Abstract
The extent to which leaf senescence is induced by nitrogen deficiency or by sugar accumulation varies between natural accessions of Arabidopsis (Arabidopsis thaliana). Analysis of senescence in plants of the Bay-0 × Shahdara recombinant inbred line (RIL) population revealed a large variation in developmental senescence of the whole leaf rosette, which was in agreement with the extent to which glucose (Glc) induced senescence in the different lines. To determine the regulatory basis of genetic differences in the Glc response, we investigated changes in gene expression using Complete Arabidopsis Transcriptome MicroArray (CATMA) analysis. Genes whose regulation did not depend on the genetic background, as well as genes whose regulation was specific to individual RILs, were identified. In RIL 310, a line that does not show the typical senescence response to Glc, stress response genes, especially those responding to cold stress, were induced by Glc. We therefore tested whether cold acclimation delays senescence by reducing sugar sensitivity. In cold-acclimated plants, leaf senescence was severely delayed and Glc did not induce the typical senescence response. Together, our results suggest that cold acclimation extends rosette longevity by affecting metabolic regulation of senescence, thereby allowing vernalization-dependent plants to survive the winter period. The role of functional chloroplasts and of nitrogen and phosphate availability in this regulation is discussed.
The timing of leaf senescence is an important life history trait. Early onset of senescence could severely impair photosynthetic carbon gain, whereas late senescence could inhibit senescence-dependent nutrient recycling (Himelblau and Amasino, 2001), which is important for the growth of young leaves and for fruit and seed formation (Levey and Wingler, 2005; Wingler et al., 2005). When plants are aging, their shoot carbon-to-nitrogen ratio increases as a result of nitrogen dilution. Physiological studies have shown that peaks in sugar content in tobacco (Nicotiana tabacum) and Arabidopsis (Arabidopsis thaliana) leaves coincide with the onset of senescence (Masclaux et al., 2000; Diaz et al., 2005). In addition, an external supply of Glc results in leaf yellowing (Wingler et al., 2004), indicating an induction of senescence by sugar accumulation. Affymetrix GeneChip analysis of gene expression has confirmed that Glc induces changes in gene expression that are typical of developmental leaf senescence (Pourtau et al., 2006).
In Arabidopsis, considerable variation in the regulation of senescence can be found in accessions from different geographic origins (Levey and Wingler, 2005; Luquez et al., 2006). The genetic basis of this variation can be studied using recombinant inbred lines (RILs) for quantitative trait loci (QTL) analysis. Several QTL for leaf yellowing and senescence-dependent anthocyanin accumulation have been found using a Bay-0 × Shahdara (Sha) RIL population (Diaz et al., 2006). Additional QTL for leaf and rosette longevity were detected by Luquez et al. (2006) using a Landsberg erecta × Cape Verde Islands RIL population. Phenotypic variation in the RILs typically exceeds that of the parental lines, making it possible to obtain lines with extreme phenotypes. Similar to mutants, these RILs can be used to determine the impact of genetic variation on the regulation of processes, such as leaf senescence.
Using RILs from the Bay-0 × Sha population with different senescence phenotypes, we have shown that amino acid and sugar content can be used as markers for the timing and extent of senescence of the first six leaves during nitrogen starvation (Diaz et al., 2005). Variation was also found in the RIL response to Glc and in the Glc response of the parental lines (Wingler et al., 2006). However, senescence of the first six leaves during nitrogen starvation was inversely related to the whole-rosette senescence phenotype induced by Glc and not related to plant longevity and flowering time (Diaz et al., 2005), suggesting that senescence of the first leaves is regulated in a different manner from senescence of leaves formed later during development. Some RILs, including RIL 310, were identified that did not show the typical senescence induction by Glc, suggesting that the Bay-0 × Sha population is an ideal tool for determining the basis of metabolic regulation of senescence.
Natural variation has also been described for freezing tolerance of Arabidopsis accessions. QTL analysis using a Landsberg erecta × Cape Verde Islands population showed that allelic variation in a transcription factor gene, C-repeat binding factor (CBF) 2, which is involved in the regulation of cold acclimation, underlies a major QTL for freezing tolerance (Alonso-Blanco et al., 2005). This natural variation in cold acclimation could also affect the regulation of leaf senescence: During cold acclimation, sugars (including Glc, Fru, Suc, and raffinose) accumulate (Cook et al., 2004). This accumulation could theoretically induce senescence. On the other hand, cold-acclimated leaves do not show the typical repression of photosynthesis by sugars (Strand et al., 1997). In addition, increased flux through the Suc biosynthetic pathway reduces inhibition of photosynthesis in the cold (Strand et al., 2003). It was therefore conceivable that cold acclimation could interact with metabolic regulation of senescence by inhibiting the induction of senescence by sugar accumulation. This regulatory interaction could, for example, increase rosette longevity of biennial and winter annual plants that require vernalization for flowering.
To investigate the basis of natural variation in the metabolic regulation of senescence, we determined the effect of sugar supply on metabolite content and on gene expression in RILs with contrasting sugar response. Complete Arabidopsis Transcriptome MicroArray (CATMA) analysis indicated that cold-responsive genes were induced by Glc in the sugar-insensitive RIL 310. We also investigated the impact of cold acclimation on the regulation of senescence.
