Abstract
There is emerging evidence that the proteolytic machinery of plants plays important roles in defense against pathogens. The oomycete pathogen Phytophthora infestans, the agent of the devastating late blight disease of tomato (Lycopersicon esculentum) and potato (Solanum tuberosum), has evolved an arsenal of protease inhibitors to overcome the action of host proteases. Previously, we described a family of 14 Kazal-like extracellular serine protease inhibitors from P. infestans. Among these, EPI1 and EPI10 bind and inhibit the pathogenesis-related (PR) P69B subtilisin-like serine protease of tomato. Here, we describe EPIC1 to EPIC4, a new family of P. infestans secreted proteins with similarity to cystatin-like protease inhibitor domains. Among these, the epiC1 and epiC2 genes lacked orthologs in Phytophthora sojae and Phytophthora ramorum, were relatively fast-evolving within P. infestans, and were up-regulated during infection of tomato, suggesting a role during P. infestans-host interactions. Biochemical functional analyses revealed that EPIC2B interacts with and inhibits a novel papain-like extracellular cysteine protease, termed Phytophthora Inhibited Protease 1 (PIP1). Characterization of PIP1 revealed that it is a PR protein closely related to Rcr3, a tomato apoplastic cysteine protease that functions in fungal resistance. Altogether, this and earlier studies suggest that interplay between host proteases of diverse catalytic families and pathogen inhibitors is a general defense-counterdefense process in plant-pathogen interactions.
There is emerging evidence that the proteolytic machinery of plants plays important and diverse roles in defense against pathogens. For example, several apoplastic proteases have been linked to defense. Rcr3, a secreted Cys protease from tomato (Lycopersicon esculentum), is required for resistance mediated by the Cf-2 receptor-like protein against strains of the fungus Cladosporium fulvum that carry the Avr2 avirulence gene (Kruger et al., 2002). An apoplastic aspartic protease (AP) CDR1 from Arabidopsis (Arabidopsis thaliana) activates defense signaling probably by generating an unknown mobile endogenous peptide elicitor (Xia et al., 2004). The multiple roles of proteases in plant defense are also reflected in their involvement in the hypersensitive response (HR), a form of programmed cell death (PCD) that is associated with resistance to pathogens (D'Silva et al., 1998; Solomon et al., 1999; Mosolov et al., 2001; Chichkova et al., 2004). Proteasome complexes involved in the ubiquitin-mediated protein degradation pathway have been implicated in PCD and disease resistance (Suty et al., 2003; Tor et al., 2003; Zeng et al., 2004). Inhibition of PCD using caspase inhibitors pointed to a role of plant proteases with caspase-like activity in the HR (D'Silva et al., 1998; Solomon et al., 1999; Chichkova et al., 2004; van der Hoorn and Jones, 2004). Plant vacuolar processing enzymes (VPEs) were recently identified to be Cys proteases with caspase-like activity (cleavage after Asp residues) that are essential for virus-induced HR and virus resistance (Hatsugai et al., 2004; Rojo et al., 2004). Interestingly, knockout of VPEγ in Arabidopsis results in increased susceptibility to Botrytis cinerea and Turnip mosaic virus, suggesting that VPEs not only contribute to resistance to avirulent pathogens but also to basal defense to pathogens during susceptible interactions (Rojo et al., 2004). A multitude of other proteases from various plants are up-regulated during infection by pathogens, suggesting a potential role in plant defense (Tornero et al., 1997; Avrova et al., 1999, 2004; Liu et al., 2001; Guevara et al., 2002; Zhao et al., 2003; Tian et al., 2004). One example is the pathogenesis-related (PR) protein P69B of tomato, an apoplastic subtilisin-like Ser protease that accumulates upon infection by multiple plant pathogens, including citrus exocortis viroid, the bacterium Pseudomonas syringae, and the oomycete Phytophthora infestans (Tornero et al., 1997; Zhao et al., 2003; Tian et al., 2004). Despite overwhelming evidence indicating that plant proteases are important components of defense responses, their precise mechanisms of action remain poorly understood. Proteases could contribute to defense in several different ways, acting at the levels of perception, signaling, and execution (van der Hoorn and Jones, 2004).
Plants and plant pathogens have coevolved diverse defense and counterdefense strategies in their arms race for survival (Stahl and Bishop, 2000). For example, plants use the PR proteins β-1,3-endoglucanases to damage pathogen cell walls, either rendering the pathogen more susceptible to other plant defense responses or releasing oligosaccharide elicitors that activate plant defenses (Rose et al., 2002). The oomycete pathogen Phytophthora sojae has evolved a counterdefense mechanism to overcome the action of β-1,3-endoglucanases. P. sojae secretes glucanase inhibitor proteins that suppress the activity of a soybean (Glycine max) β-1,3-endoglucanase (Rose et al., 2002). An analogous plant enzyme-pathogen inhibitor coevolution appears to involve plant proteases and pathogen protease inhibitors (Tian et al., 2004, 2005; Rooney et al., 2005). A diverse family of Kazal-like extracellular Ser protease inhibitors with at least 35 members has been described in five plant pathogenic oomycetes: P. infestans, P. sojae, Phytophthora ramorum, Phytophthora brassicae, and the downy mildew Plasmopara halstedii (Tian et al., 2004). In P. infestans, two Kazal-like inhibitors, EPI1 and EPI10, among a total of 14, were found to bind and inhibit the PR P69B subtilisin-like Ser protease of the host plant tomato (Tian et al., 2004, 2005). Inhibition of P69B by two structurally different protease inhibitors of P. infestans suggests that EPI1 and EPI10 function in counterdefense (Tian et al., 2004, 2005).
P. infestans causes late blight, a reemerging and ravaging disease of potato (Solanum tuberosum) and tomato (Birch and Whisson, 2001; Smart and Fry, 2001; Ristaino, 2002; Shattock, 2002; Kamoun and Smart, 2005). During infection of host plants by this pathogen, various host proteases are potentially involved in defense. Besides Ser proteases, such as P69B, other catalytic types of proteases are up-regulated during infection by P. infestans. These include an extracellular AP, as well as two Cys proteases, CYP and StCathB, from potato (Avrova et al., 1999, 2004; Guevara et al., 2002). These protease genes are differentially induced in potato plants with various levels of resistance (Avrova et al., 1999, 2004; Guevara et al., 2002). For instance, StCathB is rapidly induced during R gene-mediated resistance but its expression level is gradually increased in a potato cultivar with partial resistance (Avrova et al., 2004). The induction of AP expression is higher and faster in a resistant potato cultivar than in a susceptible one (Guevara et al., 2002). Differential induction in plants with variable levels of resistance suggests that the activity of these proteases could modulate defense. It is possible that P. infestans has evolved additional counterdefense protease inhibitors, besides the EPI Kazal-like Ser protease inhibitors, to target different catalytic classes of proteases.
In our laboratory, we have been annotating and characterizing secreted proteins from P. infestans to identify and functionally analyze candidate disease effectors (Bos et al., 2003; Torto et al., 2003; Huitema et al., 2004; Tian et al., 2004, 2005; Armstrong et al., 2005; Liu et al., 2005; Kamoun, 2006). Motif searches of P. infestans unigenes with predicted signal peptides revealed a novel family of putative protease inhibitors with cystatin-like domains (EPIC1 to EPIC4, Inter-Pro IPR000010 and/or IPR003243, MEROPS family I25). Among these, the epiC1 and epiC2 genes lacked orthologs in P. sojae and P. ramorum, were relatively fast-evolving within P. infestans, and were up-regulated during infection of tomato, suggesting a role during P. infestans-host interactions. In this study, we performed biochemical functional analysis of EPIC1 and EPIC2. EPIC2B interacts with and inhibits a novel papain-like extracellular Cys protease, termed Phytophthora Inhibited Protease 1 (PIP1). Characterization of PIP1 revealed that it is a PR protein closely related to Rcr3, a tomato apoplastic Cys protease that functions in fungal resistance and is targeted by the protease inhibitor Avr2 of C. fulvum (Kruger et al., 2002; Rooney et al., 2005). Altogether, this and earlier studies suggest that interplay between host proteases of diverse catalytic families and pathogen inhibitors is a general defense-counterdefense process in plant-pathogen interactions.
RESULTS
Cystatin-Like Protease Inhibitors in P. infestans
We mined currently available expressed sequence tags (ESTs) and whole-genome shotgun sequences of P. infestans (Kamoun et al., 1999; Randall et al., 2005) using two methods: (1) the PexFinder algorithm to identify genes encoding putative extracellular proteins (Torto et al., 2003); and (2) similarity and protein motif searches to annotate the predicted extracellular proteins (see “Materials and Methods”). Among 1,294 P. infestans unigenes with predicted signal peptides, a total of five different sequences were found to encode putative protease inhibitors with similarity to cystatin-like domains (Inter-Pro domain IPR000010 or IPR003243, MEROPS family I25). The unigenes were termed epiC1-4 (extracellular protease inhibitor with cystatin-like domain; Table I). Four of the sequences corresponded to cDNAs and one to a random genomic sequence. We confirmed the sequence of the full cDNA inserts of clones PH050H7 and PA050F5 corresponding to epiC1 and epiC4, respectively. The sequences of epiC2B and epiC3 were confirmed by sequencing the PCR products amplified from cDNAs of P. infestans strain 90128 using the primers listed in Table II. We failed to amplify epiC2A from P. infestans 90128. The open reading frames (ORFs) of epiC2A and epiC2B are very similar and their predicted proteins diverge in only eight out of 125 amino acids. Considering that the epiC2A sequence was obtained from genomic reads of P. infestans strain T30-4, epiC2A and epiC2B could correspond to alleles of the same gene or could be closely related paralogs.
