Abstract
Phospholipase A2s (PLA2s) are key enzymes that catalyze the hydrolysis of membrane phospholipids to release bioactive lipids that play an important role in normal cellular homeostasis. Under certain circumstances, disrupted production of key lipid mediators may adversely impact physiological processes, leading to pathological conditions such as inflammation and cancer. In particular, cytosolic PLA2α (cPLA2α) has a high selectivity for liberating arachidonic acid (AA) that is subsequently metabolized by a panel of downstream enzymes for eicosanoid production. Although concentrations of free AA are maintained at low levels in resting cells, alterations in AA production, often resulting from dysregulation of cPLA2α activity, are observed in transformed cells. In this review, we summarize recent evidence that cPLA2α plays a role in the pathogenesis of many human cancers. Much of this evidence has been accumulated from functional studies using cPLA2α-deficient mice, as well as mechanistic studies in cell culture. We also discuss the potential contribution of cPLA2α and AA to apoptosis, and the regulatory mechanisms leading to aberrant expression of cPLA2α.
Keywords: Phospholipase A2, Arachidonic acid, Colon Cancer, Apoptosis, Eicosanoids, Sphingomyelinase
1. Overview
Phospholipase A2s (PLA2) represent an important superfamily of enzymes that catalyze the hydrolysis of membrane phospholipids at the sn-2 position to release lipid second messengers that play a fundamental role in inflammation and cancer. There are three major classes of PLA2s: secretory (sPLA2s), Ca2+-independent (iPLA2s), and cytosolic PLA2s (cPLA2s). In addition, there is a closely related group of proteins referred to as the platelet activating factor acetylhydrolases (lipoprotein-associated PLA2s). The characteristics of the PLA2s vary in terms of protein structure, tissue distribution, pH optima, Ca2+-dependency and substrate specificities, thereby conferring their distinct cellular functions. While a comprehensive discussion of PLA2s can be found in several excellent review articles [1-4], we will highlight studies that focus on the role of the PLA2s in the production of arachidonic acid (AA), and how this may relate to cancer pathogenesis.
In recent years, a number of studies have examined the regulatory mechanisms that control the activity of cPLA2α during tumorigenesis. This interest has been stimulated by the high selectivity of cPLA2α on membrane phospholipids containing AA (5.8.11.14: eicosatetraenoic acid). Free AA is rapidly metabolized by the cyclooxygenases (COX), lipoxygenases (LOX) and cytochrome P450s to yield a wide spectrum of eicosanoid metabolites (Figure 1). Because of the potent effects exerted by these bioactive lipids, the enzymatic release of AA from membrane phospholipids must be tightly controlled and its intracellular concentrations maintained at low levels in resting cells [5]. In many types of cancers, however, dysregulation of cPLA2α activation, as well as induction of downstream AA metabolizing enzymes, are often observed. This metabolic imbalance accounts for the high levels of proliferative eicosanoids, such as prostaglandin E2 (PGE2), that are commonly found in tumor cells. These alterations have provided the rationale for the use of non-steroidal anti-inflammatory drugs (NSAIDs) as chemoprevention agents against certain forms of cancer [6]. Despite their clinical effectiveness, however, NSAID treatment is often associated with adverse side effects, including gastrointestinal bleeding and cardiovascular toxicity [7]. Based on these adverse effects, other components of the AA cascade, including cPLA2α, have been considered as potential targets for therapy and chemoprevention. Although studies correlating increased levels of cPLA2α and PGE2 suggest that removing the “source” of proliferative eicosanoids may be effective in blocking tumorigenesis, there are many issues that must be resolved. In recent years, a by-product of AA, lysophosphatidic acid (LPA; 1-acyl-3-phosphoglycerol), and its metabolite, platelet-activating factor (PAF; 1-O-alkyl-2-acetyl-sn-glycero-3-phosphocholine), have also stimulated interest as a result of their potent effects on inflammation and mitogenesis [8, 9]. The role of these bioactive lipids in tumorigenesis have been reviewed extensively elsewhere, and thus will not be discussed further [8, 10].
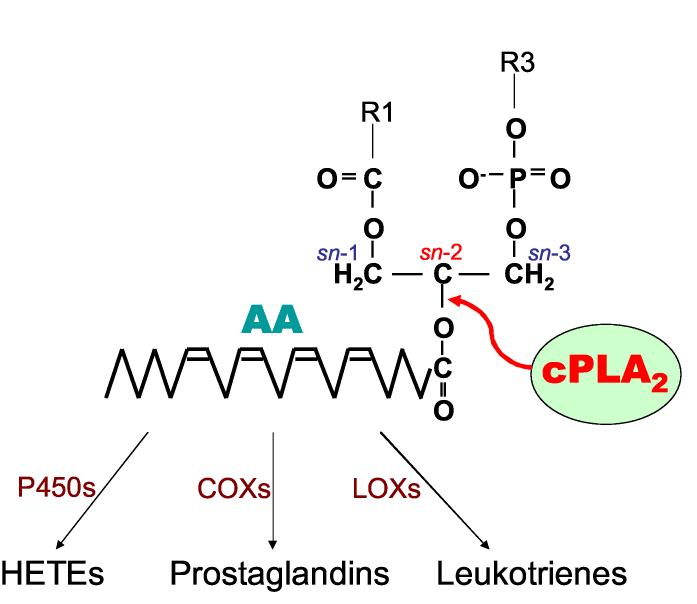
Figure 1.
Catalytic action of cPLA2α at sn-2 position of membrane phospholipids. Free AA is metabolized into a variety of eicosanoids including prostaglandins and leukotrines
In the following review, we will summarize studies demonstrating the involvement of the PLA2s, in particular cPLA2α, in the pathogenesis of cancer, both in animal models and in humans. In addition, we will focus on the contribution of the PLA2s and AA to apoptosis, and how the intracellular balance of AA is altered in transformed cells.
2. PLA2s and human cancers
Secretory phospholipse A2
In one of the first studies to associate the PLA2s with cancer, MacPhee et al. [11] reported that the gene, Pla2g2a, encoding the group IIA PLA2 (sPLA2IIA), was localized within a chromosomal region associated with the modifier of Min (Mom-1) locus. Mom-1 locus was first identified by Dietrich et al. [12] using quantitative trait loci (QTL) mapping in an attempt to clarify phenotypic variations found among individuals inheriting adenomatous polyposis coli (Apc) gene mutations [12]. It was further shown that inactivating mutations of Pla2g2a enhanced tumorigenesis in the small intestine of ApcMin mice [11], supporting the suggestion that Pla2g2a may represent a tumor modifier. These results provided the impetus for several studies to identify a human homologue of Pla2g2a with inactivating mutations. For example, Praml et al. [13] mapped the chromosomal position of a Mom-1 candidate to a region between 1p35 and 1p36.1 that is commonly lost in human cancers. Interestingly, this chromosomal region was also found to harbor loss of heterozygosity (LOH) in 48% of human sporadic colon cancers [13]. Riggins et al. [14] also tested the possibility that the human Pla2g2a homologue was in fact the tumor suppressor gene that maps to chromosome 1p. Using a set of polymorphisms, they found that 31% of colorectal cancers (CRC) lost a Pla2g2a allele. However, they found no somatic mutations within the remaining allele, and concluded that although Pla2g2a was located within a chromosomal region of common loss, it was unlikely to represent the 1p35 tumor suppressor [14]. Spirio et al. [15] then tested the possibility that a mutation in the human homologue might account for a more severe tumor phenotype among individuals with attenuated familial adenomatous polyposis (FAP), a heritable form of colon cancer. Three PLA2 genes (Pla2g2a, Pla2g2c, Pla2g5) were sequenced from subjects presenting clinically with a wide range of polyp sizes. There was no stratification of disease severity among the patient subgroups, however, indicating that mutations of the human homologue of the mouse Pla2g2a gene were not likely to be responsible for the phenotypic variations typically observed among FAP patients [15]. Tomlinson et al. [16] reached a similar conclusion regarding Pla2g2a in 70 FAP patients from 20 families, although they did report the presence of several polymorphisms and variant forms of the gene. Kennedy et al. [17] examined sPLA2IIA mRNA and protein levels in normal mucosa, as well as in duodenal and colorectal adenomas, from FAP patients. They found a significant increase in the expression of sPLA2IIA in adenomatous tissue relative to the normal intestine, a result that is in direct contrast to the earlier studies in ApcMin mice [17]. Furthermore, there were no mutations found in the adenomas, suggesting that the effect of sPLA2IIA may be species-dependent [17]. Although overexpression of sPLA2IIA has been found in other human tumors, including prostate, Barrett's adenocarcinoma and signet ring carcinoma [17-21], the role of sPLA2IIA in cancer remains ambiguous.