RESULTS
Senescence of the Leaf Rosette
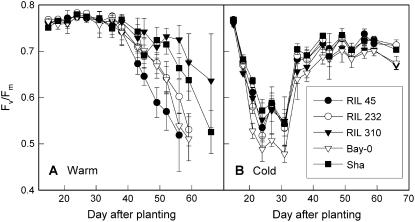
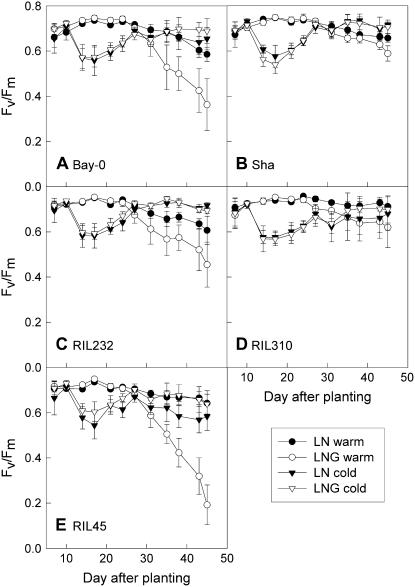
RILs with contrasting senescence phenotypes have previously been identified in the Bay-0 × Sha population based on the senescence of their first six leaves during nitrogen starvation (Diaz et al., 2005). Here, their whole-rosette senescence phenotypes, reflecting plant longevity, were characterized (Fig. 1A). When plants were grown in compost at normal (warm) growth temperatures, the rosettes of RIL 45 senesced very fast, resulting in early plant death. This line also flowered first. In contrast, whole-rosette senescence and flowering were delayed in RIL 310. Visual senescence phenotypes were confirmed by determination of photosynthetic efficiency (Fv/Fm; Fig. 2A). In RIL 45, Fv/Fm declined first, followed by RIL 232 and Bay-0, then Sha, and, finally, RIL 310. Similar results were obtained when plants were grown on agar medium containing 2% Glc to induce senescence (Fig. 1B). RIL 45 showed the strongest senescence, whereas RIL 310 stayed green in the presence of Glc. The lack of accelerated senescence in the presence of Glc in RIL 310 is unusual for Arabidopsis, but we have found a similar phenotype in nine additional RILs of the Bay-0 × Sha population. At a later stage in development, RIL 310 did senesce in the presence of Glc, but not more than in the absence of Glc. In agreement with the phenotype described by Diaz et al. (2005), RIL 310 often showed early senescence of the first leaves in the absence of Glc, but, at a later stage, the plants recovered and formed dark-green leaves that senesced late.
Figure 1.
Senescence phenotype in response to cold or Glc treatment. A, RIL 45, RIL 232, and RIL 310 with contrasting senescence phenotypes and their parental accessions (Bay-0 and Sha) were grown for 44 d in compost at 18°C (night)/22°C (day) cycles (warm) or at 5°C (cold). B, Plants were grown for 25 d on low-nitrogen agar medium in the absence (LN) or with addition of 2% Glc (LNG) under the warm conditions.
Figure 2.
Senescence-dependent decline in maximal photosynthetic efficiency (Fv/Fm) in RIL 45, RIL 232, and RIL 310 with contrasting senescence phenotypes and their parental accessions (Bay-0 and Sha). Fv/Fm was determined by fluorescence imaging in plants grown at 18°C (night)/22°C (day) cycles throughout the experiment (A; warm) and in plants that were transferred to 5°C (B; cold) on day 16. Data are means and sd of at least three plants.
Typical Arabidopsis Genes That Are Regulated by Glc during Senescence
The difference in senescence response to Glc between the RILs indicates genetic variation in the regulation of senescence. To identify genes that are commonly regulated, independent of the genetic background, we used CATMA analysis to determine changes in gene expression in response to Glc. CATMA combines a large dynamic range with good sensitivity and high specificity (Hilson et al., 2004; Allemeersch et al., 2005). Two biological replicates per treatment were analyzed. In addition, dye swaps were included for technical replication (see “Materials and Methods” for statistical analysis). All genes showing a statistically significant response to Glc were further analyzed; 119 genes were identified that showed statistically significant regulation by Glc in all three RILs (Supplemental Table S1). Apart from one gene (At1g73260), which was repressed in RIL 45, but induced in RIL 232 and RIL 310, all of these genes were regulated in the same direction in the three RILs. Comparison with a previous Affymetrix GeneChip experiment analyzing the effect of Glc in the accession Wassilewskija (Ws)-2 (Pourtau et al., 2006) confirms that there is common regulation of these genes in Arabidopsis independent of genetic background. Only two genes (At3g15460 and At3g56950) were slightly induced (2.2- and 2.5-fold) in Ws-2, but repressed in the other lines.
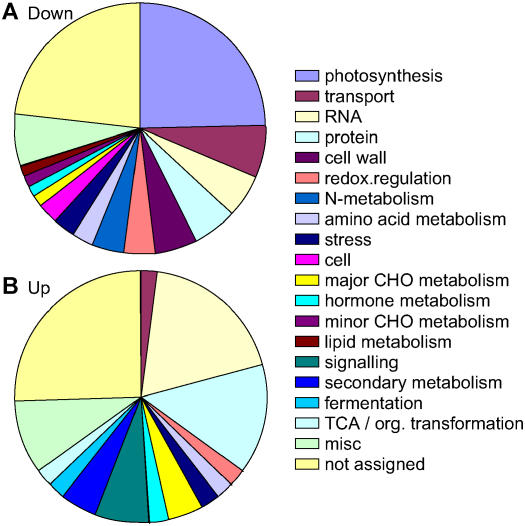
The set of coregulated genes can be used to determine gene functions that are up- or down-regulated during Glc-induced senescence (Fig. 3). About one-fourth of down-regulated genes were photosynthetic genes, mainly those involved in the light reactions, but also genes for Calvin cycle enzymes. In particular, genes encoding PSI proteins were overrepresented in the group of down-regulated genes compared to all pathways. There were also more genes for transport and cell wall composition down- than up-regulated. Pathways overrepresented in the group of up-regulated genes include several transcription factor genes in the RNA bin and genes involved in signaling. Genes for protein synthesis, degradation, and modification and for secondary metabolism were also induced by Glc. Only three stress genes (two down-regulated and one up-regulated) were found in the list of genes showing common regulation in all RILs.
Figure 3.
Function of genes affected by Glc independent of the genetic background. Biological functions of genes that are down-regulated (A) or up-regulated (B) in all three RILs (RIL 45, RIL 232, and RIL 310) are shown according to the The Institute for Genomic Research ontologies (see Supplemental Table S1 for list of genes).
It was surprising that typical senescence-associated genes (SAGs), such as SAG12 and production of anthocyanin pigment 2 (PAP2), were also induced in RIL 310 despite the lack of the typical Glc response. Indeed, induction of SAG12 was only statistically significant in one of the biological replicates. In addition, the regulation of typical SAGs in RIL 310 varied between different experiments. Whereas induction of SAG12 and PAP2 by Glc was confirmed for RIL 45, RIL 232, and the parental lines Bay-0 and Sha in an independent experiment, SAG12 was expressed in the absence of Glc in RIL 310 (Supplemental Fig. S1). This is in agreement with chlorosis of the first leaves often observed in the absence of Glc in RIL 310 (Fig. 1B).