Table I.
Predicted cystatin-like extracellular protease inhibitors from the oomycete plant pathogens P. infestans, P. sojae, and P. ramorum
| Species | Protein | GenBank Accession No. | Signal Peptide | Expression Stage |
|---|---|---|---|---|
| P. infestans | EPIC1 | AY935250 | Yes | Infected tomato, zoospores, germinating cysts |
| P. infestans | EPIC2A | AY935251 | Yes | Genomic sequence |
| P. infestans | EPIC2B | AY935252 | Yes | Infected tomato |
| P. infestans | EPIC3 | AY935253 | Yes | Infected tomato, germinating cysts |
| P. infestans | EPIC4 | AY935254 | Yes | Infected tomato; mycelium, nonsporulating growth, nitrogen starvation; mating culture; ungerminated sporangia |
| P. sojae | PsEPIC3 | 324122233a | Yes | Genomic sequence |
| P. sojae | PsEPIC4 | 275924893a | Yes | Genomic sequence |
| P. sojae | PsEPIC5 | 324093992a | Yes | Genomic sequence |
| P. sojae | PsEPIC6 | 275780911a | Yes | Genomic sequence |
| P. ramorum | PrEPIC3 | 324535088a | Yes | Genomic sequence |
| P. ramorum | PrEPIC4 | 324478325a | Yes | Genomic sequence |
| P. ramorum | PrEPIC5 | 324204607a | Yes | Genomic sequence |
| P. ramorum | PrEPIC7 | 303543465a | Yes | Genomic sequence |
Ti (trace identifier) number from the National Center for Biotechnology Information Trace Archive (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi).
Table II.
Primers used in this study
The letters in uppercase represent gene-specific nucleotide sequence. The letters in lowercase represent added nucleotides for the convenience of cloning. The restriction sites used for cloning epiC1 and epiC2B are underlined.
| Genes | Primers |
|---|---|
| epiC1 | F: 5′-gcggaattcCCAAGTGGACGGCGGATACT-3′ |
| R: 5′-gcggcggccgcggtacCTACTTAACTGGGGTAATCGACGTCAC-3′ | |
| epiC2B | F: 5′-gcggaattcCCAACTGAACGGATACTCAAAG-3′ |
| R: 5′-gcggcggccgcggtaccTTAGTTGGCGGGCGTAATCGAC-3′ | |
| epiC3 | F: 5′-gcgatcgATGGCTTTCACTCGTTCCATC-3′ |
| R: 5′-gcggcggccgcggtaccTTATCCTTGGTACTCTGCAGGCGT-3′ | |
| epiC4 | F: 5′-gcgaagcttCGCGACGTCGTTACTATCGGC-3′ |
| R: 5′-gcggcggccgctctagaCTATACGAAATCAGAAAACATAGC-3′ | |
| Pip1 | F: 5′-gcgggatccATGGCTTCCAATTTTTTCCTCAAG-3′ |
| R: 5′-gcgactagtTCAgtgatggtgatggtgatgAGCAGTAGGGAACGACGCAACCTTTGC-3′ |
The four epiC cDNAs encoded predicted proteins ranging from 125 to 172 amino acids (Fig. 1A). EPIC1 to 4 were predicted to have signal peptides with a significant mean S value over 0.9 based on SignalP Version 2.0 analyses (Nielsen et al., 1997; Nielsen and Krogh, 1998). They possessed all the signature sequences of cystatin-like protease inhibitors, including the conserved Gly residue in the N-terminal trunk, the highly conserved Gln-Xaa-Val-Xaa-Gly motif in the first binding loop (with Xaa representing any amino acid), and the conserved aromatic residue Trp in the second binding loop (Turk and Bode, 1991; Nagata et al., 2000). EPIC4 was larger than the other three proteins with a Ser-rich region of 44 residues at the C terminus.
Figure 1.
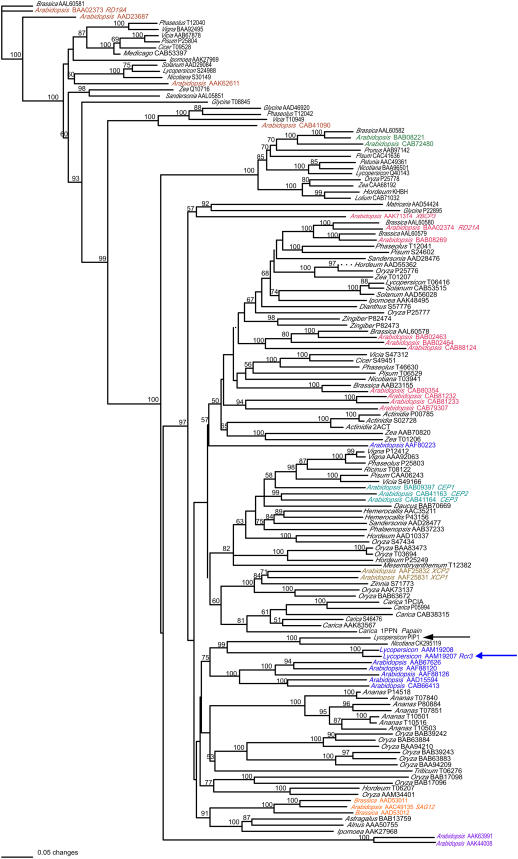
Cystatin-like extracellular protease inhibitors in Phytophthora spp. A, Sequence alignment of P. infestans EPIC1 to EPIC4 proteins with chicken egg white cystatin (CHKCYS, P01038), a type member of cystatin-like Cys protease inhibitors. The proposed active sites of cystatins, including the N-terminal trunk (NT), first binding loop (L1), and second binding loop (L2), are shown. Amino acid residues that define cystatins are marked with asterisks. The predicted signal peptides are shown in italics. B, Phylogenetic relationships of 13 cystatin-like extracellular proteinase inhibitors from P. infestans, P. sojae, and P. ramorum. The neighbor-joining tree was generated as described in “Materials and Methods” using the sequences listed in Table I. The tree was rooted with chicken egg white cystatin (CHKCYS, P01038). Bootstrap values were obtained with 1,000 replications, and values higher than 50% are shown. The length of the branches reflects weighted amino acid substitutions, and the scale bar indicates 10% weighted sequence divergence. The two clusters of orthologous genes identified in the three species are indicated by vertical lines.
Phylogenetic Relationships among Phytophthora EPICs
We used the amino acid sequences of the EPIC proteins to search for cystatin-like protease inhibitors in P. sojae and P. ramorum by performing TBLASTN searches against whole-genome shotgun sequences of P. sojae and P. ramorum. A total of four sequences from each genome were identified (Table I). All eight sequences encoded predicted proteins with putative signal peptides and similarity to cystatin-like domains.
The phylogenetic relationships among the 13 EPIC proteins from the three plant pathogenic Phytophthora species were investigated. Multiple alignments were generated and a phylogenetic tree was constructed using the neighbor-joining method. The phylogeny pointed to several clusters of related proteins with 100% bootstrap replication values (Fig. 1B). In particular, the tree topology indicated that EPIC3 and EPIC4 are highly conserved in the three oomycetes and their corresponding genes could be orthologous. In sharp contrast, EPIC1 and EPIC2 are unique to P. infestans and are missing from P. sojae and P. ramorum, suggesting that these genes evolved relatively recently in the P. infestans lineage, possibly through duplication of epiC3.
We also used reciprocal BLAST searches (all-versus-all method; Tatusov et al., 1997) to independently determine putative clusters of orthologous epiC genes in the three genomes. The two epiC3 and epiC4 clusters identified in the phylogenetic tree showed unambiguous 1:1:1 relationships (one ortholog in each species), and high similarity among proteins from the same cluster was evident through their entire sequence (data not shown). These analyses also confirmed that EPIC1 and EPIC2 are closely related and lack corresponding sequences in P. sojae and P. ramorum. The best matches to the EPIC1 and EPIC2 proteins were members of the EPIC3 cluster, again supporting the possibility that epiC1 and epiC2 evolved in P. infestans through duplication of epiC3.
The epiC1 and epiC2B Genes Are Up-Regulated during Infection of Tomato
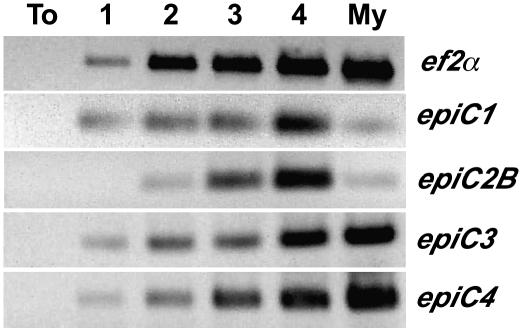
We used semiquantitative reverse transcription (RT)-PCR to examine the expression of the four epiC genes during infection of tomato by P. infestans and in mycelium cultured in vitro (Fig. 2). Transcripts for all four genes were detected during colonization of tomato. However, epiC1 and epiC2B appeared significantly up-regulated during infection compared to mycelium and relative to the constitutive elongation factor 2 α (ef2α) gene. In contrast, epiC3 and epiC4 expression patterns were similar to the constitutive ef2α gene. In summary, the up-regulation of epiC1 and epiC2B during colonization of tomato, as well as their polymorphic nature and restriction to P. infestans, led us to hypothesize that these cystatin-like inhibitors target host proteases and function during infection. We therefore proceeded to perform functional analyses of EPIC1 and EPIC2B.
