Abstract
The recent discovery of a carotenoid light-harvesting antenna in xanthorhodopsin, a retinal-based proton pump in Salinibacter ruber, made use of photoinhibition of respiration in whole cells to obtain action spectra [Balashov et al. Science 309, (2005) 2061-2064]. Here we provide further details of this phenomenon, and compare action spectra in three different systems where carotenoids have different functions or efficiencies of light-harvesting. The kinetics of light-induced inhibition of respiration in Salinibacter ruber was determined with single short flashes, and the photochemical cross section of the photoreaction was estimated. These measurements confirm that the xanthorhodopsin complex includes no more than a few, and most likely only one, carotenoid molecule, which is far less than the core complex antenna of photosynthetic bacteria. Although the total cross-section of light absorption in the purple bacterium Rhodospirillum rubrum greatly exceeds that in Salinibacter, the cross-sections are roughly equivalent in the shared wavelength range. We show further that despite interaction of bacterioruberin with archaerhodopsin, another retinal-based proton pump, there is no significant energy transfer from this carotenoid. This emphasizes the uniqueness of the salinixanthin-retinal interaction in xanthorhodopsin, and indicates that bacterioruberin in Halorubrum species has a structural or photoprotective rather than energetic role.
Keywords: xanthorhodopsin, salinixanthin, energy transfer, archaerhodopsin, bacterioruberin, Salinibacter ruber
1. Introduction
The evolution and the diversity of life on Earth depend critically on the activity of photosynthetic organisms to convert solar energy flux, in the 350-1000 nm range, into biochemically usable form. There are two known mechanisms for light-energy transduction, driven by either a) (bacterio)chlorophyll photooxidation followed by transmembrane electron transfer and in its secondary events proton transfer in photosynthetic organisms, or b) retinal photoisomerization followed by transmembrane proton [1] or chloride [2] transport. These light-energy transducing membrane systems are different in their general design. The (bacterio)chlorophyll-based photosystems contain a supramolecular organization of the photosynthetic units, with specialized reaction centers and antennas of tens to hundreds of light-harvesting chromophores [3, 4]. In contrast, the retinal-based complexes operate as functionally independent monochromophoric [5-7], or bichromophoric in the case of S. ruber [8], units. In contemporary aquatic and terrestrial ecosystems, the dominating groups are the oxygen evolving cyanobacteria, algae, and higher plants. The anoxygenic photosynthetic bacteria, and particularly cells utilizing light-energy with retinal based ion-pumps, were thought to occupy mostly other ecological niches, with extreme environmental conditions. Recent findings have shown, however, that the latter are abundant also in the oceans [9-11], and their retinal proteins have energetic functions not only in archaea and bacteria but potentially also in some eukaryotes [12]. It appears, therefore, that competition for solar energy has given rise to a variety of light-harvesting pigments, in all ecosystems [4].
The energy-transducing membranes of microorganisms that contain both respiratory and photosynthetic systems usually exhibit interdependence of the two functions, via feedback regulation mechanisms. Although the details of these mechanisms in many cases are not well established, analysis of light-induced modulation of partial reactions of the energy-transducing systems, particularly O2 gas exchange, has provided valuable information about the structural and functional organization of the relevant photosynthetic complexes in situ [13]. In the case of the archaeon Halobacterium salinarum, the light-induced transmembrane electrical potential generated by bacteriorhodopsin [14, 15] is responsible for photoinhibition of respiration [1, 5].
Recently we reported that the cells of the extremely halophilic eubacterium Salinibacter ruber display light-induced inhibition of respiratory O2 uptake rates [8], similarly to species containing either a retinal-based light driven proton pump (bacteriorhodopsin in H. salinarum) [1, 5, 16] or bacteriochlorophyll-based type II photosystems (purple bacteria [17-19]) or chlorophyll-based photosystem I (cyanobacteria [20-22] and algae [23, 24]). Unlike in bacteriorhodopsin, the action spectra of the photoprocess in S. ruber revealed tight energetic coupling of a carotenoid and the retinal in the H+-pump, xanthorhodopsin [8]. The carotenoid salinixanthin [25] is bound to this retinal protein and performs the function of light-harvesting antenna, increasing the flow of light quanta available to energize proton transport by about 2-fold [8]. Xanthorhodopsin exhibits substantial homology to bacteriorhodopsin and proteorhodopsin [8], and even greater similarity to the rhodopsin of cyanobacterium Gloeobacter violaceus [26]. Other genes in the Salinibacter genome encode a halorhodopsin-like protein [26, 27] and two sensory rhodopsins [26], but expression of only xanthorhodopsin was detected so far [8].
In this paper, we present the results of our continuing study of the xanthorhodopsin-mediated modulation of respiration rate, and compare it to analogous phenomena in photosynthetic bacteria and an archaerhodopsin-containing archaeal species. The latter contains the carotenoid bacterioruberin which interacts with the retinal protein [28], but as we report here, without antenna function.
2. Materials and Methods
Salinibacter ruber M31 (DSM 13855) was routinely grown in the slightly modified medium [29] with the concentrations of salts (per L): 195 g NaCl, 49.5 g MgSO4·7H2O, 34.6 g MgCl2·6H2O, 1.25 g CaCl2·2H2O, 5 g KCl, 0.25 g NaHCO3, 0.625 g NaBr and 2 g of yeast extract. The pH was adjusted to 7.0 by adding 10 N NaOH. A 400 ml culture was inoculated with 40 ml of 7 days old starter culture, and grown at 37 °C in 2 L flasks loosely covered with foil in an incubator shaker (Series 25, New Brunswick Scientific, NJ, USA) at 180 rpm for 2 days. Fresh medium (0.8 L) was then added to each flask 1.2 L to decrease aeration, and the culture was grown under illumination of ∼ 1 mW cm-2 (400-700 nm) for another 5 days while shaking at the same speed.
The culture of archaeon Halorubrum sp. strain A1c, isolated from the same solar salterns as S. ruber and closely related to strains AUS-1 [28] with 99% similarity in the almost complete (1385bp) 16S rRNA gene sequence to Halorubrum xinjiangense [30], was grown under the same conditions. The archaerhodopsin gene [31] was identified by PCR sequencing (Laragen, Inc., Los Angeles, CA) using primers: aR 5’ Fwd- ATGGACCCGATAGCGCTAACC and aR 3’ Rev- TCAGTCCGCGGCGGAGG.
A 2-3 day culture of the purple bacterium Rhodospirillum rubrum was grown anaerobically in Ormerud medium at 30° C and continuous light of ∼1 mW cm-2 (400-900 nm).
The membranes of S. ruber and Halorubrum sp. were isolated using osmotic shock and dialysis as described earlier [8].