Genes Whose Regulation Depends on the Genetic Background
To investigate what caused the differences in the senescence response to Glc, genes whose regulation was specific to the RILs were identified (Supplemental Table S1). For the line with an intermediate senescence phenotype, RIL 232, the number of specific genes was small (29 in total; nine genes repressed and 20 genes induced by Glc treatment). More genes were specifically regulated in the RILs with extreme senescence phenotypes; 183 genes were specifically regulated in RIL 45 and most of these genes were repressed (156 repressed, 27 induced). For RIL 310, 233 specifically regulated genes were found (98 repressed and 135 induced).
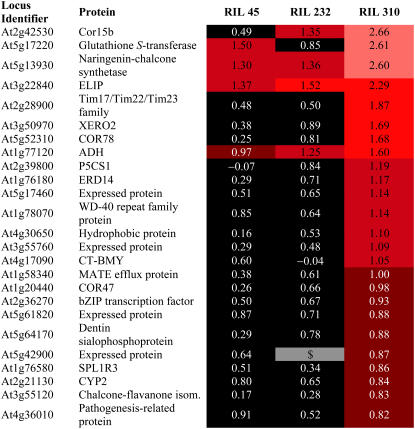
A large number of stress genes were up-regulated by Glc in RIL 310. To investigate further how RIL 310 is affected by stress, we analyzed the stress response of the genes induced by Glc in RIL 310 using Genevestigator (Zimmermann et al., 2004). Twenty-five genes that were induced by Glc in RIL 310 are typically induced by cold stress (Table I; Supplemental Table S2). The list of cold-inducible genes includes several genes of the CBF-dependent cold-response pathway. For example, At1g20440 (COR47), At2g28900, At2g42530 (COR15b), At3g50970 (XERO2), and At5g52310 (COR78) are also induced without cold treatment in plants that overexpress CBF transcription factors (Fowler and Thomashow, 2002). To ensure that Cor15b was consistently induced by Glc in RIL 310, we determined its expression by reverse transcription (RT)-PCR in plant material from an independent experiment. This experiment confirmed that this response is reproducible (Supplemental Fig. S1).
Table I.
Expression of cold-responsive genes that were induced by Glc in RIL 310
Cold-responsive genes were identified using Genevestigator (https://www.genevestigator.ethz.ch; see Supplemental Table S2 for expression of the genes under stress conditions). Data are log2 values of the Glc response. Bonferroni P values of data in black cells are >0.05. $, Missing data.
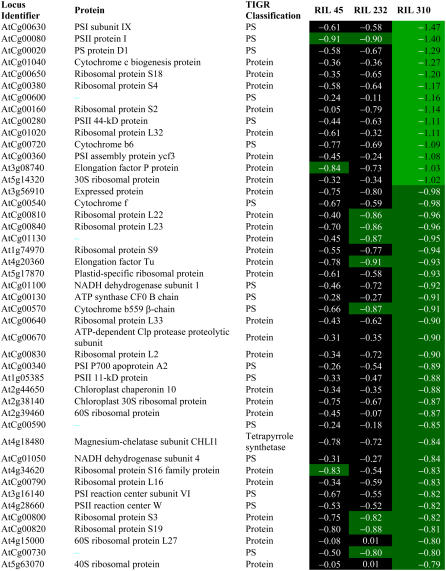
Despite its delayed senescence phenotype and high photosynthetic efficiency on Glc, an unexpectedly large number of photosynthetic genes were down-regulated by Glc in RIL 310 (Table II). Most of these genes are chloroplast-encoded genes involved in the light reactions of photosynthesis. In addition to photosynthetic genes, genes for protein synthesis in the chloroplasts, both chloroplast and nuclear encoded, were repressed by Glc.
Table II.
Expression of genes involved in processes in the chloroplasts (chloroplast-encoded genes and nuclear-encoded genes for photosynthetic proteins or chloroplast protein synthesis) that were repressed by Glc in RIL 310
Data are log2 values of the Glc response; mean values were calculated for genes represented more than once on CATMA. Bonferroni P values of data in black cells are >0.05. TIGR, The Institute for Genomic Research.
A separate group of photosynthetic genes (21 genes) was down-regulated by Glc in RIL 45 (Supplemental Table S1). In the case of RIL 45, this effect is in agreement with the strong senescence response of RIL 45 to Glc. Another interesting feature of gene regulation in RIL 45 was that three usually sugar-inducible genes involved in the response to biotic stress were repressed by Glc (At1g75040 = PR5; At2g43570, a chitinase gene; At1g73260, a Kunitz family protein).
Sugar and Amino Acid Content
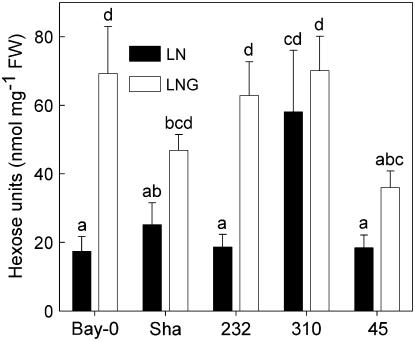
Because Glc only induces senescence in combination with low, but not high, nitrogen supply (Wingler et al., 2004), it was unclear whether differences in the regulation of senescence in the RILs were mainly due to altered sugar or nitrogen availability within the plants. To test this, we determined sugar and amino acid content in the RILs and parental lines. Glc, Fru, and Suc were affected in a similar way and are therefore presented as total sugar content (Fig. 4). Growth on Glc resulted in higher leaf sugar content in most lines, but not in RIL 310. In the absence of Glc, RIL 310 contained significantly more sugar than the other lines. On Glc-containing medium, RIL 310 contained higher amounts of sugar than the early senescing RIL 45 and similar amounts as Bay-0 and RIL 232, demonstrating that delayed senescence on Glc-containing medium in RIL 310 was not due to lower sugar content.