Figure 2.
RT-PCR analysis of epiC genes during a time course of colonization of tomato by P. infestans. Total RNA isolated from noninfected leaves (To), infected leaves of tomato, 1, 2, 3, or 4 d after inoculation with P. infestans, and from P. infestans mycelium grown in synthetic medium (My) was used in RT-PCR amplifications. Amplifications of P. infestans ef2α were used as constitutive controls to determine the relative expression of epiC genes.
EPIC1 Protein Is Abundantly Secreted in Tomato Apoplast during Infection
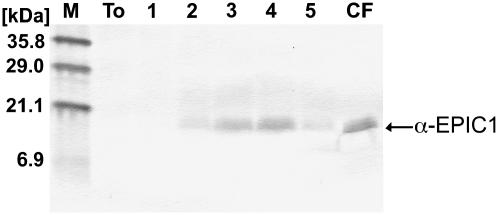
To determine the presence of the EPIC proteins in the apoplast of tomato during infection by P. infestans, we generated antisera raised against purified recombinant EPIC (rEPIC) proteins (see “Materials and Methods” for details). The EPIC2B antisera failed to react with extracts from P. infestans and infected tomato, and were not used further. In contrast, the EPIC1 antisera reacted well with rEPIC1 and detected a single band of the expected size in western-blot analyses of culture filtrates from P. infestans (data not shown). Additional western blots of intercellular fluids (10 μL of 10× concentrated extracts) collected from an infection time course of tomato (1–5 d after inoculation) indicated that the EPIC1 protein is abundantly present in infected tissue, particularly at 3 and 4 d after inoculation (Fig. 3).
Figure 3.
EPIC1 protein is abundantly secreted into the tomato apoplast during infection. Equal volumes (10 μL from 10× concentrated) of tomato intercellular fluids isolated from noninfected leaves (To), infected leaves of tomato, 1, 2, 3, 4, or 5 d after inoculation with P. infestans, and P. infestans culture filtrate (CF) were used in western-blot analyses with EPIC1 antisera. The size in kD of the molecular mass markers is shown on the left.
EPIC2B Interacts with PIP1, a Tomato Papain-Like Cys Protease
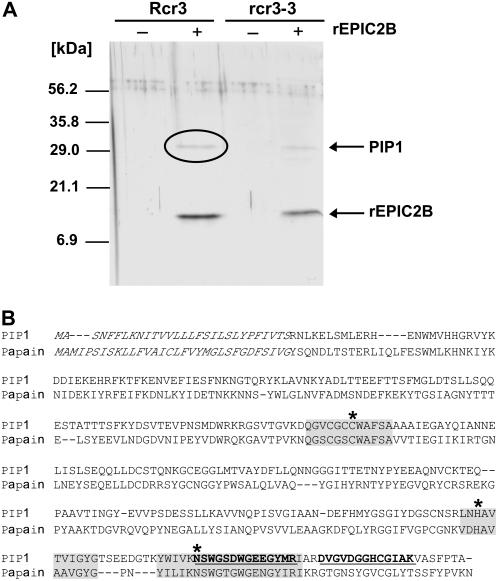
To identify the plant proteases targeted by the rEPIC proteins, we performed coimmunoprecipitation on tomato intercellular fluids incubated with rEPICs using FLAG antibody covalently linked agarose beads. To simultaneously test the possibility that the EPICs interact with the Rcr3 Cys protease of tomato, we used intercellular fluids isolated from 12-week-old tomato plants of two different genotypes: a line carrying the Rcr3pim gene and the rcr3-3 mutant, which contains a premature stop codon in the Rcr3pim coding sequence (Kruger et al., 2002). We could not detect rEPIC1 or any other proteins after coimmunoprecipitation, probably because rEPIC1 was degraded in tomato extracts (data not shown). On the other hand, rEPIC2B coimmunoprecipitation was successful and resulted in the recovery of rEPIC2B and another protein of approximately 30 kD (Fig. 4A). The rEPIC2B-interacting protein was pulled down from intercellular fluids isolated from both Rcr3pim and rcr3-3 tomato lines, excluding the possibility that this protein is Rcr3 (Fig. 4A).
Figure 4.
EPIC2B interacts with PIP1, a tomato Cys protease of the C1A papain family. A, Coimmunoprecipitation of rEPIC2B and PIP1 using FLAG antisera. Eluates from coimmunoprecipitation of rEPIC2B with proteins in intercellular fluids from Rcr3pim and rcr3-3 (a premature termination mutant of Rcr3pim) tomato were run on SDS-PAGE gel, followed by staining with silver nitrate. Rcr3 or rcr3-3 indicates whether the intercellular fluids were obtained from Rcr3pim or rcr3-3 plants. rEPIC2B indicates whether or not rEPIC2B was added to the reaction mix. The band indicated by a circle was cored from a replicate gel stained with Colloidal Coomassie Blue, used in tandem mass spectrometry, and identified as the papain-like Cys protease PIP1. The size in kD of the molecular mass markers is shown on the left. B, Sequence alignment of PIP1 with papain of papaya (Carica papaya; GenBank gi:67642), the type member of C1A Cys proteases. The predicted signal peptide sequences are shown in italics. The two peptides of PIP1 sequenced by tandem mass spectrometry are underlined in bold. The conserved protease catalytic triad residues (C154, H287, and N309) predicted based on the consensus patterns of eukaryotic thiol (Cys) protease active sites (PROSITE: PDOC00126) are shown by asterisks. The conserved sequences around catalytic residues are shaded.
To identify the rEPIC2B-interacting protein, the band obtained from coimmunoprecipitation with extracts of the Rcr3pim plants was cored from the gel and analyzed by tandem mass spectrometry. Two peptides of 13 and 14 amino acids were sequenced and matched several tomato ESTs in GenBank dBEST database. All the matching ESTs corresponded to the consensus sequence TC118154 in the tomato Gene Index of The Institute of Genome Research. Based on the putative coding sequence of TC118154, primers were designed to amplify the whole ORF from tomato cDNAs (Table II), and the PCR product was sequenced to completion. The cDNA sequence encodes a protein of 345 amino acids, with only one residue differing from the TC118154 predicted protein sequence (V87 versus I87; Fig. 4B; GenBank accession no. CK295119). The protein was named PIP1. Similarity searches indicated that PIP1 is a papain-like Cys protease (subfamily C1A, MEROPS peptidase database, http://merops.sanger.ac.uk/), with conserved protease catalytic triad residues (C154, H287, and N309) based on the consensus pattern of eukaryotic thiol (Cys) protease active sites (PROSITE entry PDOC00126).
Confirmation of EPIC2B Interaction with PIP1
To independently confirm the coimmunoprecipitation experiment, we transiently expressed PIP1 in the apoplast of Nicotiana benthamiana plants. A construct with the entire ORF of PIP1 fused to the His epitope tag at the C terminus was transiently expressed in N. benthamiana plants by agroinfiltration. We extracted intercellular fluids from infiltrated leaves of N. benthamiana and performed western blot with His antisera to detect PIP1-His (Fig. 5A). A distinct band of approximately 30 kD was detected in intercellular fluids from leaves infiltrated with Agrobacterium tumefaciens carrying the plasmid pCB302-PIP1-His but not from leaves infiltrated with A. tumefaciens containing the empty binary vector, indicating that the mature form of PIP1-His was successfully expressed.
Figure 5.
Confirmation of rEPIC2B binding to PIP1. A, Expression of PIP1-His in N. benthamiana using agroinfiltration. Intercellular fluids were isolated from N. benthamiana leaves infiltrated with A. tumefaciens containing the binary vector pCB302-PIP1-His (PIP1+) or pCB302-3 (PIP1−). A. tumefaciens carrying the gene silencing suppressor P19 was always coinfiltrated to enhance protein expression. The intercellular fluids were used in western blot with His antisera. The size in kD of the molecular mass markers is shown on the left. B, Coimmunoprecipitation of rEPIC2B and PIP1-His using FLAG antisera. Eluates from coimmunoprecipitations of rEPIC2B and rEPIC1 with proteins in intercellular fluids from N. benthamiana leaves infiltrated with A. tumefaciens carrying the binary vector pCB302-PIP1-His (PIP1+) or pCB302-3 (PIP1−). The + and − signs refer to the presence or absence of PIP1, respectively. The small molecular mass bands correspond to rEPIC2B and rEPIC1. The approximately 30-kD band that was pulled down with rEPIC2B corresponds to PIP1-His. The size in kD of the molecular mass markers is shown on the left.