Light-induced oxygen gas exchange was measured with a bare-platinum electrode as described previously [20, 33]. A sample of 20 μl cell suspension, with optical density of 0.1-0.2 at 525 nm, was placed on a 6 mm platinum disc in a groove of 0.65 mm depth, covered by cellophane membrane forming the assay microchamber. The surrounding chamber of ∼30 ml with a reference Ag/AgCl electrode was filled up by a solution of 3.4 M NaCl, 50 mM KC1 and 50 mM Na-phosphate buffer, pH 7.0, to the level of the membrane, retaining free air-space over its surface. The deposited cells were allowed to settle for 1-2 hours before the measurements. The samples in the setup were illuminated by one of three kinds of light sources: a monochromator, a “cool light” projection system through interference or cut-off filters with a fiber-optic illuminator OVS-2 (Russia), and an ISS-100-3M (Russia) xenon flash tube generating 1.8 μs flashes via a range of interference filters. The intensities of the light beams on the electrode were varied by neutral density filters and measured by a calibrated thermoelement, RTN-20S (Russia). Polarographic signals were detected by amplifier I37 (Russia) and recorded at ∼0.1 s time resolution. Under typical experimental conditions, the rate of O2 uptake in the dark by 20 μl assay of S. ruber cells with the optical density of 0.1 at 522 nm was 1.0-1.5 pmoles/s. For convenience of comparison of light-induced changes of respiration the initial dark rates were taken as 100%.
The action spectra measurements were carried out using cycles of intermittent 1 min illumination, followed by 2-3 min dark period between two successive wavelengths at 5-10 nm intervals. The spectral half-width of the monochromatic beam was 4-12 nm. Its intensity varied in the ranges 0.1-1 nE cm-2 s-1 in the spectral range of 400-900 nm. Action spectra were corrected for a small nonlinearity in the light energy-dependent curves by estimation of the energies at different wavelengths producing the same photochemical yields at a control wavelength, usually at 570 nm. Correction for time-dependent changes during scanning was routinely introduced by measuring the ratio of signals at the given wavelength versus the signal at 570 nm. The spectra were corrected also for the spectral coefficient of light reflection by the platinum surface through the settled cells. The relative error of spectral efficiency was ≤5%.
Absorption spectra were measured on Shimadzu 1601 UV-Vis spectrophotometer.
3. Results and Discussion
3. 1. Light modulation of respiration in whole cells of S. ruber under continuous illumination: kinetics, components of the action spectra, effects of inhibitors.
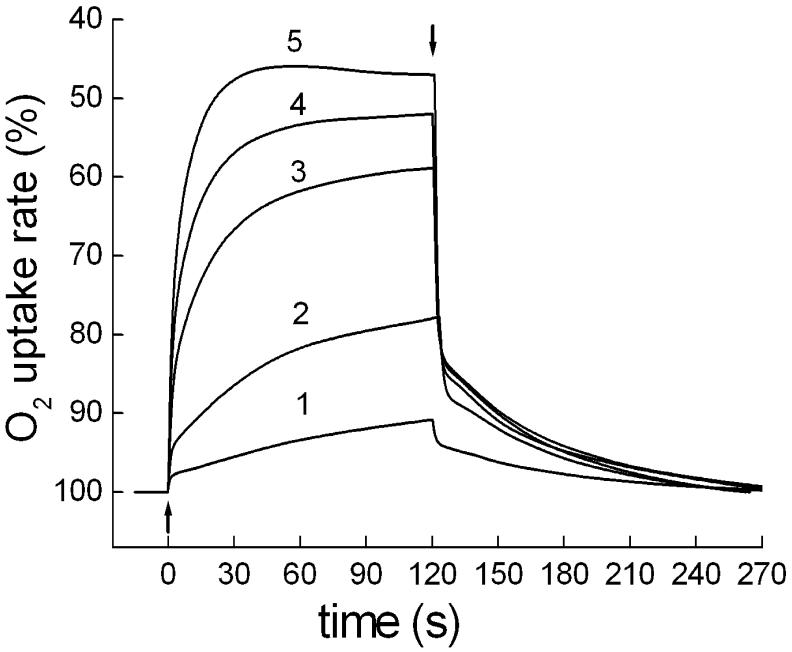
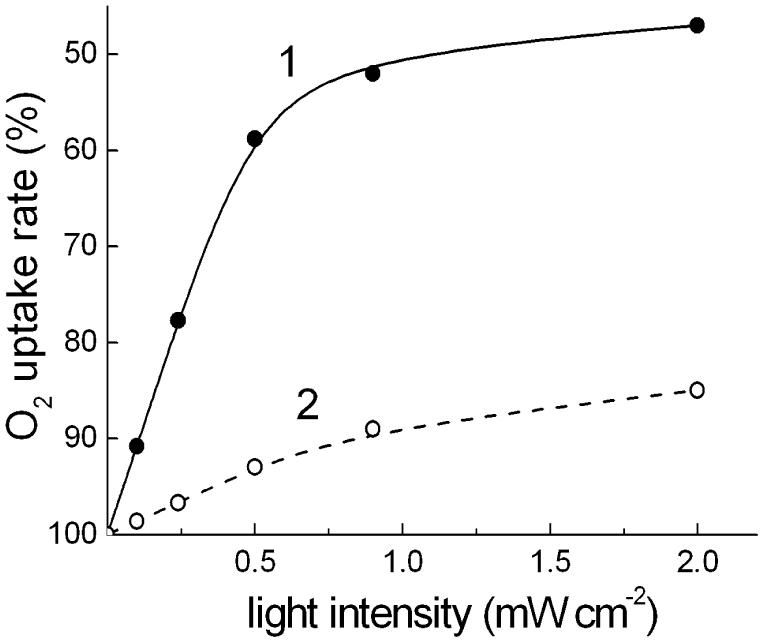
Xanthorhodopsin-containing cells of S. ruber exhibit light-induced reversible decrease of O2 uptake rate (Fig. 1). Typically, the photoinhibition of respiration in S. ruber saturates at light intensities of ≥ 2 mW cm-2 in the spectral range 400-600 nm, similarly to H. salinarum [5], and the maximal depression of the O2 uptake rate is 40-50% (Fig. 2). This is a 2-3 times greater effect than in the bacteriorhodopsin-containing archaeon [5, 16]. Measurements of cells with different contents of xanthorhodopsin (as a result of variations in the culture growth conditions or strain features) revealed that the light-saturated level of photoinhibition is less variable than the initial slope of the light-saturation curves, which in early-harvested cultures sometimes have a more or less marked S-like shape (data not shown). Like bacteriorhodopsin in H. salinarum, the expression of xanthorhodopsin was stimulated by illumination and low aeration of the culture, although a systematical study of the dependence was not done.