Cytosolic phospholipase A2
Alterations in the levels and functional activity of cPLA2α have also been associated with cancer pathogenesis. Several of these studies are summarized below in Table 1. Overexpression of cPLA2α has been reported in a variety of human cancers and tumor cell lines such as cholangiocarcinomas (SG231) and non-small lung squamous carcinoma (NSCLC) [22-24]. In Barrett's esophagus, however, cPLA2α mRNA expression was found in only 18% of adenocarcinomas, and an inverse association was reported between cPLA2α expression and depth of tumor infiltration, vascular invasion and perineural invasiveness [20]. Interestingly, cPLA2α expression in NSCLC was associated with the presence of oncogenic Ras mutations [23]. Furthermore, constitutively active Ras was sufficient to induce the expression of both cPLA2α as well as COX-2 in normal lung epithelial cells [25], providing a potential mechanism for the high levels of PGE2 often found in this form of lung cancer.
Table 1.
cPLA2α status in human cancers and cell lines
| Organ | Tumor type / cell line |
cPLA2α status | Reference(s) | |
|---|---|---|---|---|
| mRNA | Protein | |||
| Bile duct | Cholangiocarcinoma | ↑ 5-fold | [20] | |
| Lung | NSCLC | ↑ | [22] | |
| Small intestine |
Adenocarcinoma | ↑ 48% | [23] | |
| ↑ 35% | [24] | |||
| ↑ 35% | [23] | |||
| Adenocarcionoma | ↑ 50% | [25] | ||
| Colon | ↑ 60% | [26] | ||
| ↓ 84.6% | [27] | |||
| ↑ 4/5 | [28] | |||
| Stromas | - | [29] | ||
| Esophagus | Adenocarcinoma | ↑ 18% | [16] | |
| Pancreas | BxPC3, Capan2, Cfpac1, L3.6p1 |
↑ 44% | [30] | |
In one of the earliest studies to evaluate cPLA2α status in human CRC, Soydan et al. [26] examined the levels of cPLA2α in surgically excised colon tumors and matched normal tissue. Using Western analysis, they reported increased levels of cPLA2α in a subset of tumors (6 of 17) with a concomitant increase in functional activity [26]. The same group also examined cPLA2α levels in human stomach tumors but found no increase, even when COX-2 levels were highly induced [27]. Osterstrom et al. [28] examined 42 primary colorectal cancers for expression of cPLA2α using DNA dot-blots. Samples were further stratified for the presence of K-ras mutations. Although there was an increase in cPLA2α expression, the levels varied widely and there was no correlation with K-ras mutational status [28]. Using semi-quantitative RT-PCR, Dong et al. [29] found an opposite result in five human colorectal cancers, wherein cPLA2α expression was reduced despite increased COX-2 expression in 4/5 tumors. In this study, the mRNA expression data was confirmed by immunohistochemical staining (IHC) of tumor tissue and adjacent normal epithelium [29]. These data revealed intense perinuclear staining of cPLA2α within the normal colonic epithelium, a result that is in agreement with an earlier study in CHO cells showing subcellular localization of activated cPLA2α within the endoplasmic reticular membrane and nuclear envelope [30].
To further explore the potential for coordinated regulation of cPLA2α and COX-2 expression in human colon tumors, our laboratory analyzed a total of 27 human colorectal tumors for cPLA2α and COX-2 expression using IHC and correlated these findings with apoptotic index [31]. The absence of cPLA2α was reported in 84.6% (11/13) of the COX-2-overexpressing tumors, suggesting that COX-2 and cPLA2α expression are not synchronously overexpressed in a significant percentage of human CRC. These findings suggest that up-regulation of COX-2 without a corresponding increase in cPLA2α activity may lead to diminished AA levels, an outcome that would be further exacerbated by its enhanced utilization. This in turn would lead to an imbalance in the cellular pools of AA and the potential loss of several key apoptotic pathways, perhaps conferring a survival advantage that may facilitate tumorigenesis. In a subsequent study, Wendum et al. [32] used IHC to examine cPLA2α and COX-2 expression in a total of 48 colorectal adenocarcinomas and found an opposite result. Overexpression of cPLA2α was found in superficial stromal cells, corresponding to fibroblasts and myofibroblasts. Interestingly, there was correspondence between the number of cPLA2α and COX-2 expressing cells and cPLA2α-positive stromal cells correlated with high microvessel density and VEGF expression [32], leading to speculation that cPLA2α may regulate COX-induced angiogenesis via production of AA. More recently, this group tested cPLA2α and COX-2 expression by IHC in 65 human CRC specimens [33]. Although cPLA2α expression was not correlated with microsatellite instability, histological pattern or tumor stage, approximately half of the cases showed significant cPLA2α expression that was associated with COX-2 elevation [33].
3. Genetic deletion of cPLA2α in mice
Phenotypes of cPLA2α knockout mice
An important milestone in our understanding of cPLA2α function was the generation of cPLA2α-deficient mice, accomplished in 1997 [34, 35]. These heterozygous and nullizygous mice, created by homologous recombination, have provided a useful in vivo experimental model for studying the physiological and pathogenetic roles of the enzyme. The cPLA2α knockout mice are characterized by a normal lifespan and generally do not display obvious phenotypic defects, with the exception of compromised production of leukotrienes, prostaglandins and PAF [34, 35]. Suboptimal production of prostaglandins, most notably prostaglandin F2α (PGF2α), may underlie the difficulties in embryogenesis and implantation observed in the nullizygous mice [36]. Several additional phenotypic alterations include a failure to negatively regulate insulin-like growth factor-1 (IGF-1) signaling in the skeletal muscle of the heart, and reduced aquoporin-1 expression in proximal tubules [37, 38]. Additional studies in cPLA2α-deficient mice show that its absence results in resistance to inflammation, demonstrated by suppression of ischemia-reperfusion injury of the brain [34], collagen-induced arthritis [39], inflammatory bone resorption [40], pulmonary inflammatory responses [41, 42], HCl- or endotoxin-induced acute respiratory distress syndrome [42], and experimental autoimmune encephalomyelitis [43]. In contrast, the absence of cPLA2α has been shown to exacerbate autoimmune diseases such as type-1 diabetes [44]. In one such study, it was shown that non-obese diabetic (NOD) mice deficient in cPLA2α developed severe insulitis and a higher incidence of diabetes compared to their wild-type littermates [44]. Furthermore, peritoneal macrophages isolated from NOD mice with a cPLA2α deficiency produced significantly more TNF-α in response to LPS treatment, suggesting that cPLA2α may be involved in a negative feedback loop regulating TNF-α synthesis [44].