To confirm that rEPIC2B binds PIP1, we performed coimmunoprecipitation experiments on PIP1-His apoplastic fluids of N. benthamiana incubated with rEPICs. The FLAG antibody agarose bead immunoprecipitations were successful and resulted in the recovery of the rEPIC proteins in all samples (Fig. 5B). In addition, a 30-kD protein corresponding to PIP1-His was recovered with rEPIC2B from PIP1-expressing extracts but not from the control sample, confirming the identity of PIP1 as a protein that interacts with EPIC2B. Interaction between rEPIC1 and PIP1 was not detected in this experiment.
EPIC2B Inhibits PIP1 and Other Apoplastic Cys Proteases of Tomato
We performed a series of experiments to determine whether EPIC2B inhibits tomato proteases, particularly PIP1. For these experiments, we performed Cys protease activity profiling (van der Hoorn et al., 2004) using DCG-04, a biotinylated analog of the irreversible Cys protease inhibitor E-64 that can be detected with streptavidin (Greenbaum et al., 2000). First, we examined whether rEPIC2B can inhibit the Cys proteases present in tomato intercellular fluids. DCG-04 labeling of tomato intercellular fluid proteins revealed three major bands of approximately 30 kD, 36 kD, and 40 kD, with the 30-kD band showing the highest intensity (Fig. 6A). Pretreatment of the samples with E-64 eliminated almost all DCG-04 labeling, indicating that the Cys proteases were specifically labeled. On the other hand, pretreatment with rEPIC2B completely inhibited the labeling of the 40-kD proteases and partially inhibited the 30-kD and 36-kD proteases. These experiments were performed in the presence of the Ser protease inhibitor rEPI1 (a potent inhibitor of the tomato Ser protease P69B) that was shown to increase the stability of rEPIC2B in tomato extracts (data not shown). As expected, control pretreatment with rEPI1 only did not interfere with DCG-04 labeling. This experiment indicates that EPIC2B inhibits DCG-04 labeling of tomato apoplastic Cys proteases.
Figure 6.
rEPIC2B inhibits apoplastic tomato proteases including PIP1. A, EPIC2B inhibits apoplastic Cys proteases of tomato. Intercellular fluids of tomato were preincubated with E-64 (1.02 μm) and combinations of EPIC2B (2.00 μm) and EPI1 (2.00 μm) for 1 h before DCG-04 was added to the final concentration of 0.22 μm to label Cys proteases with biotin. Following labeling, proteins were acetone precipitated, and biotin-labeled proteases were captured with magnetic streptavidin beads and analyzed by SDS-PAGE and streptavidin western blotting. The compositions of the reaction mixes are indicated by the ± signs at the top. Molecular mass markers are shown on the left. Biotin-labeled proteases are indicated by dots on the right. Black dot, gray dot, and white dot indicate biotin-labeled protease bands of 30 kD, 36 kD, and 40 kD, respectively. B, PIP1 has Cys protease activity. Intercellular fluids of N. benthamiana expressing PIP1-His were first incubated with 0.22 μm DCG-04 for 2 h. Proteins were acetone precipitated, and biotin-labeled proteases were captured either by magnetic streptavidin beads (lane 1) or by magnetic Ni-NTA beads to purify His-tagged proteins (lane 2). Extracted proteins were separated by SDS-PAGE and subjected to western blotting with streptavidin. The molecular masses of the markers in lane M are shown on the left. The band corresponding to the DCG-04-labeled PIP1-His is indicated with an arrow. C, EPIC2B is an inhibitor of PIP1. Time-course labeling of PIP1-His with DCG-04 was performed in the absence or presence of rEPIC2B. Intercellular fluids of N. benthamiana expressing PIP1-His were preincubated with rEPIC2B (2,240 nm) or the irreversible protease inhibitor E-64 (1,120 nm) prior to labeling with DCG-04 (220 nm). The labeling reactions were stopped at five time points (10, 20, 30, 60, and 120 min, respectively) by adding ice-cold acetone. PIP1-His was recovered from precipitated proteins by magnetic Ni-NTA beads and analyzed by SDS-PAGE and streptavidin western blotting. Note that rEPIC2B competitively inhibits binding of DCG-04 to PIP1 at the 30-min labeling time point but not later, suggesting that it is a reversible inhibitor. Approximate molecular masses of the labeled PIP1-His proteins are shown on the left side.
To determine whether PIP1 is a functional Cys protease, we performed DCG-04 activity profiling on intercellular fluids obtained from N. benthamiana leaves expressing PIP1-His by agroinfiltration (Fig. 5A). Following DCG-04 labeling, half the sample was used to capture biotinylated proteins using streptavidin beads, while the other half was used to capture HIS-tagged proteins (PIP1-His) using nickel-nitrilotriacetic acid (Ni-NTA) magnetic beads. At least five biotin-labeled bands ranging from 30 to 50 kD were detected in the streptavidin-captured N. benthamiana intercellular fluids (Fig. 6B). In contrast, a single biotin-labeled approximately 30-kD protein corresponding to PIP1-His was recovered with the Ni-NTA beads. This band was absent in Ni-NTA-captured proteins from intercellular fluids of N. benthamiana infiltrated with A. tumefaciens carrying the empty vector plasmid (data not shown). This experiment suggests that PIP1-His was successfully labeled with DCG-04 and, therefore, has Cys protease activity.
To determine whether rEPIC2B inhibits PIP1, we performed a time-course labeling experiment of PIP1-His with DCG-04 in the absence or presence of rEPIC2B. In the absence of inhibitors, biotinylated PIP1-His became clearly detectable 30 min after addition of DCG-04 and reached a maximal level at 120 min (Fig. 6C). In contrast, preincubation with rEPIC2B blocked DCG-04 labeling of PIP1-His at 30 min and reduced labeling at 60 to 120 min. The observation that increased incubation times with DCG-04 resulted in some level of PIP1-His labeling suggests that inhibition by EPIC2B is reversible. As expected, preincubation with the irreversible inhibitor E-64 resulted in complete blockage of DCG-04 labeling of PIP1-His for up to 120 min. These results indicate that EPIC2B inhibits PIP1, although it probably binds to PIP1 in a reversible fashion.
PIP1 Is Closely Related to Rcr3
Papain-like Cys proteases (C1A) are widely distributed in plant species and form complex families. For example, in Arabidopsis, there are 30 proteins homologous to the C1A subfamily of proteases (Beers et al., 2004). A phylogenetic analysis of 138 C1A plant proteases generated by Beers et al. (2004) revealed eight different groups designated C1A-1 to C1A-8. To investigate the relationship between PIP1 and other Cys proteases of plants, we reconstructed the Beers et al. (2004) tree but included PIP1 and a putative C1A protease of N. benthamiana (GenBank accession no. CK295119), which shows 70% identity to PIP1. Remarkably, PIP1 turned out to be very closely related to the tomato Cys protease Rcr3 in the C1A-5 group (Fig. 7; Supplemental Fig. S1). PIP1, N. benthamiana CK295119, and the two forms of Rcr3 (Kruger et al., 2002) grouped together in highly significant cluster (99% bootstrapping support).
Figure 7.
PIP1 is closely related to tomato defense-related Cys protease Rcr3. A neighbor-joining phylogenetic tree was constructed with 138 plant papain-like C1A proteases described by Beers et al. (2004), PIP1, and a putative C1A protease encoded by N. benthamiana EST CK295119. Bootstrap values were obtained with 1,000 replications, and the values higher than 50% are shown. The length of the branches reflects weighted amino acid substitutions, and the scale bar indicates 5% weighted sequence divergence. The genus name and the GenBank accession number are given for all sequences. The sequences that have been assigned to eight tentative groups (C1A-1–C1A-8; Beers et al., 2004) are color-coded. PIP1 and tomato Rcr3 are indicated with arrows.
Pip1 Is Tightly Linked to Rcr3 in Chromosome 2
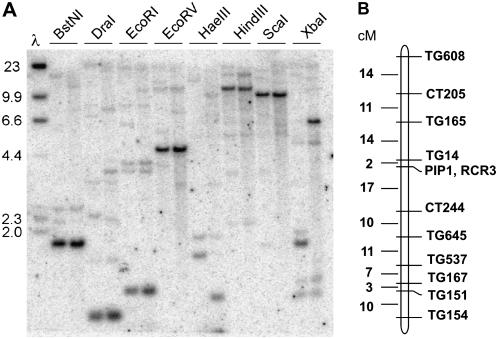
To determine the occurrence of Pip1-like sequences in the tomato genome, we hybridized genomic DNA blots of tomato with a probe corresponding to the Pip1 cDNA (Fig. 8A). A single strong hybridizing band was detected with all eight restriction enzymes tested, suggesting that Pip1 is a single-copy gene. However, several weakly hybridizing bands were also observed, indicating that multiple Pip1-like sequences occur in the tomato genome. Sequential hybridization of the blot with a gene-specific probe for the Rcr3 gene indicated that none of these bands corresponds to Rcr3.
Figure 8.
Copy number of Pip1 and map position of papain-like Cys proteases in tomato. A, Occurrence of Pip1-like sequences in tomato. A Southern blot containing tomato genomic DNA digested with the eight restriction enzymes indicated above the lanes was hybridized with a probe corresponding to the entire coding sequence of Pip1. The left lane of each pair contains genomic DNA from tomato cv Sun1642, and the right lane of each pair contains DNA from S. pimpinellifolium LA1589. Molecular mass markers are indicated. B, Molecular genetic map of tomato chromosome 2, and the position of Pip1 and Rcr3 on this map. The map was derived from a population of 100 F2 plants derived from a cross between tomato cv Sun1642 and S. pimpinellifolium LA1589. No plants were obtained that were recombinant between the two genes.