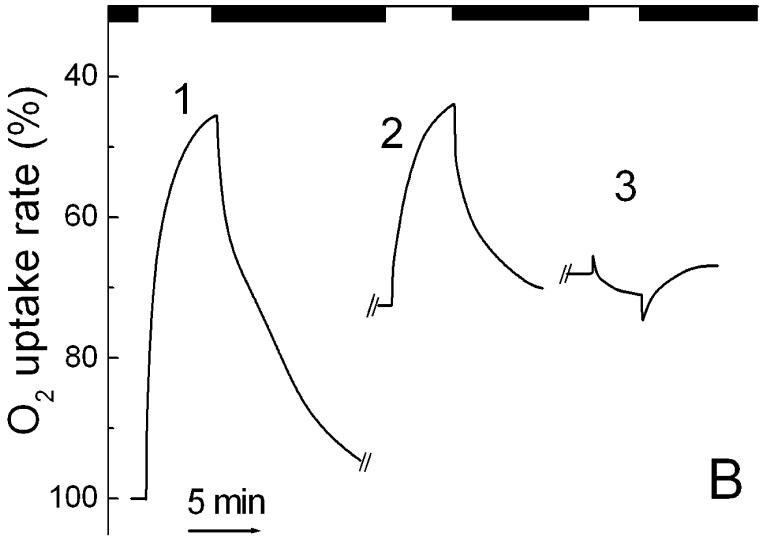
Fig. 1.
Light-induced changes of respiratory O2 uptake rates in cells of S. ruber: 1 through 5, kinetic curves of reversible photoinhibition of respiration upon illumination with 400-650 nm of 0.1, 0.24, 0.5, 0.9 and 2 mW cm-2, respectively. Up and down arrows show light on and off, respectively.
Fig. 2.
Light-saturation curve of the photoinhibition of respiration in cells of: 1, Salinibacter ruber; 2, Halorubrum sp. strain A1c.
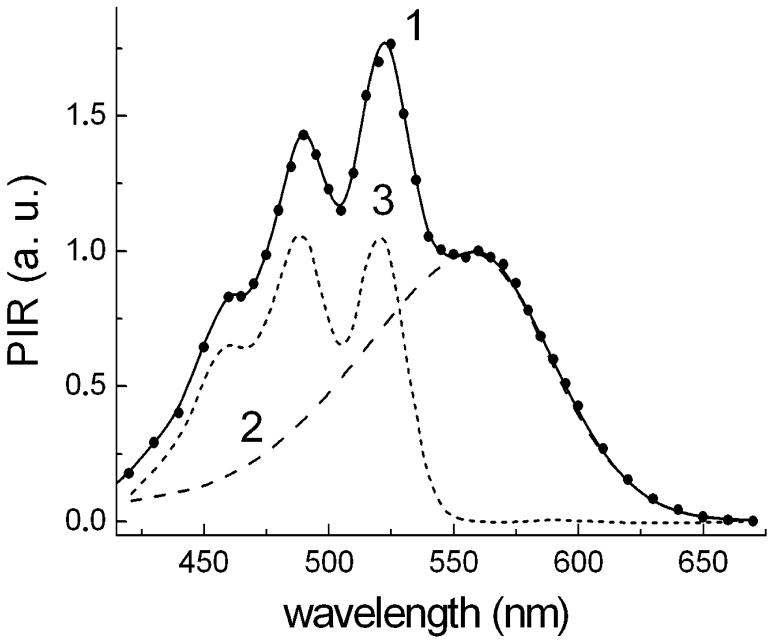
The wavelength dependence of the relative efficiency of monochromatic light in photoinhibition of respiration (action spectrum) is shown in Fig. 3. It is characterized by the band at 560 nm from the retinal, and sharper bands at 522 nm, 490 nm as well as a shoulder at 460 nm which correspond the absorption maxima of the salinixanthin. The action spectrum almost coincides with the earlier published one [8], but this spectrum was obtained under narrower spectral bandwidth of excitation (4 nm versus 6 nm), which slightly improved the resolution of the carotenoid bands and the overall accuracy of the spectrum. The action spectrum can be deconvoluted into two components, the broad band with maximum at 559 nm from the retinal chromophore and a set of four sharp vibronic bands at 522, 489, 460 and 432 nm from the absorption spectrum of bound salinixanthin (see Figure legend for the relative intensities and half-widths of the bands). The positions of the sharp bands coincide with the corresponding bands in the absorption spectrum of the Salinibacter ruber cell membranes. This coincidence indicates that light absorbed by salinixanthin is transferred to the retinal chromophore, and used for electrogenic proton transport [8]. It should be noted that the carotenoid bands in the action spectrum are narrower and better resolved than the bands in the absorption spectrum of cells or cell membranes. The reason for that is that cell membranes in most cases contain additional salinixanthin not bound to xanthorhodopsin, which exhibits poorly resolved and substantially broader bands. Only upon binding to xanthorhodopsin does salinixanthin exhibit a well-resolved spectrum as in the action spectrum in Fig. 3. The amplitudes of the two main carotenoid bands in the action spectrum are roughly the same as the amplitude of the retinal band. The extinction of salinixanthin is about 3 times larger than that of the retinal band [8, 25]. With these assumptions, the efficiency of energy transfer from salinixanthin to the retinal chromophore can be estimated at 33%.
Fig. 3.
A. Action spectrum of light-induced photoinhibition of respiration (PIR) in suspension of S. ruber cells and its components: 1, action spectrum measured with a spectral half-band of 4 nm; the amplitude was normalized to 1 at 560 nm; 2, retinal chromophore component of the action spectrum. At λ > 560 nm it coincided with the action spectrum. This part can be approximated well by a Gaussian band with maximum at 559 nm and half width of 2150 cm-1. The part of the spectrum at λ 560 nm was approximated by a spectrum of bacteriorhodopsin shifted 9 nm to shorter wavelengths. 3, carotenoid component of the action spectrum obtained as a difference “spectrum 1 minus spectrum 2”. This spectrum can be very well approximated by a sum of four Gaussian bands at 522 nm (A=1.03; Δν½ = 860 cm-1), 490 nm (A=1.0, Δν½ = 1110 cm-1), 460 nm (A = 0.62, Δν½ = 1430 cm-1), 432 nm (A = 0.17, Δν½ = 1490 cm-1).
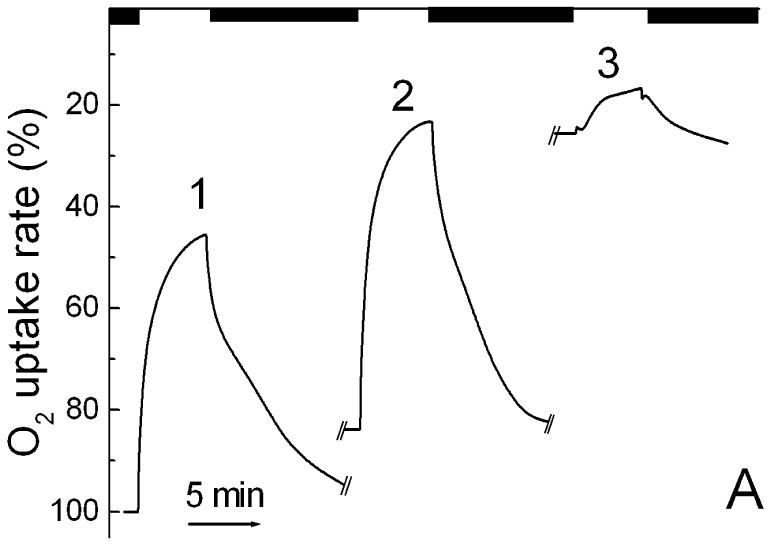
In experiments with metabolic inhibitors and poisons, we find that respiration in S. ruber cells is rather resistant to cyanide, maybe at least partly from a low permeability across the cell wall. Cyanide inhibited the dark rate by only ∼20% at 10 mM concentration. It did not affect, or even slightly increased, photoinhibition of respiration (Fig. 4A). In contrast, the rather unspecific inhibitor of the cellular energy metabolism, ortho-phenanthroline, strongly inhibited both the respiratory rates and the amplitude of the photoinhibition. Pretreatment of the cells with the inhibitor of H+-ATPase, DCCD (1 hr incubation at 0.5 mM, followed by washing with buffer), decreased both dark respiration and photoinhibition by 30-40% (Fig. 4B). There was no reversal of the light-induced signal to yield photostimulation of respiration, as was observed in H. salinarum [5]. In both control (see [8]) and DCCD-treated (Fig. 4B) cells, the uncoupler of phosphorylation, CCCP, strongly suppressed the photoinhibition. This effect of CCCP suggests that the photoinhibition of respiration is produced by the light-induced electrochemical proton gradient as in the haloarchaea [5] and partly in the purple bacteria [17, 18].