Figure 4.
Effect of Glc on sugar content (sum of Glc, Fru, and Suc) during leaf senescence in RIL 45, RIL 232, and RIL 310 with contrasting senescence phenotypes and their parental accessions (Bay-0 and Sha). Sugar content is the means and sd of two to three samples. LN (black bars), Plants grown on low-nitrogen medium in the absence of Glc; LNG (white bars), plants grown on low-nitrogen medium with addition of 2% Glc. Different letters indicate statistically different content (ANOVA; P < 0.05).
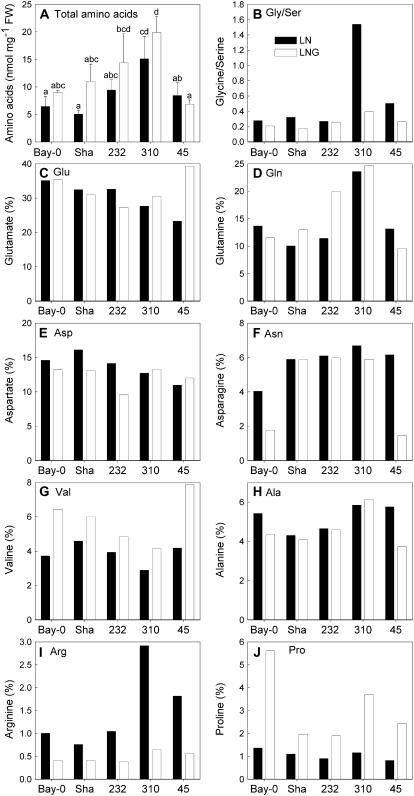
Growth in the presence of Glc resulted in a slight, but insignificant, increase in amino acid content in Bay-0, Sha, RIL 310, and RIL 232 (Fig. 5). The relative content of Glu and Asp and of minor amino acids (data not shown) largely followed the changes in total amino acids and was thus not affected to a great extent by Glc treatment. There were, however, clear effects on the proportion of Arg and Pro. Whereas the relative content of Arg decreased in all lines with sugar feeding, Pro accumulated. There was also an increase in the proportion of Val in all lines, whereas Asn was reduced after growth on Glc in the more senescent lines Bay-0 and RIL 45.
Figure 5.
Effect of Glc on amino acid content during leaf senescence in RIL 45, RIL 232, and RIL 310 with contrasting senescence phenotypes and their parental accessions (Bay-0 and Sha). A, Total amino acid content are means and sd of three to four samples. Different letters indicate statistically different content (ANOVA; P < 0.05). B to J, For individual amino acids, their proportion relative to the total amino acid content is shown. LN (black bars), Plants grown on low-nitrogen medium in the absence of Glc; LNG (white bars), plants grown on low-nitrogen medium with addition of 2% Glc.
In RIL 310, the total amino acid content was higher than in the other lines. This effect was statistically significant in comparison with all lines but RIL 232. The most striking difference between RIL 310 and the other lines was an increased Gly-to-Ser ratio on medium without Glc, which could indicate increased rates of photorespiration. High ammonium content in RIL 310 (data not shown) supports this assumption. In the absence of Glc, Arg content was also increased in RIL 310. In addition, RIL 310 contained more Gln than the other lines. Overall, the higher total amino acid content and increase in Gln suggest that internal nitrogen availability was high in RIL 310.
Comparison of the Effects of Glc Feeding and Nitrogen Starvation
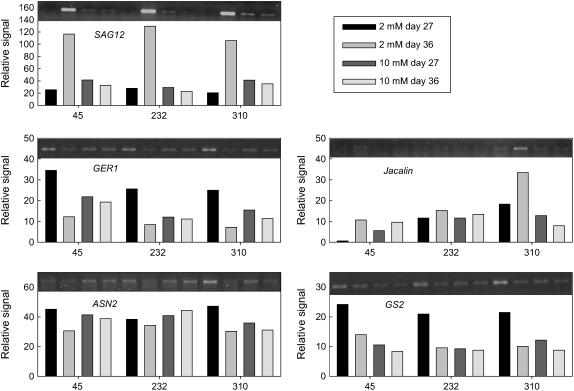
In addition to sugar accumulation, nitrogen starvation can induce leaf senescence and regulation of senescence mainly depends on the relative availability of nitrogen and carbon (Pourtau et al., 2004; Diaz et al., 2005). The shift from Arg to Pro in the presence of Glc (Fig. 5) could indicate nitrogen deprivation (Verma and Zhang, 1999). To test whether nitrogen deficiency can elicit similar effects as Glc feeding, we determined gene expression in plants grown with low (2 mm nitrate) or high (10 mm nitrate) nitrogen supply (Fig. 6). Induction of the senescence marker SAG12 confirmed that senescence was induced by low nitrogen supply. The germin gene GER1, the Asn synthetase gene ASN2, and the Gln synthetase gene GS2, which were down-regulated by Glc in the CATMA experiment (Supplemental Table S1), were repressed as plants senesced at low nitrogen supply. At high nitrogen supply, expression of GS2 was low and did not change as plants aged. In addition to these common changes in gene expression, line-specific effects were also apparent. A jacalin gene that was specifically induced by Glc in RIL 310 (Supplemental Table S1; Supplemental Fig. S1) and not in the other lines was also induced during senescence at low nitrogen supply in RIL 310. These results indicate that low nitrogen supply elicits comparable changes in gene expression as Glc feeding.
Figure 6.
Gene expression in RIL 45, RIL 232, and RIL 310 with contrasting senescence phenotypes after growth for 27 or 36 d at 2 or 10 mm nitrate. Gene expression was determined by RT-PCR and quantification of the PCR products. Genes were SAG12 (At5g45890), GER1 (At1g72610), a jacalin gene (At2g39330), ASN2 (At5g65010), and GS2 (At5g35630).