Kruger et al. (2002) showed that Rcr3 occurs in chromosome 2 of tomato among a cluster of five papain-like Cys proteases. To further determine the relationship between Pip1 and Rcr3, we genetically mapped these two genes in a population derived from a cross between tomato cv Sun1642 and Solanum pimpinellifolium accession LA1589 (van der Knaap and Tanksley, 2001). The results indicated that Pip1 and Rcr3 are tightly linked, and map near marker TG14 on chromosome 2 of tomato (Fig. 8B).
PIP1 Is a PR Protein
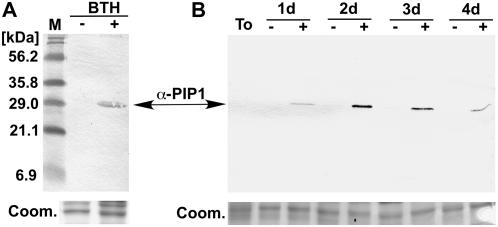
We used the GenBank accession numbers of ESTs that perfectly match the Pip1 sequence to search the literature for studies that examined expression of Pip1 during interaction with pathogens. Two studies on differential expression of tomato genes during defense included Pip1 ESTs and their corresponding cDNA clones (GenBank accession nos. BG352024 and AI780572; Xiao et al., 2001; Zhao et al., 2003). Interestingly, Zhao et al. (2003) showed that the Pip1 cDNA AI780572 is coregulated with a large number of PR protein genes, including PR1, PR2, PR3, and PR7. To test whether PIP1 is a PR protein, we generated antisera against a peptide specific to PIP1 (see “Materials and Methods”). Western blots of intercellular fluids isolated from water- and benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester (BTH)-treated tomato plants using PIP1 antisera revealed that PIP1 is highly induced by BTH treatment (Fig. 9A). We also examined the extent to which PIP1 is induced during colonization by P. infestans. Western blots of intercellular fluids isolated from tomato leaves over a time course of infection revealed that PIP1 accumulates as early as 1 d after inoculation (Fig. 9B). In contrast, PIP1 could not be detected in intercellular fluids from mock-inoculated leaves. These results indicate that PIP1 is an apoplastic protein that is induced by the salicylic acid (SA) analog BTH and during infection by P. infestans, suggesting that PIP1 is a PR protein.
Figure 9.
PIP1 is induced by pathogen-related treatments. A, PIP1 is induced by the SA analog BTH. Equal volumes of intercellular fluids isolated from water-treated (BTH−) and BTH-treated (BTH+) tomato plants were used in western-blot analyses with an antisera raised against a peptide specific to PIP1 (α-PIP1). Molecular mass markers are shown on the left. The approximately 30-kD PIP1 band is indicated with an arrow. A replicate SDS-PAGE gel stained with Coomassie Brilliant Blue was used as a loading control, and a section of this gel was shown (Coom.). B, PIP1 is induced during infection by P. infestans. Equal volumes of tomato intercellular fluids isolated from untreated tomato leaves (To), and detached tomato leaves sprayed with water (−) or a zoospore suspension of P. infestans 90128 (+) and harvested at 1, 2, 3, and 4 d postinoculation were used in western-blot analyses with PIP1 antisera (α-PIP1). A replicate SDS-PAGE gel stained with Coomassie Brilliant Blue was used as a loading control and a section of this gel was shown (Coom.).
DISCUSSION
A number of tomato and potato proteases of different catalytic types have been implicated in defense against the late blight pathogen P. infestans (Avrova et al., 1999, 2004; Guevara et al., 2002; Tian et al., 2004, 2005). One of these, P69B, a subtilisin-like PR protein of tomato, is targeted by two Kazal Ser protease inhibitors, EPI1 and EPI10, of P. infestans, suggesting that inhibition of defense proteases constitutes a virulence mechanism in this pathogen (Tian et al., 2004, 2005). In this study, we describe the cystatin-like protease inhibitors EPIC1 to EPIC4, a novel family of protease inhibitors in P. infestans that was identified based on motif annotation of putative secreted proteins. We hypothesized that EPIC1 and EPIC2 may play a role during host interactions. Indeed, EPIC2B was shown to bind and inhibit PIP1, an apoplastic PR protein of tomato with significant similarity to papain-like Cys proteases. Among all characterized plant Cys proteases, PIP1 is closely related to tomato Rcr3, another apoplastic Cys protease that is required for Cf-2 mediated resistance to C. fulvum and is inhibited by the fungal Avr2 protein (Kruger et al., 2002; Rooney et al., 2005). Together with the recent report that C. fulvum Avr2 inhibits the tomato protease Rcr3 (Rooney et al., 2005), this and our earlier studies (Tian et al., 2004, 2005) raise the possibility that interplay between host proteases and pathogen inhibitors is a general defense-counterdefense process in plant-pathogen interactions.
Phylogenomic analyses have been proposed to augment other selection criteria to identify and prioritize candidate effector genes in P. infestans (Liu et al., 2005). We performed comparative genomic analyses of epiC genes by comparing P. infestans sequences to the draft genome sequences of P. sojae and P. ramorum. These analyses revealed that epiC1 and epiC2 are fast evolving in Phytophthora. Rapid rates of evolution may have been driven by functional divergence following speciation and are consistent with a role in virulence for these genes. Indeed, of the four genes, epiC1 and epiC2 were the only ones that were up-regulated during infection, suggesting that they may target plant proteases. These analyses of rates of evolution in protease inhibitor genes helped us to discriminate between models of coevolution between the inhibitors and their target proteases. Inhibitor and proteases involved in a dynamic “arms race” coevolution, such as in interactions between pathogens and hosts, are more likely to be fast evolving than those involved in evolutionarily stable interactions. Thus, we exploited the evolutionary analyses to devise specific hypotheses regarding the function of the inhibitors, as well as prioritize candidate genes among different members of the EPIC family. Functional analyses of EPIC2B confirmed that this protein targets a plant protease. The epiC3 and epiC4 genes evolved at slower rates and have evident orthologs in the three species. We hypothesize that these genes, which are constitutively expressed in P. infestans, are likely to fulfill conserved, possibly basic, cellular functions in Phytophthora. Future functional analyses of EPIC3 and EPIC4 will test this hypothesis.
In this study, we showed that PIP1, a tomato apoplastic C1A Cys protease that is targeted by the inhibitor EPIC2B, is induced by the SA analog BTH and by P. infestans. These results confirm previous observations that Pip1 is differentially regulated under defense-related conditions (Xiao et al., 2001; Zhao et al., 2003). Pip1, identified as cDNA BG352024 (also called P271), is highly induced in transgenic tomato plants overexpressing the R gene Pto and during the resistance (incompatible) response observed on Pto tomato plants infected with P. syringae pv tomato carrying the avirulence gene avrPto (Xiao et al., 2001). Pip1, identified as cDNA AI780572, was also shown to be coregulated with a large number of SA-induced PR protein genes, including PR1, PR2, PR3, and PR7 (Zhao et al., 2003). Altogether, these independent findings confirm our finding that PIP1 is a bona fide PR protein.
The exact contribution of PIP1 to plant defenses remains to be demonstrated. The closely related Cys protease Rcr3 is required for the ability of the receptor-like protein Cf-2 to trigger cell death and fungal resistance (Kruger et al., 2002; Rooney et al., 2005). Nonetheless, the mechanism through which Rcr3 contributes to Cf-2-mediated signaling is unknown. The small secreted protein Avr2 of C. fulvum is a protease inhibitor that targets Rcr3 and triggers Cf-2-mediated HR. However, E-64, another inhibitor of Rcr3, fails to trigger HR in Cf-2 tomato, suggesting that the protease inhibition and HR induction activities of Avr2 are distinct. However, the finding that similar Cys proteases of tomato are targeted by two phylogenetically distant pathogens, an oomycete and a fungus, suggest that PIP1, Rcr3, and possibly the other papain-like proteases in the Rcr3 gene cluster play important roles in defense and confirm the importance of proteases in apoplastic defense of tomato. Based on the models described by van der Hoorn and Jones (2004), PIP1 and Rcr3 could function in different aspects of plant defense, including perception of invading microbes, mediation of defense signaling, and execution of defense responses. To distinguish between these different hypotheses, we have initiated experiments toward identifying the role of PIP1 in defense.