Small intestine
The deletion of cPLA2α in mice has provided fundamental new insights into its potential role in the pathogenesis of cancer. In the first study to directly test the role of cPLA2α in cancer, Takaku et al. [45] introduced the cPLA2α deletion into Apc⊿716 mice, a model of human FAP. At 10 weeks of age, the size of small intestinal polyps was reduced by approximately 11-fold, although there were no significant differences in the total numbers of polyps [45]. This result provided the first genetic evidence that cPLA2α plays a key role in the expansion of intestinal polyps without affecting the initiation of tumors [45]. In contrast to these striking findings in the small intestine, however, colon polyp growth was unaffected by the absence of cPLA2α [45]. Hong et al. [46] performed a similar study using ApcMin mice. Consistent with the results of the earlier study with Apc⊿716 mice, the deletion of cPLA2α was found to repress the growth of polyps in the small intestine. In contrast to the earlier study, the number of tumors was also reduced by 83% in the compound mutants [46]. Interestingly, the observed suppression of intestinal tumors in cPLA2α-null mice is consistent with earlier findings in COX-2 knockout mice crossed with ApcMin mice [47], suggesting that cPLA2α is the predominant source of AA for COX-2 within the small intestinal epithelium [46]. It should also be noted that the ApcMin study [46] showed a trend towards larger numbers and/or size of polyps within the colon, suggesting an organ-specific role of cPLA2α in modulating intestinal tumorigenesis.
Colon
The reported trend in colon tumorigenesis observed in ApcMin mice [45] was investigated in greater detail using a murine model of colon carcinogenesis. In this study [48], cPLA2α wild-type, heterozygous and nullizygous mice were administered azoxymethane (AOM), an organospecific methylating carcinogen that targets the distal colon [49-51]. AOM treatment of cPLA2α-deficient mice increased the number of colon tumors at twenty-four weeks after carcinogen exposure relative to wild-type controls, reflected by an increased tumor multiplicity (7.2-fold) and tumor volume (5.5-fold) [48]. The enhanced tumor sensitivity might be explained, in part, by compromised apoptosis, evidenced by the significant reduction (up to 50%) in the number of TUNEL-positive cells observed within the colonic epithelium of heterozygous and nullizygous mice [48]. Interestingly, reduced levels of apoptosis corresponded with diminished production of the cell death mediator, ceramide [48]. A key observation is that enhanced tumorigenesis occurred despite a significant reduction in PGE2 production, even in COX-2 overexpressing tumors.
Lung
The effects of cPLA2α deletion have also been tested in a lung cancer model [52]. cPLA2α-null mice on a genetic background of intermediate sensitivity to lung tumorigenesis (B6/129) were treated with six injections of the lung carcinogen, urethane. Nineteen weeks after carcinogen exposure, cPLA2α-null mice developed 43% fewer tumors compared to their wild-type littermates. Although the steady-state levels of PGE2 and PGI2 in lung tissue from wild-type or cPLA2α-null mice were similar, the levels of both prostaglandins were significantly elevated in tumors harvested from wild-type mice, an increase that was significantly attenuated in cPLA2α-null tumors [52]. The authors conclude that cPLA2α may play a role in lung carcinogenesis that is mediated through the production of prostaglandins [52], an outcome that is consistent with earlier studies using ApcMin and Apc⊿716 compound mutant mice [46].
4. cPLA2α and apoptosis
A number of recent studies have attempted to determine how cPLA2α activation influences apoptotic signaling [53]. These studies have generated data that are consistent with a mechanism in which AA derived from cPLA2α is cytotoxic. Upon addition of exogenous AA to cells in culture, a range of apoptotic responses are observed including alterations in mitochondrial membrane integrity, caspase activation and DNA fragmentation [54]. It has further been demonstrated that AA can directly activate the sphingomyelinase (SMase)-ceramide pathway, which has also been associated with cell death [55]. However, the relative contribution of endogenous AA, or its downstream metabolites, to cell death remains obscure. For example, the 15-LOX product, 15-HETE, and several cytochrome P450 metabolic products, including 12-EET and 19-HETE, can induce apoptosis when applied directly to cells. The apoptotic effects of these cytochrome P450-derived eicosanoids have been described recently in an excellent review by Cederbaum and Caro [56] and thus will not be further discussed. Instead, we will focus primarily on potential mechanisms by which the activation of cPLA2α and the resulting accumulation of intracellular AA promote cellular apoptosis.
TNF-α
The cytotoxic effects elicited by the pro-inflammatory cytokine, tumor necrosis factor-α (TNF-α), require the induction of cPLA2α gene expression and its functional activation, leading to AA release [57]. Additional evidence for the role of cPLA2α in the biological actions of TNF-α has been obtained with the use of pharmacologic or genetic inhibition [58-61]. In a recent study by Dong et al. [29], inhibition of cPLA2α activity by arachidonyl trifluoromethyl ketone (AACOCF3) in young adult mouse colon cells (YAMC) was found to protect colonocytes from TNF-α-induced apoptosis [29]. In a subsequent study, Dong et al. [31] produced a dose-dependent inhibition in both the levels and functional activity of cPLA2α in a human colon tumor cell line (HT-29) with the use of antisense oligonucleotides. As predicted, treatment of HT-29 cells with TNF-α was less effective in activating caspase-3 when cPLA2α expression was blocked, indicating that its activity is required for signaling by TNF-α. These results are consistent with findings in other tumor cell lines, including those derived from breast, skin and leukemia cells [59, 61].
Accumulation of free AA
The intracellular levels of AA are regulated through distinct and non-overlapping mechanisms. In resting cells, low concentrations of AA are tightly maintained by basal levels of rapid catabolism, and also by membrane phospholipid recycling via the actions of the arachidonyl-CoA transferase (CoA-T) (Lands cycle) [62]. In addition, esterification of AA by fatty acid CoA ligase (FACL) 4 also contributes to the maintenance of low AA levels [63]. On the other hand, stimulus-induced release of AA by cPLA2α results in its rapid metabolism by induced COX-2, thereby limiting the intracellular AA pools [62]. In fact, the chemopreventive benefits of the NSAIDs, including the COX-2 selective inhibitors, may result, in part, from their ability to cause a buildup of AA [64].
Several recent studies have attempted to establish the importance of the Lands cycle in apoptosis. For example, pharmacological inhibition of CoA-T and CoA-independent transacylase activity resulted in a buildup of AA in leukemia cells followed rapidly by cell death [65-67]. Interestingly, Perez et al. [66] reported that in phagocytes, pharmacological inhibition of COXs or LOXs did not enhance the rate of apoptosis induced by inhibition of the Lands cycle, suggesting that AA-derived eicosanoid metabolites may not be directly involved in this process. Furthermore, in a recent study by Chalimoniuk et al. [68], nitric oxide stimulation of neuronal cells caused the activation of cPLA2α with a concomitant reduction in CoA-T activity. This in turn resulted in inhibition of the Lands cycle [68]. These results suggest that reactive oxygen species induced by nitric oxide are capable of elevating intracellular AA through distinct and non-overlapping mechanisms [68].