Effect of Cold Acclimation
Because leaves that have developed in the cold do not show the typical down-regulation of photosynthesis in response to sugar accumulation (Strand et al., 1997), it was possible that induction of the cold acclimation pathway in RIL 310 (Table I) had resulted in decreased sugar sensitivity and thus delayed senescence. To determine whether cold acclimation inhibits Glc-induced senescence, the RILs and parental lines were grown at 5°C in compost and on agar medium. In all lines, cold treatment delayed senescence in compost (Fig. 1A). Transfer to 5°C resulted in an initial decline in Fv/Fm in compost (Fig. 2B), as well as in agar-grown plants (Fig. 7), but, as plants acclimated, Fv/Fm recovered. Whereas Glc-induced senescence was apparent from the decline in Fv/Fm in plants grown at warm temperatures (especially RIL 45 and Bay-0), Glc did not induce senescence in any of the lines at 5°C over the same period of time (Fig. 7). Some effect of Glc on senescence became apparent at a very late stage (from day 60 onward; data not shown), but, at this stage, the agar medium had started to dry out, making it difficult to study physiological effects.
Figure 7.
Effect of cold acclimation on Glc-induced senescence in RIL 45, RIL 232, and RIL 310 with contrasting senescence phenotypes and their parental accessions (Bay-0 and Sha). Maximal photosynthetic efficiency (Fv/Fm) was determined by fluorescence imaging in plants grown on low-nitrogen medium without (LN; black symbols) or with (LNG; white symbols) addition of 2% Glc. Plants were either grown at 18°C (night)/22°C (day) cycles throughout the experiment (warm; circles) or were transferred to 5°C (cold; triangles) on day 11. Data are means and sd of at least three plants.
DISCUSSION
Whole-rosette senescence varied considerably between lines, but similar line-specific differences were found after growth in compost and on Glc-containing agar medium (Fig. 1). Fv/Fm data (Fig. 7) confirmed our previous observation that senescence is very strongly induced by Glc in RIL 45, but not in RIL 310, with RIL 232 showing an intermediate phenotype (Diaz et al., 2005). Surprisingly, the whole-rosette senescence phenotype is opposite to senescence of the first six leaves of plants grown at low nitrogen supply (Diaz et al., 2005). Whereas the first six leaves of RIL 310 senesce early in the absence of Glc, this line then continues to produce dark-green, late-senescing leaves and flowers late. In contrast, the first leaves of RIL 45 senesce late, but this line produces fewer leaves and flowers early, resulting in a reduced lifespan. The contrasting senescence characteristics of the first leaves and leaves formed later during development show that the regulation of senescence can vary depending on leaf position. Zentgraf et al. (2004) found that gene expression is not only influenced by leaf age, but also by plant age, indicating that leaves of different positions senesce in different ways. This may also help explain the conundrum of why an inverse relationship was found between QTL for senescence of the first six leaves and carbohydrate content (Calenge et al., 2006), despite the ability of sugars to induce senescence.
Effect of the Genetic Background on the Regulation of Gene Expression
Leaf senescence is a complex process that can be triggered by a range of environmental and age-related factors. It is therefore not surprising that, depending on the cause of leaf senescence, different pathways are induced. Buchanan-Wollaston et al. (2005), for example, identified differences in gene expression between developmental leaf senescence and dark/starvation-induced senescence. Gene expression during developmental and dark-induced senescence was also analyzed by van der Graaff et al. (2006), who found differences in the regulation of genes for transporters, suggesting that catabolite mobilization pathways vary between different forms of senescence. On the other hand, there seems to be good agreement between changes in gene expression during Glc-induced and developmental leaf senescence (Pourtau et al., 2006).
So far, gene expression profiling during senescence has focused on single accessions. Our results on the variation of senescence between the RILs and parental lines (Fig. 1) do, however, suggest that regulation of senescence can vary significantly, not only in response to different environmental conditions, but also depending on the genetic background. In an experiment comparing gene expression with and without salicylic acid treatment in several Arabidopsis accessions, Kliebenstein et al. (2006) found over 6,000 genes that were differentially expressed between at least one pair of different Arabidopsis accessions. Differentially expressed genes were enriched in those involved in biotic and abiotic responses, stress, and signal transduction, suggesting that evolutionary differences in response to the environment are particularly common. It was therefore not surprising that we found differences in the response to Glc in the three RILs with contrasting senescence phenotypes (Supplemental Fig. S1).
We also identified genes that were regulated in the same way in all lines. These can serve as a core set of genes that respond to sugars during senescence (Fig. 3). For RIL 45, RIL 232, and the parental accessions Bay-0 and Sha, the response of these genes was confirmed in an independent experiment (Supplemental Fig. S1). However, some variation in gene expression in the absence of Glc was found in RIL 310, which, when grown without sugar, often showed chlorosis of the first leaves. Especially for the up-regulated genes, there was also a good agreement of CATMA results shown here with the Affymetrix GeneChip data on sugar-induced senescence in the Ws-2 accession. Although the dynamic range of CATMA is larger than that of the Affymetrix ATH1 GeneChip (Allemeersch et al., 2005), we found that up-regulated genes were more strongly induced in the Affymetrix analysis of Ws-2 than on CATMA. For example, the senescence markers PAP2 (At1g66390) and SAG12 (At5g45890) were up-regulated over 300-fold and over 900-fold, respectively, on the Affymetrix GeneChip, but only 9- to 18-fold and 2- to 3.5-fold on CATMA. These differences in the extent of induction could be due to differences between the array platforms, the accessions used, or the slight differences in the extent of senescence at the time of harvest.
Variation in the Regulation of Leaf Senescence Is Reflected in Changes in Amino Acid Content
Senescence is not induced in Arabidopsis plants grown on Glc in the presence of high nitrogen supply (Wingler et al., 2004), suggesting that it is controlled by the carbon-to-nitrogen ratio and not by sugars alone (Paul and Pellny, 2003; Gibson, 2005; Masclaux-Daubresse et al., 2005). Because changes in gene expression that are caused by Glc can also be induced by growth at low nitrogen supply (Fig. 6), we expected Glc feeding to have an impact on the availability of nitrogen, as reflected in amino acid content (Fig. 5). A shift from Arg to Pro in the presence of Glc was found here and suggests that Glc feeding led to nitrogen deprivation. At low nitrogen supply or during stress, Δ1-pyrroline-5-carboxylate synthetase (P5CS), a rate-limiting enzyme in the synthesis of Pro from Glu, is induced, resulting in increased Pro formation and reduced Glu availability for Arg synthesis (Verma and Zhang, 1999). In addition to increased Pro synthesis, down-regulation of the first enzyme for Pro degradation, Pro dehydrogenase, by sugars could also result in Pro accumulation (Hellmann et al., 2000).