Several PR proteins, such as glucanases, chitinases, and proteases, are hydrolytic enzymes. Plant pathogens have evolved various mechanisms to protect their cells from the activity of these PR proteins (O'Connell and Ride, 1990; Siegrist and Kauss, 1990; Punja, 2004; van den Burg et al., 2004). To evade attack by host chitinases during plant colonization, the fungus Colletotrichum lagenarium deacetylates its cell wall chitin to render the polymer chain more resistant to chitinases, resulting in partial protection of the cell wall (Siegrist and Kauss, 1990; Punja, 2004). In a related fungal plant pathogen, Colletotrichum lindemuthianum, the cell wall within infection structures, such as infection pegs and young hyphae, differs from other structures by lacking chitin (O'Connell and Ride, 1990; Punja, 2004). In C. fulvum, the secreted AVR4 protein binds to chitin, possibly shielding it from chitinases (van den Burg et al., 2004). Pathogens may also evade the defenses mediated by PR proteins by suppressing the induction of PR protein genes or releasing subthreshold amounts of SA pathway elicitors, resulting in low levels of PR protein accumulation (Krebs and Grumet, 1993; Zhao et al., 2003; Punja, 2004). For example, the plant pathogenic bacterium P. syringae suppresses SA-dependent induction of PR protein genes by activating the antagonistic JA signaling pathway with the phytotoxin coronatine (Kloek et al., 2001; Zhao et al., 2003; Abramovitch and Martin, 2004). Similar to other plant pathogens, plant pathogenic oomycetes, such as Phytophthora, have also evolved mechanisms to escape the enzymatic activity of PR proteins. Oomycetes contain little chitin in their cell wall and are therefore resistant to plant chitinases (Kamoun, 2003). Besides the EPI and EPIC protease inhibitors of P. infestans that target tomato proteases, the glucanase inhibitor proteins of P. sojae inhibit a soybean β-1,3-glucanase belonging to the PR-2 class (Rose et al., 2002). Therefore, at least three different PR proteins are targeted by secreted inhibitors of Phytophthora, suggesting that inhibition of plant defense enzymes is a common counterdefense mechanism in these organisms.
In the future, we will continue to dissect the patterns of inhibition of tomato proteases during the late blight disease and determine the specific role of the host and pathogen proteins that mediate these cascades. Genetic experiments involving both the pathogen and the host are required to test the predictions of the hypothetical mode of actions of the defense proteases PIP1 and P69B that are targeted by P. infestans.
MATERIALS AND METHODS
Sequence and Phylogenetic Analyses
Similarity searches were performed locally on Apple Macintosh OS X workstations using BLAST (Altschul et al., 1997), and the similarity search programs implemented in the BLOCKS (Henikoff et al., 2000), pfam (Bateman et al., 2002), SMART (Letunic et al., 2002), and Inter-Pro (Apweiler et al., 2001) databases. The examined sequence databases included GenBank nonredundant, dBEST, and TraceDB (Karsch-Mizrachi and Ouellette, 2001), PFGD (http://www.pfgd.org; Randall et al., 2005), and the whole-genome shotgun sequences of Phytophthora sojae and Phytophthora ramorum (available at GenBank TraceDB Archive). The sequences described in this study were deposited in GenBank under the accession numbers listed in Table I.
Multiple sequence alignments were generated using ClustalX (Thompson et al., 1997). Phylogenetic trees were constructed using the neighbor-joining method implemented in PAUP Version 4.0b10 (Sinauer Associates) with 1,000 bootstrap replications. Clusters of orthologous epiC genes from Phytophthora infestans, P. sojae, and P. ramorum were determined using the pairwise reciprocal BLAST searches described by Tatusov et al. (1997).
Phytophthora Strain and Culture Conditions
P. infestans isolate 90128 (A2 mating type, race 1.3.4.7.8.9.10.11) was routinely grown on rye agar medium supplemented with 2% Suc (Caten and Jinks, 1968). For RNA extractions and preparation of culture filtrates, plugs of mycelium were transferred to modified Plich medium (Kamoun et al., 1993) and grown for 2 weeks before harvesting.
Bacterial Strains and Plasmids
Escherichia coli XL1-Blue or DH5α and Agrobacterium tumefaciens GV3101 were used in this study, and routinely grown in LB medium (Sambrook et al., 1989) at 37°C and 28°C, respectively.
Plasmids pFLAG-EPIC1 and pFLAG-EPIC2B were constructed by cloning PCR-amplified DNA fragments corresponding to the mature sequences of EPIC1 and EPIC2B into the EcoRI and KpnI sites of pFLAG-ATS (Sigma), a vector that allows secreted expression in E. coli. The oligonucleotides epiC1-F/R and epiC2B-F/R (Table II) were used to amplify mature EPIC1 and EPIC2B sequences, respectively. The N-terminal sequence of the processed recombinant proteins FLAG-EPIC1 (rEPIC1) and FLAG-EPIC2B (rEPIC2B) are “DYKDDDDKvkllensQVDGGYSKKE…” and “DYKDDDDKvkllensQLNGYSKKEV…,” respectively. The FLAG epitope sequence is underlined, and the first 10 amino acids of mature EPIC1 and EPIC2B are shown in bold.
Plasmid pCB302-PIP1-His was constructed using the methods described by Tian et al. (2005) and is derived from the A. tumefaciens binary vector pCB302-3 (Xiang et al., 1999). pCB302-PIP1-His contains the ORF of the Pip1 cDNA (GenBank accession no. DQ631808) fused to the His tag at the C terminus.
Plant Growth and Assays
Tomato (Lycopersicon esculentum) and Nicotiana benthamiana plants were grown in pots at 25°C, 60% humidity, under 16-h-light/8-h-dark cycle. Tomato cultivar Ohio 7814 was used in this study except when mentioned. BTH treatment and infection of tomato leaves by P. infestans were performed using 3-week-old and 8-week-old Ohio 7814 tomato plants, respectively, following the methods described earlier (Tian et al., 2004). Transient expression of PIP1 in N. benthamiana was performed by agroinfiltration as described elsewhere (Tian et al., 2005). To enhance gene expression, all constructs were coinfiltrated with pJL3-p19-55 obtained from J. Lindbo (The Ohio State University). pJL3-p19-55 is an A. tumefaciens binary vector expressing the P19 protein of Tomato bushy stunt virus, a suppressor of posttranscriptional gene silencing in N. benthamiana (Voinnet et al., 2003). Intercellular (apoplastic) fluids were prepared from tomato and N. benthamiana leaves according to the method of de Wit and Spikman (1982). For tomato leaves, a 0.24 m sorbitol solution was used as extraction buffer. For leaves from N. benthamiana, a solution of 300 mm NaCl, 50 mm NaPO4 (pH 7) was used as extraction buffer (Kruger et al., 2002). The intercellular fluids were filter sterilized (0.45 μm), and were used immediately or stored at −20°C.
Expression and Purification of Recombinant Proteins
Expression of FLAG-EPIC1 (rEPIC1) and FLAG-EPIC2B (rEPIC2B) in E. coli using plasmids pFLAG-EPIC1 and pFLAG-EPIC2B was conducted as described previously (Kamoun et al., 1997; Tian et al., 2004). Purification of FLAG-EPIC1 and FLAG-EPIC2B was performed as described earlier (Kamoun et al., 1997; Tian et al., 2004). Protein concentrations were determined using the Bio-Rad protein assay. To determine purity, 0.5 μg of purified protein was run on a SDS-PAGE gel, followed by staining with silver nitrate.
RNA Isolation and Semiquantitative RT-PCR
RNA isolation and semiquantitative RT-PCR were performed as described elsewhere (Torto et al., 2003; Tian et al., 2004). The oligonucleotides epiC1-F/R, epiC2B-F/R, epiC3-F/R, and epiC4-F/R used to amplify epiC transcripts are listed in Table II. Amplifications of P. infestans ef2α gene using the primer pair described previously (Torto et al., 2002) were used as constitutive controls to determine the relative transcript levels of epiC genes.
SDS-PAGE and Western-Blot Analyses
Proteins were subjected to 10% to 15% SDS-PAGE as described previously (Sambrook et al., 1989). Following electrophoresis, proteins were visualized by silver nitrate staining following the method of Merril et al. (1981), Coomassie Brilliant Blue staining (Sambrook et al., 1989), Colloidal Coomassie Blue staining (Anderson et al., 1991), or the proteins were transferred to supported nitrocellulose membranes (Bio-Rad Laboratories) using a Mini Trans-Blot apparatus (Bio-Rad Laboratories). Detection of antigen-antibody complexes was carried out with a western-blot alkaline phosphatase kit (Bio-Rad Laboratories). Antisera to PIP1 were produced by immunizing rabbits with the peptide H2N-INGYEVVPSDESSL-COOH, conjugated with keyhole limpet hemocyanin via added Cys residue at the N-terminal region. The peptide sequence was chosen for its highly antigenic characteristics and specificity for PIP1 among GenBank sequences. Selection of the PIP1 peptide sequence, peptide synthesis and conjugation, as well as antisera production were performed by Sigma Genosys. In western-blot analyses of tomato intercellular fluids, the antisera to PIP1 reacted only with a single band of around 30 kD. Antisera to EPIC1 and EPIC2B were raised by immunizing rabbits with recombinant proteins rEPIC1 and rEPIC2B by Cocalico Biologicals. The EPIC1 antisera reacted with rEPIC1 and identified a single band of similar size in P. infestans culture filtrate and intercellular fluids from infected tomato. Monoclonal anti-FLAG M2 antibody and anti-polyHis antibody were purchased from Sigma.
Coimmunoprecipitation
Coimmunoprecipitations of rEPIC1 and rEPIC2B with intercellular fluid proteins from tomato and N. benthamiana were performed using the FLAG-tagged Protein Immunoprecipitation kit (Sigma) following the manufacturer's instructions. Purified rEPIC1 and rEPIC2B were preincubated with intercellular fluids for 30 min at 25°C. Forty microliters anti-flag M2 resin was added and incubated at 4°C for 2 h with gentle shaking. The precipitated protein complexes were eluted in 60 μL of FLAG peptide solution (150 ng/μL) and were analyzed by SDS-PAGE.