Fig. 4.
Effects of metabolic poisons on light-induced changes of respiratory O2 uptake rates in S. ruber. A. 1, light-induced changes of respiration at 2 mW cm-2 of white light; 2, shift of dark level and light-induced changes of respiration in the cells after addition of 10 mM KCN; 3, after subsequent addition of 10 mM of o-phenanthroline. B. 1, light-induced changes of respiration in control cells; 2, in the cells pretreated for 1 h with 0.5 mM DCCD; 3, and after subsequent addition of 1 μM CCCP. The breaks in the traces A and B correspond to ∼20-30 min of equilibration of O2 concentration in the microchamber.
3.2. Flash-induced photoinhibition of respiration in S. ruber: estimation of the photochemical cross section.
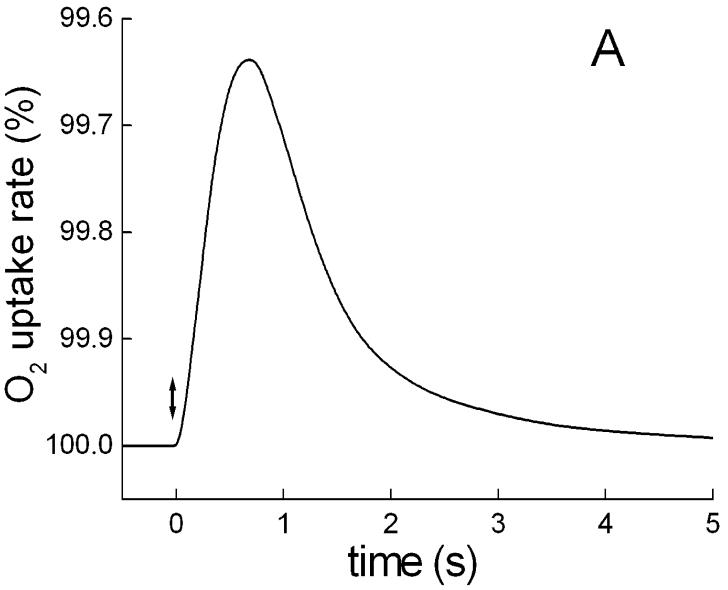
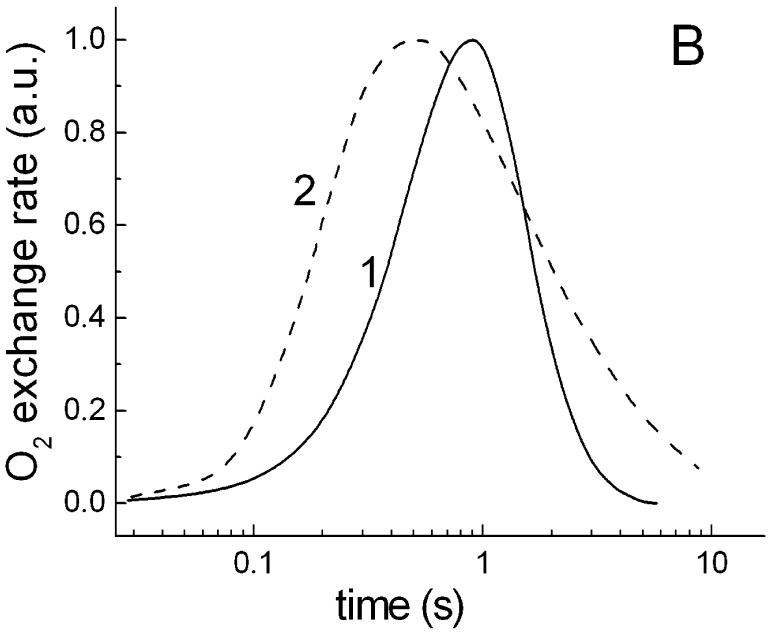
Single flashes of 2 μs duration produced transient photoinhibition of respiration which arose with half-times of ∼0.2 s and decayed with ∼1.2 s (Fig. 5A). These changes are also abolished by CCCP. Presumably, the first (rise) phase corresponds mainly to a limiting step in the generation of the proton gradient and its interaction with respiratory electron transport chain, whereas the second (decay) phase displays apparently the kinetics of relaxation of the photoinduced proton gradient to a dark level. Comparison of this kinetics with flash-induced pulses of photoinhibition of respiration in the purple bacterium Rhodospirillum rubrum (Fig. 5B), measured with the same set-up and time constant of the amplifier, showed that the photoprocess in S. ruber develops considerably slower than in the photosynthetic bacterium. The differences in the initial kinetic phases of the species might be caused by direct competition of the respiratory chain with the photosynthetic electron transport chain for a common pool of electron carriers. This type of interaction of the two chains explains also why the flash-induced photoinhibition in photosynthetic bacteria is insensitive to the uncoupler CCCP (data not shown, cf [17, 18]).
Fig. 5.
Typical kinetic curves of a single 2 μs flash-induced O2 exchange rates in S. ruber and in photosynthetic bacteria. A. Time course of flash-induced pulse of inhibition of respiration in cells of S. ruber grown in the standard medium. B. Normalized kinetics of flash-induced O2 exchange rates in: 1, S. ruber; 2, Rhodospirillum rubrum. The different measurements were carried out with the same set-up at ∼0.1 time constant and displayed with a logarithmic time scale for convenience of comparison.