The late-senescing line RIL 310 had the highest amino acid content of all lines (Fig. 5) and also high protein content (data not shown). In particular, high content of Arg (on medium without Glc) and Gln (on both media) suggests that nitrogen storage capacity is high in RIL 310. This could explain why this line does not respond to Glc in the same way as the other lines, despite containing large amounts of Glc, Fru, and Suc (Fig. 4). Although, the relative Pro content (as a proportion of total amino acids) was higher in Bay-0 than in RIL 310 after growth on Glc, the absolute Pro content was highest in RIL 310 due to the overall increased amino acid content in this line. In addition to a high carbon-to-nitrogen ratio, stress conditions, such as cold stress or drought, induce Pro synthesis by induction of P5CS (Svensson et al., 2006). In our experiment, the P5CS1 gene (At2g39800) was induced in RIL 310 by Glc (Table I), suggesting a link between sugar and cold-response pathways.
The Gly-to-Ser ratio can serve as a predictive marker of senescence (Diaz et al., 2005). Similar to the high Gly-to-Ser ratio found in the first six leaves in RIL 310, the ratio was also increased on low nitrogen medium without Glc in the experiment shown here. In addition, RIL 310 had a high ammonium content in the absence of Glc (data not shown), indicating enhanced rates of photorespiration, possibly due to impaired photosynthetic function (Wingler et al., 2000). This would be in agreement with chlorosis and necrosis in RIL 310 in the absence of Glc (Fig. 1). In the presence of Glc, the Gly-to-Ser ratio decreased in agreement with vigorous growth on Glc and the late-senescence phenotype.
Interaction between Chloroplast Function and Expression of Cold-Responsive Genes
In the presence of Glc, RIL 310 had the highest chlorophyll content (Fig. 1) and functional chloroplasts, as indicated by the Fv/Fm values (Table III; Fig. 7). The down-regulation of genes for photosynthesis, plastid protein synthesis, and chloroplast-encoded genes in RIL 310 (Table II) is therefore difficult to interpret. However, synthesis of chloroplast-encoded proteins is not necessarily related to transcript abundance. In Chlamydomonas, a drop of 90% in chloroplast transcript did not affect synthesis of chloroplast proteins (Eberhard et al., 2002). It is therefore feasible that down-regulation of chloroplast gene expression in RIL 310 reflects regulatory interactions that do not necessarily have an impact on photosynthetic function.
Table III.
Growth stage (according to Boyes et al., 2001) and maximal photosynthetic efficiency (Fv/Fm) a day before the harvest for CATMA analysis
LN, Plants grown on low-nitrogen medium in the absence of Glc. LNG, Plants grown on low-nitrogen medium in the presence of Glc.
| Growth Stage | Fv/Fm | |
|---|---|---|
| RIL 45 LN | 6.00–6.50 | 0.720 ± 0.019 |
| RIL 45 LNG | 6.90 | 0.703 ± 0.028 |
| RIL 232 LN | 3.90 | 0.743 ± 0.017 |
| RIL 232 LNG | 5.10 | 0.726 ± 0.025 |
| RIL 310 LN | 3.90 | 0.728 ± 0.032 |
| RIL 310 LNG | 3.90 | 0.758 ± 0.015 |
In RIL 310, cold-response genes were induced by Glc (Table I; Supplemental Fig. S1). Complex interactions exist between chloroplast function and cold acclimation. Functional chloroplasts are required for cold acclimation and also for regulation of the majority of cold-responsive genes, probably because the induction of cold acclimation depends on PSII excitation pressure (Ensminger et al., 2006). Using barley (Hordeum vulgare) mutants in chloroplast development, Svensson et al. (2006) showed that only 11% of genes regulated by cold treatment in wild-type barley were also regulated to a similar extent in mutants in chloroplast development. Maintained chloroplast function could explain why cold-responsive genes were more strongly induced by Glc in RIL 310 than in the other lines, which showed stronger senescence-dependent decline in photosynthesis. In barley, up-regulation of genes under CBF control is, however, not chloroplast dependent (Svensson et al., 2006). Nine genes involved in redox regulation were also specifically regulated in RIL 310 (Supplemental Table S1). This could, for example, indicate induction of the cold response due to oxidative stress (Prasad et al., 1994; Svensson et al., 2006) or a redox signal. Furthermore, induction of some of the cold-responsive genes by Glc in all lines may suggest that cold-dependent sugar accumulation could be part of a feed-forward loop in the cold acclimation pathway. It is possible that this sugar effect is mediated by changes in amino acid content, such as Pro or Arg (Fig. 5).
Cold Acclimation Interacts with the Metabolic Regulation of Senescence
Our results show that senescence is severely delayed in plants grown at 5°C, both in compost and on Glc-containing agar medium (Figs. 1, 2, and 7). This is in agreement with the observation that leaves that have developed in the cold do not show repression of photosynthesis, despite sugar accumulation (Strand et al., 1997). In winter annuals, which germinate in autumn and require vernalization in winter for flowering in spring, it is particularly important that sugar-induced senescence is inhibited to allow the plants to resume growth in the spring. Although both parental accessions, Bay-0 and Sha, are early flowering and do not require vernalization, flowering in RIL 310 is vernalization dependent (C. Masclaux and A. Wingler, unpublished data) and Sha has a functional FRIGIDA allele (Loudet et al., 2002), demonstrating that the genetic basis for a winter-annual life cycle is present in the Bay-0 × Sha population.
Transfer of seedlings to 5°C resulted in a temporary decline in Fv/Fm in all lines, showing that, without acclimation, RIL 310 was not more cold tolerant than the other lines. In addition, the presence of Glc had no impact on the extent of the decline in Fv/Fm. This suggests that only part of the typical cold acclimation response was induced by Glc in RIL 310, resulting in decreased sugar sensitivity, but not in increased cold hardening. Because changes in amino acid content could be involved in this regulation, it would be interesting to determine the effect of cold acclimation on senescence in response to varied nitrogen availability.