A recent report by the Prescott laboratory [69] shows that overexpression of FACL4 and COX-2 synergistically inhibit apoptosis in colon cancer cells by reducing free AA. In addition, increased FACL4 expression was highly correlated with well (or moderately) differentiated adenocarcinomas, suggesting the possibility that selective FACL4 inhibitors may be useful for colon cancer chemoprevention [69]. In a related study, triacsin C was used to inhibit FACL4 in human embryonic kidney cells (HEK293). Apoptosis induced by the addition of exogenous AA or inhibition of COX-2 was amplified by treatment with the FACL4 inhibitor [69]. Additional studies using human lung carcinoma cells have demonstrated a synergistic reduction in cell viability upon treatment with a combination of inhibitors targeted against FACL4, the COXs and the Lands cycle, providing further evidence that intracellular accumulation of AA contributes to cell death [65].
Role of AA in mitochondrial-mediated apoptosis
Emerging evidence has shown that elevated intracellular AA can induce cell death via the mitochondrial-mediated apoptosis pathway [54]. Specifically, AA has been shown to affect mitochondrial membrane permeabilization (MMP) [54]. While MMP may involve either the inner or outer mitochondrial membranes, the outcome is always associated with cell death and release of cytochrome c [70]. The Bcl-2 family of proteins are responsible for the integrity of the outer membrane, and the inner mitochondrial membrane is regulated by the redox status of vicinal thiols and the mitochondrial permeability transition pore (PTP) [70]. An extended opening of the PTP has been reported in a rat hepatoma (MH1C1) cell line upon treatment with either TNF-α or exogenous AA, followed by release of cytochrome c and apoptosis [71]. Penzo et al. [72] extended these observations using the Ca2+ ionophore (A23187) in MH1C1 cells to directly stimulate the activity of cPLA2α and its subsequent release of AA. In this study, apoptosis was observed in approximately 30% of cells treated with Ca2+ alone, while the addition of the non-specific COX and LOX inhibitors, indomethacin and MK866, respectively, increased apoptosis by up to 2-fold [72, 73].
There are other mechanisms by which AA may influence cell death. For example, the anti-apoptotic protein, Bcl-2, is overexpressed in a number of human tumors, and elevated levels of Bcl-2 are associated with resistance to apoptosis [70]. As shown by Sun et al. [74], colon tumor cells that stably overexpress COX-2 were resistant to apoptosis induction by NSAIDs and 5-FU. Interestingly, COX-2 expressing cells had markedly elevated Bcl-2 expression, an effect that occurred independently of PGE2. These observations provide a mechanism by which some colon tumor cells acquire resistance to apoptosis following treatment with the COX-2 inhibitors, NS-398 and sulindac. Of note, the LOX inhibitors may have a dual function in altering the balance of both anti- and pro-apoptotic proteins. For example, treatment of breast cancer cells with the LOX inhibitors, Rev-5901, baicalein and MK-886, induced mitochondria-mediated apoptosis, in part by reducing the expression level of Bcl-2, while increasing the pro-apoptotic protein, Bax [75]. Similarly, alterations to the Bcl-2/Bax ratio have been reported following treatment of gastric and pancreatic cancer cells with LOX inhibitors [75, 76], suggesting that the mechanisms that underlie the anti-tumorigenic properties of this class of molecules may be complex.
Recently, several research groups have undertaken a novel strategy to overcome NSAID-resistance to mitochondrial-mediated (intrinsic) apoptosis by combining NSAID treatment with Fas or TRAIL to stimulate membrane receptor-mediated apoptosis via the extrinsic pathway [77, 78]. . In one such study, Sinicrope et al. [78] demonstrated that forced expression of Bcl-2 in colon cancer cells reduced cytochrome c release. This effect was associated with resistance to apoptosis in response to the COX-2 inhibitor, sulindac sulfide. Treatment of these colon tumor cells with HA 14-1, a specific Bcl-2 inhibitor, restored cytochrome c release and apoptosis [78]. Moreover, simultaneous treatment with a combination of HA 14-1 and TRAIL markedly enhanced sulindac sulfide-mediated apoptosis [78]. This latter result is important and demonstrates the possibility of exploiting alternative mechanisms for targeting NSAID-resistant tumor cells.
Activation of the SMase-ceramide pathway by AA
The cytotoxicity of AA may also be mediated through its ability to increase ceramide levels. Ceramide, a complex family of lipid molecules that is comprised of a sphingosine base and a fatty acid moiety is a potent inducer of cell death. Ceramide can be synthesized through a de novo synthetic pathway that involves the condensation of serine and palmitoyl CoA [79]. Ceramide can also be generated directly from sphingomyelin by the actions of the SMases, a family of proteins consisting of three isoforms (acidic, neutral and alkaline) [79]. The activation of SMases with the resultant production of ceramide has been shown to occur in response to various stimuli, including treatment of cells with either TNF-α or NSAIDs. In an attempt to reveal the mechanism of TNF-α-mediated apoptosis, Jayadev et al. [80] identified a correlation between TNF-α-induced accumulation of AA and increased levels of ceramide in human promyelocytic leukemia cells. Furthermore, SMase activation has also been shown to occur in a number of human tumor cell lines, including breast [81], prostate [82], brain [83], and colon [84] following treatment of cells with TNF-α. A recent report by Martin et al. [77] demonstrated that an accumulation of AA caused by the COX-2 inhibitor, DuP-697, activated acidic SMase to generate ceramide-enriched caveolae within the plasma membrane outer leaflet in HT-29 colon cancer cells [77]. Subsequently, these caveolae enhanced the clustering of a TRAIL-mediated death-inducing signaling complex [77]. Similarly, Voelkel-Johnson et al. [85] showed that the addition of a membrane permeable, synthetic C6-ceramide enabled metastatic colon cancer cells to overcome their normal resistance to TRAIL-induced apoptosis, thus establishing a strong association between AA, ceramide and subsequent apoptosis.
Our laboratory and others have also shown that alterations of the SMaseceramide pathway may be associated with tumorigenesis [48, 86, 87]. For example, as noted above, Ilsley et al. [48] established an association between reduced ceramide production and diminished apoptosis in carcinogen-induced colon tumors, a finding that was restricted to cPLA2α-deficient mice. In addition, Wu et al. [87] showed that the growth inhibitory effects of omega-3 polyunsaturated fatty acids on breast tumor xenografts were correlated with elevated activity of the neutral SMase isoform. Moreover, reduced activity of the intestinal-specific alkaline SMase has been reported in human colorectal tumors [88, 89]. Decreased levels of alkaline SMase activity have also been found in human stool samples collected from CRC patients, suggesting its potential as a novel diagnostic marker for colon cancer [86]. For a detailed discussion of alkaline SMase, see the recent review by Duan [90].