Tandem Mass Spectrometry Sequencing
Tandem mass spectrometric sequencing of PIP1 was performed at the proteomics facility of The Cleveland Clinic Foundation as described previously (Tian et al., 2004).
Activity Profiling and Inhibition Assays of Cys Proteases
Activity profiling of PIP1 and other Cys proteases in tomato and N. benthamiana apoplastic fluids was performed as described elsewhere (van der Hoorn et al., 2004; Rooney et al., 2005) using DCG-04 (supplied by Michiel Leeuwenburgh, Leiden University), a biotinylated analog of the irreversible Cys protease inhibitor E-64 (Greenbaum et al., 2000). For tomato, 50 μL of intercellular fluids was diluted 10-fold in DCG-04 assay buffer (50 mm sodium acetate, 10 mm l-Cys, pH 5.0) to a final volume of 500:l. The diluted samples were preincubated with an excess of E-64 (1,020 nm; Sigma) or rEPIC2B (2 μm) for 1 h at room temperature. rEPI1 (Tian et al., 2004) at a final concentration of 2 μm was also included in the rEPIC2B reaction to improve the stability of rEPIC2B. DCG-04 (220 nm) was added to each reaction to label Cys proteases with biotin. The reaction mixtures were incubated for 5 h at room temperature. Proteins were precipitated by adding 1 mL of ice-cold acetone followed by centrifugation at 13,000g for 30 min at 4°C. The pellets were washed with ice-cold 70% (w/w) acetone, air-dried, and resuspended in 500 μL of TBS buffer (50 mm Tris/HCl, 150 mm NaCl, pH 7.5). To capture the biotinylated proteins, 20 μL of magnetic streptavidin beads (Promega) was added to each reaction and incubated for 16 h at 4°C. The magnetic beads were washed with 1 mL of TBS buffer using a magnetic stand (Promega) to remove nonspecifically bound proteins. Biotin-labeled proteins were harvested by boiling the magnetic beads in 30 μL of Laemmli SDS loading buffer, and subjected to SDS-PAGE (12%) and western blotting. Biotin-labeled proteins were detected by probing the membrane with streptavidin-horseradish peroxidase polymers (Sigma), followed by color development using the substrate 3,3′-diaminobenzidine (Sigma). For N. benthamiana intercellular fluids expressing PIP1-His, similar methods were used except that, after incubation with DCG-04, half the sample was used to capture biotinylated proteins by adding 20 μL of magnetic streptavidin beads as described above, while the other half was used to capture His-tagged PIP1 by adding 20 μL of Ni-NTA magnetic beads (Promega). Time-course labeling of PIP1 with DCG-04 in the presence or absence of inhibitors was conducted using N. benthamiana intercellular fluids expressing PIP1-His essentially as described above. Following preincubation with E-64 (1,120 nm) or EPIC2B (2,240 nm) for 1 h at room temperature, DCG-04 (220 nm) was added to the reaction for 10, 20, 30, 60, and 120 min. The biotin-labeling reactions were stopped by adding 1 mL of ice-cold acetone and precipitating the proteins immediately. Precipitated proteins were pelleted by centrifugation at 13,000g for 30 min at 4°C, washed with ice-cold 70% (w/w) acetone, air-dried, and resuspended in 500 μL of TBS buffer. PIP1-His was captured using Ni-NTA magnetic beads as described above. Biotin-labeled proteins were detected by probing the membrane with streptavidin-horseradish peroxidase.
Mapping of Pip1 and Rcr3
The fragment of the Pip1 gene used for probe labeling corresponded to the entire coding region. The gene-specific fragment of the Rcr3 gene used for probe labeling was derived from a 173-bp fragment spanning one intron. Primers used to amplify the Rcr3 gene-specific fragment were 5′-CGTCTAACTTGGACTGGAGAGAGT-3′ and 5′-CTAGTGATCCAACTGCACAAAACG-3′. Labeling and hybridization were done as described previously (van der Knaap and Tanksley, 2001). Pip1 and Rcr3 were mapped in a population of 100 F2 plants derived from a cross between tomato cv Sun1642 and Solanum pimpinellifolium LA1589 (Van der Knaap and Tanksley, 2001), and no plants were obtained that were recombinant between the two genes. The position of the genes relative to the framework map markers was determined using the software package MAPMAKER Version 2 (Lander et al., 1987).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple sequence alignment of PIP1 and RCR3pimp proteins.
Supplementary Material
Acknowledgments
We thank Zhenyu Liu and Diane Kinney for technical assistance, Jonathan D.G. Jones for providing us with the Rcr3 tomato material, John Lindbo for the P19 construct, Michiel Leeuwenburgh and Matthew Bogyo for access to DCG-04, and Mike Kinter for the mass spectrometry.
This work was supported by the U.S. Department of Agriculture National Research Initiative (project no. OHO00963–SS) and the National Science Foundation (Plant Genome grant no. DBI–0211659). Salaries and research support also were provided by state and federal funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sophien Kamoun (kamoun.1@osu.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Abramovitch RB, Martin GB (2004) Strategies used by bacterial pathogens to suppress plant defenses. Curr Opin Plant Biol 7 356–364 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 17 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NL, Esquer-Blasco R, Hofmann JP, Anderson NG (1991) A two-dimensional gel database of rat liver proteins useful in gene regulation and drug effects studies. Electrophoresis 12 907–930 [DOI] [PubMed] [Google Scholar]
- Apweiler R, Attwood TK, Bairoch A, Bateman A, Birney E, Biswas M, Bucher P, Cerutti L, Corpet F, Croning MD, et al (2001) The InterPro database, an integrated documentation resource for protein families, domains and functional sites. Nucleic Acids Res 29 37–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, Avrova AO, Rehmany AP, Bohme U, Brooks K, Cherevach I, et al (2005) An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci USA 102 7766–7771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avrova AO, Stewart HE, De Jong WD, Heilbronn J, Lyon GD, Birch PR (1999) A cysteine protease gene is expressed early in resistant potato interactions with Phytophthora infestans. Mol Plant Microbe Interact 12 1114–1119 [DOI] [PubMed] [Google Scholar]
- Avrova AO, Taleb N, Rokka V, Heilbronn J, Campbell E, Hein I, Gilroy EM, Cardle L, Bradshaw JE, Stewart HE, et al (2004) Potato oxysterol binding protein and cathepsin B are rapidly up-regulated in independent defence pathways that distinguish R gene-mediated and field resistances to Phytophthora infestans. Mol Plant Pathol 5 45–46 [DOI] [PubMed] [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer EL (2002) The Pfam protein families database. Nucleic Acids Res 30 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beers EP, Jones AM, Dickerman AW (2004) The S8 serine, C1A cysteine and A1 aspartic protease families in Arabidopsis. Phytochemistry 65 43–58 [DOI] [PubMed] [Google Scholar]
- Birch PRJ, Whisson S (2001) Phytophthora infestans enters the genomics era. Mol Plant Pathol 2 257–263 [DOI] [PubMed] [Google Scholar]
- Bos JIB, Armstrong M, Whisson SC, Torto T, Ochwo M, Birch PRJ, Kamoun S (2003) Intraspecific comparative genomics to identify avirulence genes from Phytophthora. New Phytol 159 63–72 [DOI] [PubMed] [Google Scholar]
- Caten CE, Jinks JL (1968) Spontaneous variability of single isolates of Phytophthora infestans. I. Cultural variation. Can J Bot 46 329–347 [Google Scholar]
- Chichkova NV, Kim SH, Titova ES, Kalkum M, Morozov VS, Rubtsov YP, Kalinina NO, Taliansky ME, Vartapetian AB (2004) A plant caspase-like protease activated during the hypersensitive response. Plant Cell 16 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva I, Poirier GG, Heath MC (1998) Activation of cysteine proteases in cowpea plants during the hypersensitive response—a form of programmed cell death. Exp Cell Res 245 389–399 [DOI] [PubMed] [Google Scholar]
- de Wit PJGM, Spikman G (1982) Evidence for the occurrence of race and cultivar-specific elicitors of necrosis in intercellular fluids of compatible interactions of Cladosporium fulvum and tomato. Physiol Plant Pathol 21 1–11 [Google Scholar]
- Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M (2000) Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem Biol 7 569–581 [DOI] [PubMed] [Google Scholar]
- Guevara MG, Oliva CR, Huarte M, Daleo GR (2002) An aspartic protease with antimicrobial activity is induced after infection and wounding in intercellular fluids of potato tubers. Eur J Plant Pathol 108 131–137 [Google Scholar]
- Hatsugai N, Kuroyanagi M, Yamada K, Meshi T, Tsuda S, Kondo M, Nishimura M, Hara-Nishimura I (2004) A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305 855–858 [DOI] [PubMed] [Google Scholar]
- Henikoff JG, Greene EA, Pietrokovski S, Henikoff S (2000) Increased coverage of protein families with the blocks database servers. Nucleic Acids Res 28 228–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huitema E, Bos JI, Tian M, Win J, Waugh ME, Kamoun S (2004) Linking sequence to phenotype in Phytophthora-plant interactions. Trends Microbiol 12 193–200 [DOI] [PubMed] [Google Scholar]