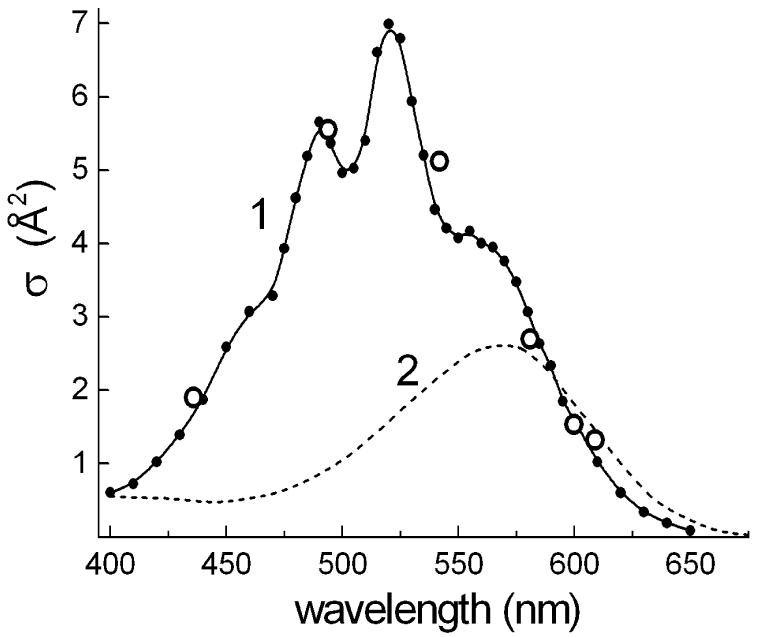
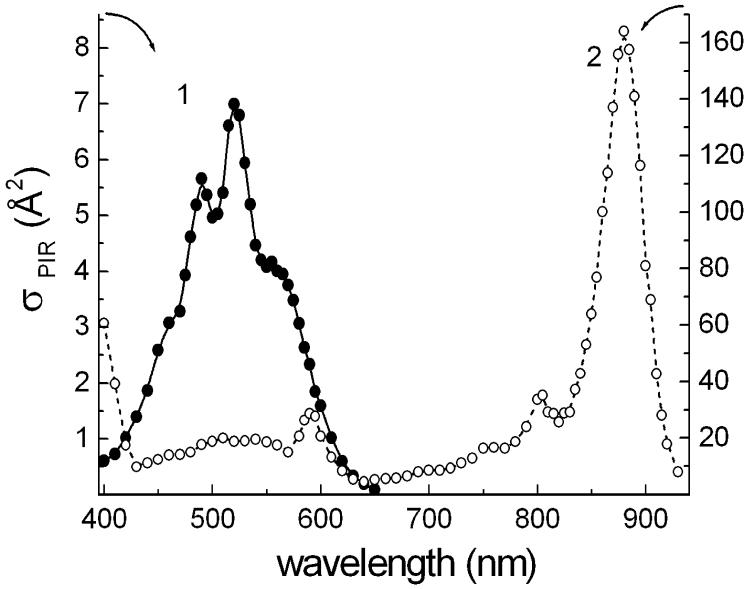
Analysis of the light-intensity dependence of the flash-induced photoinhibition can be used for determining photochemical cross-section of the photoprocess at selected wavelengths [13, 34]. It provides a way of the estimating the size of the light-harvesting antenna. Under our experimental conditions, the yields of photoinhibition (integral under the flash-induced kinetic curve) per 2 μs xenon flashes were not fully saturated even for white light in the spectral range of 400-700 nm of the lamp emission. Thus, the maximum yield was roughly estimated by extrapolating a Poissonian curve (1-exp-(σφI)) for white light flashes of varying energies. The photochemical cross-sections σ at wavelengths of interference filters with calibrated transmitted energy were calculated from the ratios of corresponding yields versus the maximum yield. As shown in Fig. 6, the values of calculated photochemical cross-sections of flash-induced photoinhibition at the selected spectral bands are well fit to the contour of the action spectrum, which can be hence defined now as an absolute action spectrum of the photoprocess. The cross-section of the retinal band at 560 nm (∼4 Å2) is 2.7 times larger than the expected value of about 1.5 Å2 for a single xanthorhodopsin molecule, i.e., the product of the absorption cross-section ∼2.4 Å2 (corresponding to the extinction coefficient, 63 mM-1 cm-1 [35]) and the quantum efficiency of the photocycle (0.64 [36, 37]) in bacteriorhodopsin. The higher experimental value might indicate an arrangement of xanthorhodopsin molecules in trimeric units in the cytoplasmic membrane of S. ruber, similarly to bacteriorhodopsin molecules in purple membranes of H. salinarum, but with energy coupling of monomers within the trimers, unlike in the latter [38]. However, we cannot exclude the possibility that the experimental value is overestimated because of errors in extrapolation from a not completely saturated light-intensity dependence.
Fig. 6.
Photochemical cross sections of the photoprocess of inhibition of respiration in cells of S. ruber. 1, absolute action spectrum of photoinhibition of respiration in the cells obtained by adjustment of the data of monochromatic light measurements with the data of estimated photochemical cross sections from energy-saturation behavior of the flash-induced photoprocess at several wavelengths, assigned by interference filters (open circles); 2, optical cross section spectrum of isolated bacteriorhodopsin molecule (evaluated from [35]).
On the other hand, the increased cross-section at 522 nm is from the additional contribution of the antenna carotenoid, salinixanthin [8] (Fig. 6), which, as follows from the deconvolution in Fig. 3, has a maximum cross-section equal to that of the retinal chromophore. The shape of the action spectrum and estimated cross-sections are consistent with the conclusion that xanthorhodopsin complex includes only a few chromophore molecules, most likely one retinal and one carotenoid molecule as an accessory light-harvesting antenna per monomeric unit [8].
3.3. Comparison of energy transfer efficiency in xanthorhodopsin with other light energy transducing complexes: light-harvesting systems of Rhodospirillum rubrum and Halorubrum strain A1c.
In xanthorhodopsin, the effective energy migration from salinixanthin to retinal increases the photochemical cross-section of the complex about 2-fold [8]. This will provide some advantage to S. ruber in competition with haloarchaea for survival in the same ecological niche under limiting light intensity, although the maximum efficiency of light-harvesting by the retinal-based photosystems is more than one order of magnitude less compared with even the smallest bacteriochlorophyll-based photosynthetic unit, complex of reaction center and LH1 antenna in purple bacteria like Rhodospirillum rubrum (Fig. 7). It should be noted however that in the spectral region of 450-650 nm the photochemical cross sections are comparable in S. ruber and R. rubrum. In the latter species, there is a lower efficiency for energy migration from spirilloxanthin to bacteriochlorophyll [39].
Fig. 7.
Comparison of absolute action spectra of photoinhibition of respiration in: 1, S. ruber; 2, R. rubrum. Note the 20-fold difference of the ordinate axis for the spectra.
S. ruber shares its habitat, the crystallizer ponds in solar salterns, with extremely halophilic archaea that numerically dominate in these environments [29]. Halorubrum species, in turn, dominate the culturable fraction of archaea in crystallizer ponds [40, 41], although culture-independent methods indicate that Walsby square archaea are numerically dominant in many hypersaline environments [42]. Our strain Halorubrum sp. A1c contains the carotenoid bacterioruberin, and the retinal protein was identified as archaerhodopsin from the gene sequence that indicated an amino acid sequence identical to that published earlier [31]. The membranes isolated from this strain exhibited a CD spectrum with the characteristic bands of bacterioruberin (data not shown) similar to those described earlier [28], confirming an earlier result from which interaction of the bacterioruberin and archaerhodopsin was proposed [28]. Thus, a question arises about the possibility of energy transfer from bacterioruberin to the chromophore of the interacting retinal protein. Earlier attempts to detect such process by measuring the relative efficiency of proton transfer upon excitation at two different wavelengths produced a negative answer [28]. Our recent results with Salinibacter ruber stimulated us to undertake a comparative study.