In contrast to nitrogen deficiency, phosphate deficiency decreases sugar sensitivity (Nielsen et al., 1998). The interactions between cold acclimation and phosphate availability are complex. Although phosphate deficiency may be responsible for the short-term inhibition of photosynthesis in the cold, phosphate availability increases during cold acclimation, probably due to changes in phosphate compartmentalization and increased Suc synthesis (Strand et al., 1999). In addition, phosphate deficiency improves cold acclimation, possibly by stimulating Suc synthesis (Hurry et al., 2000). The role of phosphate deficiency in the regulation of senescence by sugars is still unclear, but the phenotype of RIL 310 resembles that of phosphate-deficient pho1 mutants: Similar to RIL 310, pho1-2 has dark-green leaves (Delhaize and Randall, 1995), with increased content of chlorophyll, total amino acids, Pro, and sugars (Hurry et al., 2000). In addition, cold acclimation is improved in pho1-2, leading to increased expression of Calvin cycle enzymes. Phosphate analysis in the RILs confirmed that, in comparison with other RILs, RIL 310 has low phosphate content of 11.7 nmol mg−1 dry weight (O. Loudet, personal communication; see http://dbsgap.versailles.inra.fr/vnat/Documentation/33/DOC.html), which is similar to that of the phosphate-deficient pho1-1 mutant (Poirier et al., 1991).
In conclusion, our results show that leaf senescence is controlled by interactions between sugar and nitrogen signaling with the cold acclimation pathway. Chloroplast signals, as well as phosphorus availability, are likely to affect this signaling interaction.
MATERIALS AND METHODS
Plant Material
RILs with contrasting senescence phenotypes (Diaz et al., 2005) were selected from the Arabidopsis (Arabidopsis thaliana L. Heynh.) Bay-0 × Sha population (Loudet et al., 2002; Nottingham Arabidopsis Stock Centre ID N57920). For growth in compost, seeds were suspended in 0.5% low-melting agarose and pipetted onto compost (Murphy's multipurpose compost; Murphy Garden Products). For cultivation on agar plates, seeds were sterilized in commercial bleach, washed, resuspended in 0.7% low-melting agarose, and pipetted onto agar (1% [w/v]) medium with low nitrogen supply (4.7 mm nitrate) as described by Pourtau et al. (2004). After 3 to 4 d of cold treatment for stratification, plants were transferred into the growth conditions. Agar plates were placed vertically. Plants were grown in growth chambers under long-day conditions (16 h/d) at a photon flux density of 100 μmol m−2 s−1. The temperature was 22°C during the day and 18°C at night. For cold acclimation, plants were transferred to 5°C after germination in the warm conditions (on day 16 for compost-grown plants and on day 11 for agar-grown plants).
For growth under low and high nitrogen conditions, seeds were stratified and sown as described by Diaz et al. (2005). Plants were grown in short days with a photoperiod of 8 h throughout the culture. Day and night temperatures were regulated at 21°C and 17°C, respectively. Light was provided by 20 mercury-vapor bulbs, ensuring a photosynthetic photon flux density of approximately 160 μmol m−2 s−1. Pots were watered three times per week by immersion of the base of the pots in a solution containing 2 or 10 mm of nitrate as described by Loudet et al. (2003).
Determination of Chlorophyll Fluorescence
Maximal photosynthetic efficiency (Fv/Fm) was determined nondestructively for the whole rosettes of Arabidopsis plants using a pulse-modulated imaging fluorometer (FluorCam 700MF; Photon Systems Instruments) as described by Wingler et al. (2004).
CATMA Analysis
Two independent biological replicates were cultivated for 25 d on agar medium. Leaf rosettes of approximately 12 plants for each replicate were harvested at midday by removing the roots and inflorescences with a razor blade and immediately frozen in liquid nitrogen. Total RNA was extracted according to Logemann et al. (1987). At the time of harvest, development of RIL 45 was most advanced, especially in the presence of Glc, where flowering was complete (Table III). For RIL 232, flower buds were already visible on the low nitrogen plus Glc, but not on the low nitrogen medium. In RIL 310, no buds were visible on either medium. Both in RIL 45 and RIL 232, Glc induced senescence, as shown by the decline in Fv/Fm, compared to the medium without sugar. RIL 45 was generally more senescent than RIL 232.
Microarray hybridizations were performed with CATMA (Crowe et al., 2003; Hilson et al., 2004), which contains 24,576 gene-specific tags (Thareau et al., 2003) corresponding to 22,089 genes. The gene-specific tag amplicons were purified on Multiscreen plates (Millipore) and resuspended in Tris-EDTA-dimethyl sulfoxide at 100 ng μL−1. Purified probes were transferred to 1,536-well plates with a Genesis workstation (TECAN) and spotted on UltraGAPS slides (Corning) using Microgrid II (Genomic Solution). cRNAs were produced from 2 μg of total RNA with the MessageAmp aRNA kit (Ambion). Five micrograms of cRNA were reverse transcribed in the presence of 200 units of SuperScriptII (Invitrogen), cy3-dUTP, and cy5-dUTP (NEN). Samples were combined, purified, and concentrated with YM30 Microcon columns (Millipore). Slides were prehybridized for 1 h and hybridized overnight at 42°C in 25% formamide. For each comparison, one technical replication with fluorochrome reversal was performed for each pool of RNA (i.e. dye swap) to remove dye biases. Arrays were scanned on a GenePix 4000 A scanner (Axon Instruments) at constant photomultiplier tube gain and images were analyzed by GenePix Pro 3.0 (Axon Instruments).