- Kamoun S (2003) Molecular genetics of pathogenic oomycetes. Eukaryot Cell 2 191–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun S (2006) A catalogue of the effector secretome of plant pathogenic oomycetes. Annu Rev Phytopathol 44 41–60 [DOI] [PubMed] [Google Scholar]
- Kamoun S, Hraber P, Sobral B, Nuss D, Govers F (1999) Initial assessment of gene diversity for the oomycete pathogen Phytophthora infestans based on expressed sequences. Fungal Genet Biol 28 94–106 [DOI] [PubMed] [Google Scholar]
- Kamoun S, Smart CD (2005) Late blight of potato and tomato in the genomics era. Plant Dis 89 692–699 [DOI] [PubMed] [Google Scholar]
- Kamoun S, van West P, de Jong AJ, de Groot K, Vleeshouwers V, Govers F (1997) A gene encoding a protein elicitor of Phytophthora infestans is down-regulated during infection of potato. Mol Plant Microbe Interact 10 13–20 [DOI] [PubMed] [Google Scholar]
- Kamoun S, Young M, Glascock C, Tyler BM (1993) Extracellular protein elicitors from Phytophthora: host-specificity and induction of resistance to fungal and bacterial phytopathogens. Mol Plant Microbe Interact 6 15–25 [Google Scholar]
- Karsch-Mizrachi I, Ouellette BF (2001) The GenBank sequence database. Methods Biochem Anal 43 45–63 [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN (2001) Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J 26 509–522 [DOI] [PubMed] [Google Scholar]
- Krebs SL, Grumet R (1993) Characterization of celery hydrolytic enzymes produced in response to infection by Fusarium oxysporum. Physiol Mol Plant Pathol 43 193–208 [Google Scholar]
- Kruger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JD (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296 744–747 [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1 174–181 [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res 30 242–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Dammann C, Bhattacharyya MK (2001) The matrix metalloproteinase gene GmMMP2 is activated in response to pathogenic infections in soybean. Plant Physiol 127 1788–1797 [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Bos JI, Armstrong M, Whisson SC, da Cunha L, Torto-Alalibo T, Win J, Avrova AO, Wright F, Birch PR, et al (2005) Patterns of diversifying selection in the phytotoxin-like scr74 gene family of Phytophthora infestans. Mol Biol Evol 22 659–672 [DOI] [PubMed] [Google Scholar]
- Merril CR, Goldman D, Sedman SA, Ebert MH (1981) Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science 211 1437–1438 [DOI] [PubMed] [Google Scholar]
- Mosolov VV, Grigor'eva LI, Valueva TA (2001) Involvement of proteolytic enzymes and their inhibitors in plant protection. Appl Biochem Microbiol 37 115–123 [PubMed] [Google Scholar]
- Nagata K, Kudo N, Abe K, Arai S, Tanokura M (2000) Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica. Biochemistry 39 14753–14760 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int J Neural Syst 8 581–599 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Krogh A (1998) Prediction of signal peptides and signal anchors by a hidden Markov model. In Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology (ISMB 6). AAAI Press, Menlo Park, CA, pp 122–130 [PubMed]
- O'Connell RJ, Ride JP (1990) Chemical detection and ultrastructural localization of chitin in cell walls of Collectotrichum lindemuthianum. Physiol Mol Plant Pathol 37 39–53 [Google Scholar]
- Punja ZK (2004) Fungal Disease Resistance in Plants. Food Products Press, Binghamton, NY
- Randall TA, Dwyer RA, Huitema E, Beyer K, Cvitanich C, Kelkar H, Fong AM, Gates K, Roberts S, Yatzkan E, et al (2005) Large-scale gene discovery in the oomycete Phytophthora infestans reveals likely components of phytopathogenicity shared with true fungi. Mol Plant Microbe Interact 18 229–243 [DOI] [PubMed] [Google Scholar]
- Ristaino JB (2002) Tracking historic migrations of the Irish potato famine pathogen, Phytophthora infestans. Microbes Infect 4 1369–1377 [DOI] [PubMed] [Google Scholar]
- Rojo E, Martin R, Carter C, Zouhar J, Pan S, Plotnikova J, Jin H, Paneque M, Sanchez-Serrano JJ, Baker B, et al (2004) VPEgamma exhibits a caspase-like activity that contributes to defense against pathogens. Curr Biol 14 1897–1906 [DOI] [PubMed] [Google Scholar]
- Rooney HC, Van't Klooster JW, van der Hoorn RA, Joosten MH, Jones JD, de Wit PJ (2005) Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 308 1783–1786 [DOI] [PubMed] [Google Scholar]
- Rose JK, Ham KS, Darvill AG, Albersheim P (2002) Molecular cloning and characterization of glucanase inhibitor proteins: coevolution of a counterdefense mechanism by plant pathogens. Plant Cell 14 1329–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Siegrist J, Kauss H (1990) Chitin deacetylase in cucumber leaves infected by Colletotrichum lagenarium. Physiol Mol Plant Pathol 36 267–275 [Google Scholar]
- Shattock RC (2002) Phytophthora infestans: populations, pathogenicity and phenylamides. Pest Manag Sci 58 944–950 [DOI] [PubMed] [Google Scholar]
- Smart CD, Fry WE (2001) Invasions by the late blight pathogen: renewed sex and enhanced fitness. Biol Invasions 3 235–243 [Google Scholar]
- Solomon M, Belenghi B, Delledonne M, Menachem E, Levine A (1999) The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11 431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Bishop JG (2000) Plant-pathogen arms races at the molecular level. Curr Opin Plant Biol 3 299–304 [DOI] [PubMed] [Google Scholar]
- Suty L, Lequeu J, Lancon A, Etienne P, Petitot AS, Blein JP (2003) Preferential induction of 20S proteasome subunits during elicitation of plant defense reactions: towards the characterization of “plant defense proteasomes”. Int J Biochem Cell Biol 35 637–650 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ (1997) A genomic perspective on protein families. Science 278 631–637 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Benedetti B, Kamoun S (2005) A second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol 138 1785–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian M, Huitema E, Da Cunha L, Torto-Alalibo T, Kamoun S (2004) A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem 279 26370–26377 [DOI] [PubMed] [Google Scholar]
- Tor M, Yemm A, Holub E (2003) The role of proteolysis in R gene mediated defence in plants. Mol Plant Pathol 4 287–296 [DOI] [PubMed] [Google Scholar]
- Tornero P, Conejero V, Vera P (1997) Identification of a new pathogen-induced member of the subtilisin-like processing protease family from plants. J Biol Chem 272 14412–14419 [DOI] [PubMed] [Google Scholar]
- Torto TA, Li S, Styer A, Huitema E, Testa A, Gow NA, van West P, Kamoun S (2003) EST mining and functional expression assays identify extracellular effector proteins from the plant pathogen Phytophthora. Genome Res 13 1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto TA, Rauser L, Kamoun S (2002) The pipg1 gene of the oomycete Phytophthora infestans encodes a fungal-like endopolygalacturonase. Curr Genet 40 385–390 [DOI] [PubMed] [Google Scholar]
- Turk V, Bode W (1991) The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett 285 213–219 [DOI] [PubMed] [Google Scholar]
- van den Burg HA, Spronk CA, Boeren S, Kennedy MA, Vissers JP, Vuister GW, de Wit PJ, Vervoort J (2004) Binding of the AVR4 elicitor of Cladosporium fulvum to chitotriose units is facilitated by positive allosteric protein-protein interactions: The chitin-binding site of AVR4 represents a novel binding site on the folding scaffold shared between the invertebrate and the plant chitin-binding domain. J Biol Chem 279 16786–16796 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA, Jones JD (2004) The plant proteolytic machinery and its role in defence. Curr Opin Plant Biol 7 400–407 [DOI] [PubMed] [Google Scholar]
- van der Hoorn RA, Leeuwenburgh MA, Bogyo M, Joosten MH, Peck SC (2004) Activity profiling of papain-like cysteine proteases in plants. Plant Physiol 135 1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Knaap E, Tanksley SD (2001) Identification and characterization of a novel locus controlling early fruit development in tomato. Theor Appl Genet 103 353–358 [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 949–956 [DOI] [PubMed] [Google Scholar]
- Xia Y, Suzuki H, Borevitz J, Blount J, Guo Z, Patel K, Dixon RA, Lamb C (2004) An extracellular aspartic protease functions in Arabidopsis disease resistance signaling. EMBO J 23 980–988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Han P, Lutziger I, Wang K, Oliver DJ (1999) A mini binary vector series for plant transformation. Plant Mol Biol 40 711–717 [DOI] [PubMed] [Google Scholar]
- Xiao F, Tang X, Zhou JM (2001) Expression of 35S∷Pto globally activates defense-related genes in tomato plants. Plant Physiol 126 1637–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LR, Qu S, Bordeos A, Yang C, Baraoidan M, Yan H, Xie Q, Nahm BH, Leung H, Wang GL (2004) Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16 2795–2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Thilmony R, Bender CL, Schaller A, He SY, Howe GA (2003) Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J 36 485–499 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.