As in bacteriorhodopsin-containing Halobacterium salinarum [5], photoinhibition of respiration in the cells of Halorubrum sp. strain A1c was significantly lower than in S. ruber (Fig. 2). Typically, the initial slope and saturation level of the light curves for photoinhbition of respiration in the archaeon were about 6-7-fold and 3-5-fold less, compared to those in S. ruber, respectively, despite similar dark rates of respiration in the two species. The observed significant difference between the two species in the efficiency of modulation of respiration by light-driven proton pumps can be caused by different proportions of various oxidases with different sensitivities to the light-induced proton electrochemical gradient and/or by different proton gradient generation capabilities of xanthorhodopsin and archaerhodopsin.
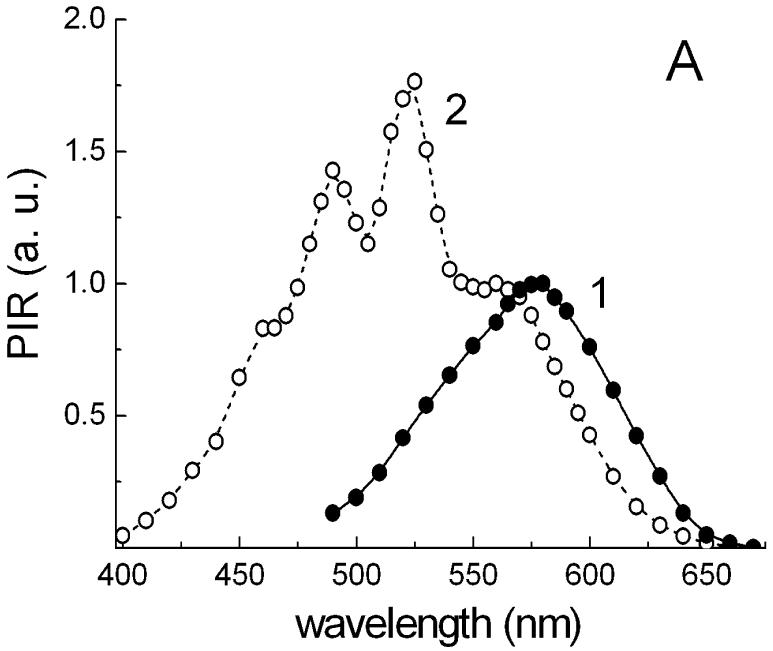
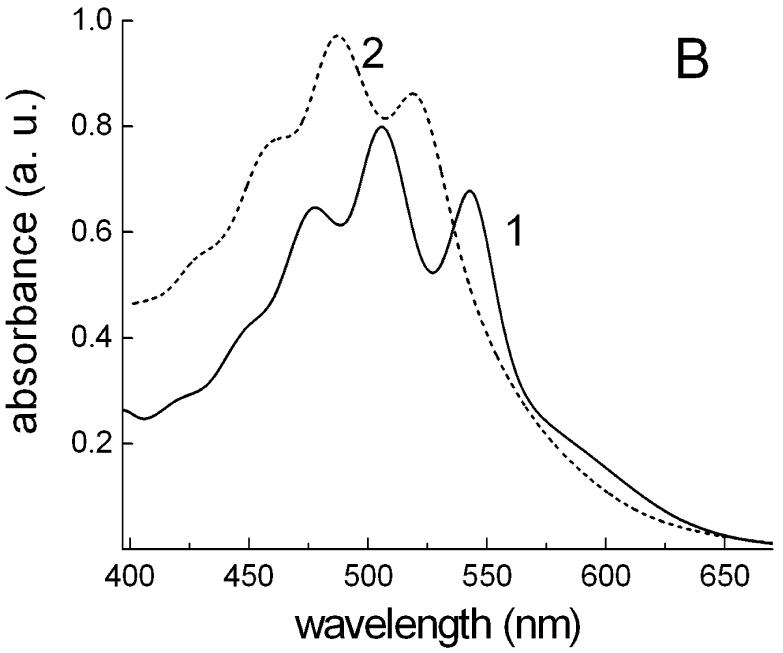
The action spectrum for Halorubrum sp. strain A1c (spectrum 1 in Fig. 8A) exhibits a single maximum at 576 nm from archaerhodopsin. Unlike the action spectrum in Salinibacter ruber (spectrum 2 in Fig. 8A), there is no significant energy transfer from the carotenoid to the retinal chromophore of archaerhodopsin, at least in this isolate. The prominent bands of bacterioruberin at 477, 506 and 543 nm in the absorption spectrum of Halorubrum cell membranes (spectrum 1 in Fig. 8B) are missing in the action spectrum. The band of retinal in the archaerhodopsin is ∼16 nm red-shifted compared to that in xanthorhodopsin, whereas the maxima of salinixanthin are close to the minima of bacterioruberin (Fig. 8B). This might indicate specific spectral tuning of the absorption maxima of xanthorhodopsin and archaerhodopsin in the coexisting cultures to maximize light absorption.
Fig. 8.
Differences of energetic coupling of carotenoids with archaerhodopsin and xanthorhodopsin. A. Normalized action spectra of photoinhibition of respiration (PIR) in cells of: 1, Halorubrum sp., strain A1c; 2, Salinibacter ruber. B. Absorption spectra of isolated cell membranes of: 1, Halorubrum sp. strain A1c; 2, Salinibacter ruber.
4. Conclusions
Recent genomic studies have revealed a widespread and continuously widening family of microbial type 1 rhodopsins [43], photoactive retinal-proteins with sensory or energy-transducing functions in phylogenetically diverse species from haloarchaea [44], proteobacteria [11, 45], cyanobacteria [46], fungi [12] and algae [47]. Among the energy-transducing light-driven proton pumps, there are a few known and rather well characterized examples, i.e., bacteriorhodopsin and archaerhodopsin of haloarchaea and proteorhodopsin of marine proteobacteria. With the exception of bacteriorhodopsin, the significance of these pumps in cellular metabolism, particularly in the proteobacteria which have not been cultured, is still poorly understood. Our earlier [8] and present study shows that analysis of light-induced modulation of respiration [13] is a useful tool for in situ characterization of the proton pump functioning in intact cells. It demonstrated that xanthorhodopsin of Salinibacter ruber is a novel pigment complex combining a carotenoid antenna, salinixanthin, with the retinal proton pump to increase light-harvesting to a broader spectral range. The light-harvesting carotenoid of xanthorhodopsin remains to this day a unique feature in the retinal based ion pumps; perhaps representing one of the early chromophore complexes that utilizes excitation energy transfer for proton translocation. Despite interaction between bacterioruberin and the retinal protein archaerhodopsin, no efficient energy transfer was found between them. This suggests that unlike in xanthorhodopsin, the function of bacterioruberin is limited to a structural and/or photoprotective role.