5. Regulation of cPLA2α expression and activity
cPLA2α expression induced by transformed cells
The regulation of cPLA2α is complex, involving both direct transcriptional activation, as well as post-translational modifications. As described above, the levels of cPLA2α are tightly regulated in order to maintain consistent production of AA as well as an optimum balance of eicosanoid metabolites [2]. In tumor cells, however, the transcriptional and post-translational regulation of cPLA2α is often dysregulated, resulting in aberrant production of downstream proliferative signals, including PGE2. This observation is well illustrated by the transcriptional activation of cPLA2α by oncogenic Ras in human lung tumor cells (NSCLC) [23]. As noted earlier, Rasactivating mutations are frequently found in a variety of tumors, including prostate, colon and lung [91]. Oncogenic transformation of Ras results in constitutive activation of the ERK and PI3K/Akt pathways, which are commonly associated with tumor promotion [91]. Blaine et al. [92] have shown that Ras induces expression of cPLA2α in NSCLC cells via direct binding of the transcription factors, Sp1 and c-jun, to the cPLA2α promoter [92]. Interestingly, it was shown in human skin-derived carcinoma cells (A431) that the translocation and phosphorylation of cPLA2α could be accomplished by another target of c-jun, epidermal growth factor receptor (EGFR) [93]. Recently, Hassan et al. [94] demonstrated in prostate cancer cells that enhanced cPLA2α activation during EGFR signaling provides a source of AA for the synthesis of 5-hydroxyeicosatetraenoic acid (5-HETE) by 5-LOX, which will subsequently increase PKC activity, resulting in cell growth. Moreover, EGFR signaling is also reported to induce COX-2 expression in human epithelial carcinoma cells (A431) [93]. Thus, it can be argued that the coordinated activation of these key pathway enzymes in transformed cells likely accounts for the high levels of PGE2 that are found in these tumors.
The tumor growth factor, TGF-β, has also been shown to enhance cPLA2α-mediated PGE2 production [95]. Although TGF-β induces tumor growth arrest in normal epithelial cells through Smad-mediated gene transcription, transformed cells acquire resistance to the anti-tumorigenic effect of TGF-β [95]. To determine whether this resistance is associated with aberrant production of PGE2, Han et al. [95] tested the effects of TGF-β in a panel of human liver cancer cells. Addition of TGF-β1 resulted in the activation of the Smad2 transcription factor, as well as phosphorylation of cPLA2α [95]. Increased levels of cPLA2α blocked TGF-β-mediated growth inhibition, an effect that was observed by the addition of cPLA2α inhibitor which otherwise induced the growth arrest [95]. Moreover, the inhibition of COX-2, PGE2 receptor 1 (EP1) or PGJ2 receptor, peroxisome proliferator-activated receptor-gamma (PPAR-γ), also restored the anti-tumorigenic effects of TGF-β, invoking multiple prostaglandin-dependent pathways for disruption of tumorigenesis [95].
There is recent evidence that cPLA2α activity may also be involved in cell-cell contact within endothelial HUVEC cells. In the following study, pharmacological inhibition of cPLA2α with pyrrolidine-1 was found to inhibit HUVEC proliferation, while exogenously added AA reversed this effect [96]. Interestingly, a confluence-dependent redistribution of cPLA2α to the distal Golgi was observed, an effect that was associated with inhibition of activity and reduced cell proliferation in the HUVECs. cPLA2α activity was restored by its cellular relocation to the cytoplasm and perinuclear region in response to mechanical wounding of the confluent monolayers, suggesting a novel role for cPLA2α in angiogenesis [96].
Transcriptional suppression of cPLA2α
cPLA2α expression can also be down-regulated at the level of gene transcription. A recent report by D'Orazi et al. [97] describes the suppression of cPLA2α expression by a transcriptional co-repressor, homeodomain-interacting protein kinase-2 (HIPK2). HIPK2 is a unique transcriptional regulator with kinase activity that exhibits anti-tumorigenic properties through its ability to directly activate p53 [98, 99]. Silencing of HIPK2 by RNAi in colon cancer cells (RKO) enhanced cPLA2α expression, and its increased activity was associated with elevated production of PGE2 [97]. Moreover, RKO cells lacking HIPK2 resulted in enhanced tumor formation in nude mice [97]. Interestingly, it was also shown that an inverse relationship between the levels of HIPK2 and cPLA2α occurred in colon tumors from FAP patients [97]. This novel observation may provide insight into earlier findings of reduced cPLA2α expression in a subset of human colon cancers [29, 58].
Regulation by AA metabolites
There is also evidence that AA and its metabolites can directly influence the levels of cPLA2α, raising the possibility of a positive feedback mechanism for the production of eicosanoids in transformed cells. As shown by Hughes-Fulford et al. [100], AA added exogenously to human prostate tumor cells (PC-3) induced the transcriptional activation of cPLA2α and COX-2, followed by a subsequent dose-dependent increase in the synthesis of PGE2 [100]. In addition, treatment of PC-3 cells with a COX-2 inhibitor, flurbiprofen, blocked the up-regulation of both cPLA2α and COX-2 [100]. Although the latter finding suggests that PGE2 may be able to induce cPLA2α expression, the role of PGE2 in this effect is unknown. In fact, it was shown in mouse lung fibroblasts that treatment of cells with various prostaglandins, including PGE2, PGJ2 and PGF2 induced COX-2, but not cPLA2α, expression [101]. Only a slight increase in the cytokine-induced expression of cPLA2α was observed by the addition of PGF2α [101]. These conflicting results suggest that the regulation of cPLA2α by metabolites of AA may not only be tissue-specific, but also context-specific.
6. Conclusions and future directions
Activation of cPLA2α is the rate-limiting step in the generation of AA. As such, its catalytic activity plays a central role in a variety of cellular processes, including inflammation, proliferation and apoptosis. As highlighted in this review, alterations in the levels of cPLA2α expression and functional activity are a common feature in tumors. In fact, because of its critical role in controlling the formation of prostaglandins, cPLA2α has been proposed as a potential target for cancer chemoprevention, perhaps even as a substitute in some cases for the NSAIDs and COX-2 inhibitors. However, there is still much to learn with respect to how the cPLA2α-AA cascade elicits such opposing cellular responses, ranging from robust cellular proliferation to apoptosis. In tumor tissue, the effects of cPLA2α remain enigmatic. For example, the expression levels of cPLA2α in human colorectal tumors are highly variable, and it is not clear under what conditions cPLA2α activity is associated with cancer-promoting or suppressing effects. Considering these observations, it may be more appropriate to consider cPLA2α status as a potential diagnostic marker, around which individualized treatment strategies might be designed.
