito M, Soga T, Hiyama H, Matsumoto M, Takasaki J, Kamohara M, et al. Proc Natl Acad Sci USA. 2006;103:4140–4145. doi: 10.1073/pnas.0508881103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Negri L, Lattanzi R, Giannini E, De Felice M, Colucci A, Melchiorri P. Br J Pharmacol. 2004;142:181–191. doi: 10.1038/sj.bjp.0705686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 18.Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, et al. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- 19.Maywood ES, Reddy AB, Wong GK, O'Neill JS, O'Brien JA, McMahon DG, Harmar AJ, Okamura H, Hastings MH. Curr Biol. 2006;16:599–605. doi: 10.1016/j.cub.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 20.Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 21.Masumoto KH, Nagano M, Takashima N, Hayasaka N, Hiyama H, Matsumoto S, Inouye ST, Shigeyoshi Y. Eur J Neurosci. 2006;23:2959–2970. doi: 10.1111/j.1460-9568.2006.04834.x. [DOI] [PubMed] [Google Scholar]
- 22.Cheng MY, Bittman EL, Hattar S, Zhou QY. BMC Neurosci. 2005;6:17. doi: 10.1186/1471-2202-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morton AJ, Wood NI, Hastings MH, Hurelbrink C, Barker RA, Maywood ES. J Neurosci. 2005;25:157–163. doi: 10.1523/JNEUROSCI.3842-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dode C, Teixeira L, Levilliers J, Fouveaut C, Bouchard P, Kottler ML, Lespinasse J, Lienhardt-Roussie A, Mathieu M, Moerman A, et al. PLoS Genet. 2006;2:e175. doi: 10.1371/journal.pgen.0020175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mrosovsky N, Edelstein K, Hastings MH, Maywood ES. J Biol Rhythms. 2001;16:471–478. doi: 10.1177/074873001129002141. [DOI] [PubMed] [Google Scholar]
- 26.Ng KL, Li JD, Cheng MY, Leslie FM, Lee AG, Zhou QY. Science. 2005;308:1923–1927. doi: 10.1126/science.1112103. [DOI] [PubMed] [Google Scholar]
- 27.Possidente B, Lumia AR, McGinnis MY, Rapp M, McEldowney S. Brain Res. 1996;713:108–113. doi: 10.1016/0006-8993(95)01490-x. [DOI] [PubMed] [Google Scholar]
- 28.Abbott A. Nature. 2003;425:896–898. doi: 10.1038/425896a. [DOI] [PubMed] [Google Scholar]
- 29.Ramirez-Solis R, Davis AC, Bradley A. Methods Enzymol. 1993;225:855–878. doi: 10.1016/0076-6879(93)25054-6. [DOI] [PubMed] [Google Scholar]
- 30.Hatfield CF, Herbert J, van Someren EJ, Hodges JR, Hastings MH. Brain. 2004;127:1061–1074. doi: 10.1093/brain/awh129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.