Acknowledgements:
This work was supported by the following grants: ARO (W911NF-06-01-0020 to S.P.B. and J.K.L.), NIH (GM29498 to J.K.L.), DOE (DEFG03-86ER13525 to J.K.L.) and Russian Fond for Basic Research (project 06-04-48668, to V.A.B.), The Molecular and Cellular Biology Program of RAS (to V.A.B.), Expert Visit NATO Grant (to V.A.B.), and the Spanish Ministry of Science grant BOS2003-05198-CO2 (to J. A.)
Footnotes
- CCCP
- carbonyl cyanide m-chlorophenyl-hydrazone
- DCCD
- N,N’-dicyclohexylcarbodiimide
- PIR
- photoinhibition of respiration
References
- [1].Oesterhelt D, Stoeckenius W. Functions of a new photoreceptor membrane. Proc. Natl. Acad. Sci. U.S.A. 1973;70:2853–2857. doi: 10.1073/pnas.70.10.2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Schobert B, Lanyi JK. Halorhodopsin is a light-driven chloride pump. J. Biol. Chem. 1982;257:10306–10313. [PubMed] [Google Scholar]
- [3].Green BR, Anderson JM, Parson WW. Photosynthetic membranes and their light-harvesting antennas. In: Green BR, Parson WW, editors. Light-Harvesting Antennas in Photosynthesis. Kluwer Academic Publishers; Dordrecht/Boston/London: 2003. pp. 1–28. [Google Scholar]
- [4].Boichenko VA. Photosynthetic units of phototrophic organisms. Biochemistry (Moscow) 2004;69:581–596. doi: 10.1023/b:biry.0000029844.31857.40. [DOI] [PubMed] [Google Scholar]
- [5].Litvin FF, Boichenko VA, Balashov SP, Dubrovskii VT. Photoinduced inhibition and stimulation of respiration in cells of Halobacterium halobium: kinetics, action spectra, relation to photoinduction of ΔpH. Biofizika. 1977;22:1062–1071. [PubMed] [Google Scholar]
- [6].Stoeckenius W, Lozier RH, Bogomolni RA. Bacteriorhodopsin and the purple membrane of Halobacteria. Biochim. Biophys. Acta. 1979;505:215–278. doi: 10.1016/0304-4173(79)90006-5. [DOI] [PubMed] [Google Scholar]
- [7].Dencher NA, Heyn MP. Bacteriorhodopsin monomers pump protons. FEBS Lett. 1979;108:307–310. doi: 10.1016/0014-5793(79)80552-9. [DOI] [PubMed] [Google Scholar]
- [8].Balashov SP, Imasheva ES, Boichenko VA, Antón J, Wang JM, Lanyi JK. Xanthorhodopsin: A proton pump with a light-harvesting carotenoid antenna. Science. 2005;309:2061–2064. doi: 10.1126/science.1118046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, Jovanovich SB, Gates CM, Feldman RA, Spudich JL, Spudich EN, DeLong EF. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. doi: 10.1126/science.289.5486.1902. [DOI] [PubMed] [Google Scholar]
- [10].Béjà O, Spudich EN, Spudich JL, Leclerc M, DeLong EF. Proteorhodopsin phototrophy in the ocean. Nature. 2001;411:786–789. doi: 10.1038/35081051. [DOI] [PubMed] [Google Scholar]
- [11].Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, Fouts DE, Levy S, Knap AH, Lomas MW, Nealson K, White O, Peterson J, Hoffman J, Parsons R, Baden-Tillson H, Pfannkoch C, Rogers Y-H, Smith HO. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- [12].Waschuk SA, Bezerra AG, Jr., Shi L, Brown LS. Leptosphaeria rhodopsin: Bacteriorhodopsin-like proton pump from a eukaryote. Proc. Natl. Acad. Sci. U.S.A. 2005;102:6879–6883. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Boichenko VA. Study of the organization of photosynthetic units by action spectra of functional activity. Biophysics. 2004;49:238–247. [PubMed] [Google Scholar]
- [14].Drachev LA, Jasaitis AA, Kaulen AD, Kondrashin AA, Liberman EA, Nemecek IB, Ostroumov SA, Semenov AY, Skulachev VP. Direct measurement of electric current generation by cytochrome oxidase, H+-ATPase and bacteriorhodopsin. Nature. 1974;249:321–324. doi: 10.1038/249321a0. [DOI] [PubMed] [Google Scholar]
- [15].Michel H, Oesterhelt D. Light-induced changes of the pH gradient and the membrane potential in Halobacterium halobium. FEBS Lett. 1980;65:175–178. doi: 10.1016/0014-5793(76)80473-5. [DOI] [PubMed] [Google Scholar]
- [16].Oesterhelt D, Krippahl G. Light inhibition of respiration in Halobacterium halobium. FEBS Lett. 1973;36:72–76. doi: 10.1016/0014-5793(73)80339-4. [DOI] [PubMed] [Google Scholar]
- [17].Vermeglio A, Carrier JM. Photoinhibition by flash and continuous light of oxygen uptake by intact photosynthetic bacteria. Biochim. Biophys. Acta. 1984;764:233–238. [Google Scholar]
- [18].Richaud P, Marrs BL, Vermeglio A. Two modes of interaction between photosynthetic and respiratory electron chains in whole cells of Rhodopseudomonas capsulate. Biochim. Biophys. Acta. 1986;850:256–263. [Google Scholar]
- [19].Boichenko VA, Makhneva ZK. Light-induced inhibition of respiration and organization of photosynthetic units in whole cells of purple bacteria. Biochemistry (Moscow) 1994;59:885–890. [Google Scholar]
- [20].Boichenko VA, Klimov VV, Mayes SR, Barber J. Characterization of the light-induced oxygen gas exchange from the IC2 mutant of Synechocystis sp PCC 6803 lacking the photosystem II 33 kDa extrinsic protein. Z. Naturforsch. 1993;48:224–233. [Google Scholar]
- [21].Boichenko VA, Klimov VV, Miyashita H, Miyachi S. Functional characteristics of chlorophyll d-predominating photosynthetic apparatus in intact cells of Acaryochloris marina. Photosynth. Res. 2000;65:269–277. doi: 10.1023/A:1010637631417. [DOI] [PubMed] [Google Scholar]
- [22].Rakhimberdieva MG, Boichenko VA, Karapetyan NV, Stadnichuk IN. Interaction of phycobilisomes with photosystem II dimers and photosystem I monomers and trimers in the cyanobacterium Spirulina platensis. Biochemistry. 2001;40:15780–15788. doi: 10.1021/bi010009t. [DOI] [PubMed] [Google Scholar]
- [23].Peltier G, Ravenel J, Vermeglio A. Inhibition of respiratory activity by saturating flashes in Chlamydomonas: evidence for a chlororespiration. Biochim. Biophys. Acta. 1987;893:83–90. [Google Scholar]