References
- 1.Akiba S, Sato T. Biol Pharm Bull. 2004;27:1174–8. doi: 10.1248/bpb.27.1174. [DOI] [PubMed] [Google Scholar]
- 2.Hirabayashi T, Murayama T, Shimizu T. Biol Pharm Bull. 2004;27:1168–73. doi: 10.1248/bpb.27.1168. [DOI] [PubMed] [Google Scholar]
- 3.Murakami M, Kudo I. J Biochem (Tokyo) 2002;131:285–92. doi: 10.1093/oxfordjournals.jbchem.a003101. [DOI] [PubMed] [Google Scholar]
- 4.Chen CH. Curr Opin Lipidol. 2004;15:337–41. doi: 10.1097/00041433-200406000-00015. [DOI] [PubMed] [Google Scholar]
- 5.Balsinde J, Balboa MA, Insel PA, Dennis EA. Annu Rev Pharmacol Toxicol. 1999;39:175–89. doi: 10.1146/annurev.pharmtox.39.1.175. [DOI] [PubMed] [Google Scholar]
- 6.Samoha S, Arber N. Oncology. 2005;69(Suppl 1):33–7. doi: 10.1159/000086630. [DOI] [PubMed] [Google Scholar]
- 7.Abir F, Alva S, Kaminski DL, Longo WE. Dis Colon Rectum. 2005;48:1471–83. doi: 10.1007/s10350-005-0015-y. [DOI] [PubMed] [Google Scholar]
- 8.Moolenaar WH. Curr Opin Cell Biol. 1995;7:203–10. doi: 10.1016/0955-0674(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 9.van Corven EJ, Groenink A, Jalink K, Eichholtz T, Moolenaar WH. Cell. 1989;59:45–54. doi: 10.1016/0092-8674(89)90868-4. [DOI] [PubMed] [Google Scholar]
- 10.Gardell SE, Dubin AE, Chun J. Trends Mol Med. 2006;12:65–75. doi: 10.1016/j.molmed.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 11.MacPhee M, Chepenik KP, Liddell RA, Nelson KK, Siracusa LD, Buchberg AM. Cell. 1995;81:957–66. doi: 10.1016/0092-8674(95)90015-2. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich WF, Lander ES, Smith JS, Moser AR, Gould KA, Luongo C, Borenstein N, Dove W. Cell. 1993;75:631–9. doi: 10.1016/0092-8674(93)90484-8. [DOI] [PubMed] [Google Scholar]
- 13.Praml C, Finke LH, Herfarth C, Schlag P, Schwab M, Amler L. Oncogene. 1995;11:1357–62. [PubMed] [Google Scholar]
- 14.Riggins GJ, Markowitz S, Wilson JK, Vogelstein B, Kinzler KW. Cancer Res. 1995;55:5184–6. [PubMed] [Google Scholar]
- 15.Spirio LN, Kutchera W, Winstead MV, Pearson B, Kaplan C, Robertson M, Lawrence E, Burt RW, Tischfield JA, Leppert MF, Prescott SM, White R. Cancer Res. 1996;56:955–8. [PubMed] [Google Scholar]
- 16.Tomlinson IP, Beck NE, Neale K, Bodmer WF. Ann Hum Genet. 1996;60(Pt 5):369–76. doi: 10.1111/j.1469-1809.1996.tb00434.x. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy BP, Soravia C, Moffat J, Xia L, Hiruki T, Collins S, Gallinger S, Bapat B. Cancer Res. 1998;58:500–3. [PubMed] [Google Scholar]
- 18.Graff JR, Konicek BW, Deddens JA, Chedid M, Hurst BM, Colligan B, Neubauer BL, Carter HW, Carter JH. Clin Cancer Res. 2001;7:3857–61. [PubMed] [Google Scholar]
- 19.Jiang J, Neubauer BL, Graff JR, Chedid M, Thomas JE, Roehm NW, Zhang S, Eckert GJ, Koch MO, Eble JN, Cheng L. Am J Pathol. 2002;160:667–71. doi: 10.1016/S0002-9440(10)64886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagorce-Pages C, Paraf F, Wendum D, Martin A, Flejou JF. Virchows Arch. 2004;444:426–35. doi: 10.1007/s00428-004-1003-7. [DOI] [PubMed] [Google Scholar]
- 21.Masuda S, Murakami M, Mitsuishi M, Komiyama K, Ishikawa Y, Ishii T, Kudo I. Biochem J. 2005;387:27–38. doi: 10.1042/BJ20041307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han C, Demetris AJ, Michalopoulos G, Shelhamer JH, Wu T. Am J Physiol Gastrointest Liver Physiol. 2002;282:G586–97. doi: 10.1152/ajpgi.00305.2001. [DOI] [PubMed] [Google Scholar]
- 23.Heasley LE, Thaler S, Nicks M, Price B, Skorecki K, Nemenoff RA. J Biol Chem. 1997;272:14501–4. doi: 10.1074/jbc.272.23.14501. [DOI] [PubMed] [Google Scholar]
- 24.Wu T, Han C, Lunz JG, 3rd, Michalopoulos G, Shelhamer JH, Demetris AJ. Hepatology. 2002;36:363–73. doi: 10.1053/jhep.2002.34743. [DOI] [PubMed] [Google Scholar]
- 25.Van Putten V, Refaat Z, Dessev C, Blaine S, Wick M, Butterfield L, Han SY, Heasley LE, Nemenoff RA. J Biol Chem. 2001;276:1226–32. doi: 10.1074/jbc.M003581200. [DOI] [PubMed] [Google Scholar]
- 26.Soydan AS, Tavares IA, Weech PK, Temblay NM, Bennett A. Eur J Cancer. 1996;32A:1781–7. doi: 10.1016/0959-8049(96)00166-9. [DOI] [PubMed] [Google Scholar]
- 27.Soydan AS, Gaffen JD, Weech PK, Tremblay NM, Kargman S, O'Neill G, Bennett A, Tavares IA. Eur J Cancer. 1997;33:1508–12. doi: 10.1016/s0959-8049(97)00168-8. [DOI] [PubMed] [Google Scholar]
- 28.Osterstrom A, Dimberg J, Fransen K, Soderkvist P. Cancer Lett. 2002;182:175–82. doi: 10.1016/s0304-3835(02)00081-2. [DOI] [PubMed] [Google Scholar]
- 29.Dong M, Guda K, Nambiar PR, Rezaie A, Belinsky GS, Lambeau G, Giardina C, Rosenberg DW. Carcinogenesis. 2003;24:307–15. doi: 10.1093/carcin/24.2.307. [DOI] [PubMed] [Google Scholar]
- 30.Schievella AR, Regier MK, Smith WL, Lin LL. J Biol Chem. 1995;270:30749–54. doi: 10.1074/jbc.270.51.30749. [DOI] [PubMed] [Google Scholar]
- 31.Dong M, Johnson M, Rezaie A, Ilsley JN, Nakanishi M, Sanders MM, Forouhar F, Levine J, Montrose DC, Giardina C, Rosenberg DW. Clin Cancer Res. 2005;11:2265–71. doi: 10.1158/1078-0432.CCR-04-1079. [DOI] [PubMed] [Google Scholar]
- 32.Wendum D, Comperat E, Boelle PY, Parc R, Masliah J, Trugnan G, Flejou JF. Mod Pathol. 2005;18:212–20. doi: 10.1038/modpathol.3800284. [DOI] [PubMed] [Google Scholar]
- 33.Panel V, Boelle PY, Ayala-Sanmartin J, Jouniaux AM, Hamelin R, Masliah J, Trugnan G, Flejou JF, Wendum D. Cancer Lett. 2006 doi: 10.1016/j.canlet.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 34.Bonventre JV, Huang Z, Taheri MR, O'Leary E, Li E, Moskowitz MA, Sapirstein A. Nature. 1997;390:622–5. doi: 10.1038/37635. [DOI] [PubMed] [Google Scholar]
- 35.Uozumi N, Kume K, Nagase T, Nakatani N, Ishii S, Tashiro F, Komagata Y, Maki K, Ikuta K, Ouchi Y, Miyazaki J, Shimizu T. Nature. 1997;390:618–22. doi: 10.1038/37622. [DOI] [PubMed] [Google Scholar]
- 36.Song H, Lim H, Paria BC, Matsumoto H, Swift LL, Morrow J, Bonventre JV, Dey SK. Development. 2002;129:2879–89. doi: 10.1242/dev.129.12.2879. [DOI] [PubMed] [Google Scholar]
- 37.Downey P, Sapirstein A, O'Leary E, Sun TX, Brown D, Bonventre JV. Am J Physiol Renal Physiol. 2001;280:F607–18. doi: 10.1152/ajprenal.2001.280.4.F607. [DOI] [PubMed] [Google Scholar]
- 38.Haq S, Kilter H, Michael A, Tao J, O'Leary E, Sun XM, Walters B, Bhattacharya K, Chen X, Cui L, Andreucci M, Rosenzweig A, Guerrero JL, Patten R, Liao R, Molkentin J, Picard M, Bonventre JV, Force T. Nat Med. 2003;9:944–51. doi: 10.1038/nm891. [DOI] [PubMed] [Google Scholar]
- 39.Hegen M, Sun L, Uozumi N, Kume K, Goad ME, Nickerson-Nutter CL, Shimizu T, Clark JD. J Exp Med. 2003;197:1297–302. doi: 10.1084/jem.20030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miyaura C, Inada M, Matsumoto C, Ohshiba T, Uozumi N, Shimizu T, Ito A. J Exp Med. 2003;197:1303–10. doi: 10.1084/jem.20030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meyer AM, Dwyer-Nield LD, Hurteau G, Keith RL, Ouyang Y, Freed BM, Kisley LR, Geraci MW, Bonventre JV, Nemenoff RA, Malkinson AM. Am J Physiol Lung Cell Mol Physiol. 2006 doi: 10.1152/ajplung.00182.2005. [DOI] [PubMed] [Google Scholar]
- 42.Nagase T, Uozumi N, Ishii S, Kume K, Izumi T, Ouchi Y, Shimizu T. Nat Immunol. 2000;1:42–6. doi: 10.1038/76897. [DOI] [PubMed] [Google Scholar]
- 43.Marusic S, Leach MW, Pelker JW, Azoitei ML, Uozumi N, Cui J, Shen MW, DeClercq CM, Miyashiro JS, Carito BA, Thakker P, Simmons DL, Leonard JP, Shimizu T, Clark JD. J Exp Med. 2005;202:841–51. doi: 10.1084/jem.20050665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oikawa Y, Yamato E, Tashiro F, Yamamoto M, Uozumi N, Shimada A, Shimizu T, Miyazaki J. FEBS Lett. 2005;579:3975–8. doi: 10.1016/j.febslet.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 45.Takaku K, Sonoshita M, Sasaki N, Uozumi N, Doi Y, Shimizu T, Taketo MM. J Biol Chem. 2000;275:34013–6. doi: 10.1074/jbc.C000585200. [DOI] [PubMed] [Google Scholar]
- 46.Hong KH, Bonventre JC, O'Leary E, Bonventre JV, Lander ES. Proc Natl Acad Sci U S A. 2001;98:3935–9. doi: 10.1073/pnas.051635898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oshima M, Dinchuk JE, Kargman SL, Oshima H, Hancock B, Kwong E, Trzaskos JM, Evans JF, Taketo MM. Cell. 1996;87:803–9. doi: 10.1016/s0092-8674(00)81988-1. [DOI] [PubMed] [Google Scholar]
- 48.Ilsley JN, Nakanishi M, Flynn C, Belinsky GS, De Guise S, Adib JN, Dobrowsky RT, Bonventre JV, Rosenberg DW. Cancer Res. 2005;65:2636–43. doi: 10.1158/0008-5472.CAN-04-3446. [DOI] [PubMed] [Google Scholar]
- 49.Guda K, Upender MB, Belinsky G, Flynn C, Nakanishi M, Marino JN, Ried T, Rosenberg DW. Oncogene. 2004;23:3813–21. doi: 10.1038/sj.onc.1207489. [DOI] [PubMed] [Google Scholar]
- 50.Nambiar PR, Nakanishi M, Gupta R, Cheung E, Firouzi A, Ma XJ, Flynn C, Dong M, Guda K, Levine J, Raja R, Achenie L, Rosenberg DW. Cancer Res. 2004;64:6394–401. doi: 10.1158/0008-5472.CAN-04-0933. [DOI] [PubMed] [Google Scholar]
- 51.Papanikolaou A, Wang QS, Papanikolaou D, Whiteley HE, Rosenberg DW. Carcinogenesis. 2000;21:1567–72. [PubMed] [Google Scholar]
- 52.Meyer AM, Dwyer-Nield LD, Hurteau GJ, Keith RL, O'Leary E, You M, Bonventre JV, Nemenoff RA, Malkinson AM. Carcinogenesis. 2004;25:1517–24. doi: 10.1093/carcin/bgh150. [DOI] [PubMed] [Google Scholar]
- 53.Taketo MM, Sonoshita M. Biochim Biophys Acta. 2002;1585:72–6. doi: 10.1016/s1388-1981(02)00326-8. [DOI] [PubMed] [Google Scholar]
- 54.Pompeia C, Lima T, Curi R. Cell Biochem Funct. 2003;21:97–104. doi: 10.1002/cbf.1012. [DOI] [PubMed] [Google Scholar]
- 55.Levade T, Jaffrezou JP. Biochim Biophys Acta. 1999;1438:1–17. doi: 10.1016/s1388-1981(99)00038-4. [DOI] [PubMed] [Google Scholar]
- 56.Caro AA, Cederbaum AI. Free Radic Biol Med. 2006;40:364–75. doi: 10.1016/j.freeradbiomed.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 57.Kronke M, Adam-Klages S. FEBS Lett. 2002;531:18–22. doi: 10.1016/s0014-5793(02)03407-5. [DOI] [PubMed] [Google Scholar]
- 58.Dong M, Rezaie A, Nakanishi M, Guda K, Nambiar PR, Marino JN, Sanders MM, Levine JB, Rosenberg DW. Digestive Disease Week. Vol. 1. New Orleans: 2004. p. 782. [Google Scholar]
- 59.Pirianov G, Danielsson C, Carlberg C, James SY, Colston KW. Cell Death Differ. 1999;6:890–901. doi: 10.1038/sj.cdd.4400563. [DOI] [PubMed] [Google Scholar]
- 60.Thorne TE, Voelkel-Johnson C, Casey WM, Parks LW, Laster SM. J Virol. 1996;70:8502–7. doi: 10.1128/jvi.70.12.8502-8507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu YL, Jiang XR, Newland AC, Kelsey SM. J Immunol. 1998;160:5929–35. [PubMed] [Google Scholar]
- 62.Brash AR. J Clin Invest. 2001;107:1339–45. doi: 10.1172/JCI13210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cao Y, Traer E, Zimmerman GA, McIntyre TM, Prescott SM. Genomics. 1998;49:327–30. doi: 10.1006/geno.1998.5268. [DOI] [PubMed] [Google Scholar]
- 64.Chan TA, Morin PJ, Vogelstein B, Kinzler KW. Proc Natl Acad Sci U S A. 1998;95:681–6. doi: 10.1073/pnas.95.2.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Monjazeb AM, High KP, Koumenis C, Chilton FH. Prostaglandins Leukot Essent Fatty Acids. 2005;73:463–74. doi: 10.1016/j.plefa.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 66.Perez R, Matabosch X, Llebaria A, Balboa MA, Balsinde J. J Lipid Res. 2006;47:484–91. doi: 10.1194/jlr.M500397-JLR200. [DOI] [PubMed] [Google Scholar]
- 67.Surette ME, Winkler JD, Fonteh AN, Chilton FH. Biochemistry. 1996;35:9187–96. doi: 10.1021/bi9530245. [DOI] [PubMed] [Google Scholar]