- [24].Boichenko VA. Photosynthetic generation of O2 and H2 by photosystem I-deficient Chlamydomonas mutants. Biochemistry (Moscow) 1998;63:164–170. [PubMed] [Google Scholar]
- [25].Lutnaes BF, Oren A, Liaaen-Jensen S. New C-40-carotenoid acyl glycoside as principal carotenoid in Salinibacter ruber, an extremely halophilic eubacterium. J. Nat. Prod. 2002;65:1340–1343. doi: 10.1021/np020125c. [DOI] [PubMed] [Google Scholar]
- [26].Mongodin EF, Nelson KE, Daugherty S, DeBoy RT, Wister J, Khouri H, Weidman J, Walsh DA, Papke RT, Sanchez Perez G, Sharma AK, Nesbø CL, MacLeod D, Bapteste E, Doolittle WF, Charlebois RL, Legault B, Rodriguez-Valera F. The genome of Salinibacter ruber: Convergence and gene exchange among hyperhalophilic bacteria and archaea. Proc. Natl. Acad. Sci. U.S.A. 2005;102:18147–18152. doi: 10.1073/pnas.0509073102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Peña A, Valens M, Santos F, Buczolits S, Antón J, Kämpfer P, Busse H-J, Amann R, Rosselló-Mora R. Intraspecific comparative analysis of the species Salinibacter ruber. Extremophiles. 2005;9:151–161. doi: 10.1007/s00792-005-0430-y. [DOI] [PubMed] [Google Scholar]
- [28].Mukohata Y, Ihara K, Uegaki K, Miyashita Y, Sugiyama Y. Australian Halobacteria and their retinal-protein ion pumps. Photochem. Photobiol. 1991;54:1039–1045. doi: 10.1111/j.1751-1097.1991.tb02127.x. [DOI] [PubMed] [Google Scholar]
- [29].Antón J, Rosselló-Mora R, Rodríguez-Valera F, Amann R. Extremely halophilic Bacteria in crystallizer ponds from solar salterns. Appl. Environ. Microbiol. 2000;66:3052–3057. doi: 10.1128/aem.66.7.3052-3057.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Feng J, Zhou PJ, Liu SJ. Halorubrum xinjiangense sp. nov., a novel halophile isolated from saline lakes in China. Int. J. Syst. Evol. Micrbiol. 2004;54:1789–1791. doi: 10.1099/ijs.0.63209-0. [DOI] [PubMed] [Google Scholar]
- [31].Sugiyama Y, Maeda M, Futai M, Mukohata Y. Isolation of a gene that encodes a new retinal protein, archaerhodopsin, from Halobacterium sp. aus-1. J. Biol. Chem. 1989;264:20859–20862. [PubMed] [Google Scholar]
- [32].Antón J, Oren A, Benlloch S, Rodríguez-Valera F, Amann R, Rosselló-Mora R. Salinibacter ruber gen. nov., sp. nov., a novel, extremely halophilic member of the Bacteria from saltern crystallizer ponds. Int. J. Syst. Evol. Micrbiol. 2002;52:485–491. doi: 10.1099/00207713-52-2-485. [DOI] [PubMed] [Google Scholar]
- [33].Boichenko VA. Action spectra and functional antenna sizes of Photosystems I and II in relation to the thylakoid membrane organization and pigment composition. Photosynth. Res. 1998;581:163–174. [Google Scholar]
- [34].Mauzerall D, Greenbaum N. The absolute size of a photosynthetic unit. Biochim. Biophys. Acta. 1989;974:119–140. [Google Scholar]
- [35].Oesterhelt D, Meentzen M, Schuhmann L. Reversible dissociation of the purple complex in bacteriorhodopsin and identification of 13-cis and all-trans-retinal as its chromophores. Eur. J. Biochem. 1973;40:453–463. doi: 10.1111/j.1432-1033.1973.tb03214.x. [DOI] [PubMed] [Google Scholar]
- [36].Govindjee R, Balashov SP, Ebrey TG. Quantum efficiency of the photochemical cycle of bacteriorhodopsin. Biophys. J. 1990;58:597–608. doi: 10.1016/S0006-3495(90)82403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Tittor J, Oesterhelt D. The quantum yield of bacteriorhodopsin. FEBS Lett. 1990;263:269–273. [Google Scholar]
- [38].Balashov SP, Litvin FF, Sineshchekov VA. Photochemical processes of light energy transformation in bacteriorhodopsin. In: Skulachev VP, editor. Physicochemical Biology Reviews. Harwood Academic Publishers; GmbH, UK: 1988. pp. 1–61. [Google Scholar]
- [39].Polivka T, Sundström V. Ultrafast dynamics of carotenoid excited states-from solution to natural and artificial systems. Chem. Rev. 2004;104:2021–2071. doi: 10.1021/cr020674n. [DOI] [PubMed] [Google Scholar]
- [40].Benlloch S, Acinas SG, Antón J, Luz SP, Rodríguez-Valera F. Archaeal diversity in crystallizer ponds from a solar saltern: culture versus PCR. Microb. Ecol. 2001;41:12–19. doi: 10.1007/s002480000069. [DOI] [PubMed] [Google Scholar]
- [41].Burns DG, Camakaris HM, Janssen PH, Dyall-Smith ML. Combined use of cultivation-dependent and cultivation independent methods indicates that members of most haloarchaeal groups in an Australian crystallizer pond are cultivable. Appl. Environ. Microbiol. 2004;70:5258–5265. doi: 10.1128/AEM.70.9.5258-5265.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Oren A. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol. Ecol. 2002;39:1–7. doi: 10.1111/j.1574-6941.2002.tb00900.x. [DOI] [PubMed] [Google Scholar]
- [43].Spudich JL, Jung K-H. Microbial rhodopsins: phylogenetic and functional diversity. In: Briggs WR, Spudich JL, editors. Handbook of photosensory receptors. Wiley-VCH; Darmstadt: 2005. pp. 1–23. [Google Scholar]
- [44].Frigaard N-U, Martinez A, Mincer TJ, DeLong EF. Proteorhodopsin lateral gene transfer between marine planktonic Bacteria and Archaea. Nature. 2006;439:847–850. doi: 10.1038/nature04435. [DOI] [PubMed] [Google Scholar]
- [45].Giovannoni SJ, Bibbs L, Cho J-C, Stapels MD, Desiderio R, Vergin KL, Rappé MS, Laney S, Wilhelm LJ, Tripp HJ, Mathur EJ, Barofsky DF. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature. 2005;438:82–85. doi: 10.1038/nature04032. [DOI] [PubMed] [Google Scholar]
- [46].Jung KH, Trivedi VD, Spudich JL. Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 2003;47:1513–1522. doi: 10.1046/j.1365-2958.2003.03395.x. [DOI] [PubMed] [Google Scholar]
- [47].Sineshchekov OA, Jung KH, Spudich JL. Two rhodopsins mediate phototaxis to low- and high-intensity light in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 2002;99:8689–8694. doi: 10.1073/pnas.122243399. [DOI] [PMC free article] [PubMed] [Google Scholar]