- 68.Chalimoniuk M, Glowacka J, Zabielna A, Eckert A, Strosznajder JB. Neurochem Int. 2006;48:1–8. doi: 10.1016/j.neuint.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 69.Cao Y, Pearman AT, Zimmerman GA, McIntyre TM, Prescott SM. Proc Natl Acad Sci U S A. 2000;97:11280–5. doi: 10.1073/pnas.200367597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Armstrong JS. Br J Pharmacol. 2006;147:239–48. doi: 10.1038/sj.bjp.0706556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scorrano L, Penzo D, Petronilli V, Pagano F, Bernardi P. J Biol Chem. 2001;276:12035–40. doi: 10.1074/jbc.M010603200. [DOI] [PubMed] [Google Scholar]
- 72.Penzo D, Petronilli V, Angelin A, Cusan C, Colonna R, Scorrano L, Pagano F, Prato M, Di Lisa F, Bernardi P. J Biol Chem. 2004;279:25219–25. doi: 10.1074/jbc.M310381200. [DOI] [PubMed] [Google Scholar]
- 73.Gugliucci A, Ranzato L, Scorrano L, Colonna R, Petronilli V, Cusan C, Prato M, Mancini M, Pagano F, Bernardi P. J Biol Chem. 2002;277:31789–95. doi: 10.1074/jbc.M204450200. [DOI] [PubMed] [Google Scholar]
- 74.Sun Y, Tang XM, Half E, Kuo MT, Sinicrope FA. Cancer Res. 2002;62:6323–8. [PubMed] [Google Scholar]
- 75.Tong WG, Ding XZ, Adrian TE. Biochem Biophys Res Commun. 2002;296:942–8. doi: 10.1016/s0006-291x(02)02014-4. [DOI] [PubMed] [Google Scholar]
- 76.Fan XM, Tu SP, Lam SK, Wang WP, Wu J, Wong WM, Yuen MF, Lin MC, Kung HF, Wong BC. J Gastroenterol Hepatol. 2004;19:31–7. doi: 10.1111/j.1440-1746.2004.03194.x. [DOI] [PubMed] [Google Scholar]
- 77.Martin S, Phillips DC, Szekely-Szucs K, Elghazi L, Desmots F, Houghton JA. Cancer Res. 2005;65:11447–58. doi: 10.1158/0008-5472.CAN-05-1494. [DOI] [PubMed] [Google Scholar]
- 78.Sinicrope FA, Penington RC. Mol Cancer Ther. 2005;4:1475–83. doi: 10.1158/1535-7163.MCT-05-0137. [DOI] [PubMed] [Google Scholar]
- 79.Pettus BJ, Bielawska A, Subramanian P, Wijesinghe DS, Maceyka M, Leslie CC, Evans JH, Freiberg J, Roddy P, Hannun YA, Chalfant CE. J Biol Chem. 2004;279:11320–6. doi: 10.1074/jbc.M309262200. [DOI] [PubMed] [Google Scholar]
- 80.Jayadev S, Linardic CM, Hannun YA. J Biol Chem. 1994;269:5757–63. [PubMed] [Google Scholar]
- 81.Cai Z, Bettaieb A, Mahdani NE, Legres LG, Stancou R, Masliah J, Chouaib S. J Biol Chem. 1997;272:6918–26. doi: 10.1074/jbc.272.11.6918. [DOI] [PubMed] [Google Scholar]
- 82.Condorelli F, Canonico PL, Sortino MA. Br J Pharmacol. 1999;127:75–84. doi: 10.1038/sj.bjp.0702507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sawada M, Kiyono T, Nakashima S, Shinoda J, Naganawa T, Hara S, Iwama T, Sakai N. Cell Death Differ. 2004;11:997–1008. doi: 10.1038/sj.cdd.4401438. [DOI] [PubMed] [Google Scholar]
- 84.Colell A, Coll O, Mari M, Fernandez-Checa JC, Garcia-Ruiz C. FEBS Lett. 2002;526:15–20. doi: 10.1016/s0014-5793(02)03106-x. [DOI] [PubMed] [Google Scholar]
- 85.Voelkel-Johnson C, Hannun YA, El-Zawahry A. Mol Cancer Ther. 2005;4:1320–7. doi: 10.1158/1535-7163.MCT-05-0086. [DOI] [PubMed] [Google Scholar]
- 86.Di Marzio L, Di Leo A, Cinque B, Fanini D, Agnifili A, Berloco P, Linsalata M, Lorusso D, Barone M, De Simone C, Cifone MG. Cancer Epidemiol Biomarkers Prev. 2005;14:856–62. doi: 10.1158/1055-9965.EPI-04-0434. [DOI] [PubMed] [Google Scholar]
- 87.Wu Z, Tandon R, Ziembicki J, Nagano J, Hujer KM, Miller RT, Huang C. J Lipid Res. 2005;46:1396–404. doi: 10.1194/jlr.M500071-JLR200. [DOI] [PubMed] [Google Scholar]
- 88.Duan RD, Hertervig E, Nyberg L, Hauge T, Sternby B, Lillienau J, Farooqi A, Nilsson A. Dig Dis Sci. 1996;41:1801–6. doi: 10.1007/BF02088748. [DOI] [PubMed] [Google Scholar]
- 89.Hertervig E, Nilsson A, Bjork J, Hultkrantz R, Duan RD. Br J Cancer. 1999;81:232–6. doi: 10.1038/sj.bjc.6690682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Duan RD. Biochim Biophys Acta. 2006 [Google Scholar]
- 91.Friday BB, Adjei AA. Biochim Biophys Acta. 2005;1756:127–44. doi: 10.1016/j.bbcan.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 92.Blaine SA, Wick M, Dessev C, Nemenoff RA. J Biol Chem. 2001;276:42737–43. doi: 10.1074/jbc.M107773200. [DOI] [PubMed] [Google Scholar]
- 93.Sato T, Nakajima H, Fujio K, Mori Y. Prostaglandins. 1997;53:355–69. doi: 10.1016/0090-6980(97)00036-1. [DOI] [PubMed] [Google Scholar]
- 94.Hassan S, Carraway RE. Regul Pept. 2006;133:105–14. doi: 10.1016/j.regpep.2005.09.031. [DOI] [PubMed] [Google Scholar]
- 95.Han C, Demetris AJ, Liu Y, Shelhamer JH, Wu T. J Biol Chem. 2004;279:44344–54. doi: 10.1074/jbc.M404852200. [DOI] [PubMed] [Google Scholar]
- 96.Herbert SP, Ponnambalam S, Walker JH. Mol Biol Cell. 2005;16:3800–9. doi: 10.1091/mbc.E05-02-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.D'Orazi G, Sciulli MG, Di Stefano V, Riccioni S, Frattini M, Falcioni R, Bertario L, Sacchi A, Patrignani P. Clin Cancer Res. 2006;12:735–41. doi: 10.1158/1078-0432.CCR-05-1557. [DOI] [PubMed] [Google Scholar]
- 98.D'Orazi G, Cecchinelli B, Bruno T, Manni I, Higashimoto Y, Saito S, Gostissa M, Coen S, Marchetti A, Del Sal G, Piaggio G, Fanciulli M, Appella E, Soddu S. Nat Cell Biol. 2002;4:11–9. doi: 10.1038/ncb714. [DOI] [PubMed] [Google Scholar]
- 99.Di Stefano V, Rinaldo C, Sacchi A, Soddu S, D'Orazi G. Exp Cell Res. 2004;293:311–20. doi: 10.1016/j.yexcr.2003.09.032. [DOI] [PubMed] [Google Scholar]
- 100.Hughes-Fulford M, Tjandrawinata RR, Li CF, Sayyah S. Carcinogenesis. 2005;26:1520–6. doi: 10.1093/carcin/bgi112. [DOI] [PubMed] [Google Scholar]
- 101.Vichai V, Suyarnsesthakorn C, Pittayakhajonwut D, Sriklung K, Kirtikara K. Inflamm Res. 2005;54:163–72. doi: 10.1007/s00011-004-1338-1. [DOI] [PubMed] [Google Scholar]