Statistical analysis was based on two dye swaps per comparison, one for each replicate. For each array, the raw data comprised the logarithm of median feature pixel intensity at wavelengths 635 nm (red) and 532 nm (green). No background was subtracted. In the following description, log-ratio refers to the differential expression between the different treatments. It is either log2 (red/green) or log2 (green/red) according to the experimental design. An array-by-array normalization was performed to remove systematic biases. First, we excluded spots that were considered to show badly formed features by the experimenter. Then we performed a global intensity-dependent normalization using the LOESS procedure (Yang et al., 2002) to correct the dye bias. Finally, on each block of the array, the log-ratio median was subtracted from each value of the log-ratio of the block to correct any print-tip effect on each block. To determine differentially expressed genes, we performed a paired t test on the log-ratios, assuming that the variance of the log-ratios is the same for all genes. Spots displaying extremes of variance (too small or too large) were excluded. The raw P values were adjusted by the Bonferroni method, which controls the family-wise error rate. We considered as being differentially expressed the genes with a Bonferroni P value < 5% as described in Lurin et al. (2004). Data were deposited in Array express (http://www.ebi.ac.uk/arrayexpress) according to MIAME standards (accession no. E-MEXP-387).
Pathways that are overrepresented in response to Glc compared to all pathways were determined using the Overview function of MapMan (Usadel et al., 2005). The meta-analyzer function of Genevestigator (Zimmermann et al., 2004) was used to determine the stress response of genes.
RT-PCR Analysis
For RT-PCR, plant material was harvested after growth on agar medium as for CATMA analysis, however, at day 30, due to the slightly later onset of senescence in this particular experiment. RNA was extracted by homogenizing plant material in TRIzol reagent (Invitrogen) using a FP220 ribolyzer (Q-Biogene). RNA was isolated according to the TRIzol protocol (Invitrogen). cDNA was synthesized as in Pourtau et al. (2006). PCR conditions were 5 min at 94°C, followed by cycles of 30 s at 94°C, 30 s at 55°C, and 45 s at 72°C, plus a final extension step of 5 min at 72°C. The following primer pairs were used: SAG12 (At5g45890; forward, 5′-AGGAGCAACACAAATAAAGAAAG-3′; reverse, 5′-ACGAATAGAATTGGAAATCAAAA-3′); PAP2 (At1g66390; forward, 5′-GATAAGTATGGAGAAGGCAAATG-3′; reverse, 5′-GAGGGGAAATAATGTTTTTCTTT-3′); Cor15b (At2g42530; forward, 5′-TGTTGGTACCGTCAGAGTTG-3′; reverse, 5′-AAAGCTTTCTTAGCTTCCTCAGT-3′); GER1 (Atlg72610; forward, 5′-CAGCTAGACTTGACTTAGCTCCT-3′; reverse, 5′-GAGTTCAGTGGGAAGACTGTTAG-3′); ASN2 (At5g65010; forward, 5′-ACCACGAGTTTCAGTTTACAGTT-3′; reverse, 5′-GCAGAAGTTGATTTGTTAGCTCT-3′); GS2 (At5g35630; forward, 5′-GACATTTCAGATGCTCATTACAA-3′; reverse, 5′-AATACTAGCTGTCTCGTGCTTTC-3′); a jacalin gene (At2g39330; forward, 5′-CTACTCCGACTCCTATAATTCCA-3′; reverse, 5′-AGATAATCATTGGTGTCTCGACT-3′); 18S rRNA (At3g41768 and At2g01010; forward, 5′-ATACGTGCAACAAACCC-3′; reverse, 5′-CTACCTCCCCGTGTCA-3′); and ACT2 (At3g18780; forward, 5′-GACGTGACCTTACTGATTACCTC-3′; reverse, 5′-TATCCACATCACACTTCATGATT-3′). The RT-PCR results were confirmed at least once with independently synthesized cDNA.
For plants grown on unfertilized compost and watered with 2 or 10 mm nitrate, whole rosettes were harvested at days 27 and 36 after sowing. The cycle numbers in the PCR reaction were 28 cycles for GER1 (At1g72610), 32 cycles for the jacalin gene (At2g39330), 30 cycles for ASN2 (At5g65010), 28 cycles for GS2 (At5g35630), and 30 cycles for SAG12 (At5g45890). PCR products were quantified after gel imaging using a Bio-Rad GelDoc 1000 camera (Bio-Rad) and the MultiGauge FujiFilm image analyzer (Fujifilm) and corrected using the signal obtained for 18S rRNA (At3g41768 and At2g01010; constitutive control).
Determination of Sugar and Amino Acid Content
Leaf rosettes for sugar and amino acid analysis were harvested after 30 d at midday. Roots and inflorescences were cut off and the rosettes frozen in liquid nitrogen. Sugars were extracted in hot (80°C) 80% ethanol and determined using enzymatic essays according to Stitt et al. (1989).
Amino acid content in leaf rosettes was determined after extraction in 2% 5-sulfosalicylic acid (50 mg mL−1fresh weight). Total amino acid content was assayed by the Rosen colorimetric method using Gln as a reference (Rosen, 1957) in each sample. Individual amino acid composition was determined in pooled extracts of three samples, adjusted according to their total amino acid content, by ion-exchange chromatography using an AminoTac JLC-500/V amino acid analyzer according to the manufacturer's instructions (JEOL [Europe]).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. RT-PCR results of genes responding to Glc-induced senescence.
Supplemental Table S1. CATMA results of genes that showed a statistically significant response to Glc in all lines or specifically in individual RILs.
Supplemental Table S2. Stress response of genes that were induced by Glc in RIL 310.
Supplementary Material
Acknowledgments
We thank Thushyanthi Sivagnanam (University College London) for excellent technical assistance. We also wish to thank Åsa Strand (Umeå University) for helpful suggestions concerning the interactions between cold acclimation and chloroplast function.
This work was supported by the Biotechnology and Biological Sciences Research Council (research grant no. 31/P16341) and a PhD studentship from the Natural Environment Research Council, United Kingdom. The Insititut National de la Recherche Agronomique and the Centre National de la Recherche Scientifique provided extra financial support for the Complete Arabidopsis Transcriptome MicroArray.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Astrid Wingler (a.wingler@ucl.ac.uk).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Allemeersch J, Durinck S, Vanderhaeghen R, Alard P, Maes R, Seeuws K, Bogaert T, Coddens K, Deschouwer K, Van Hummelen P, et al (2005) Benchmarking the CATMA microarray: a novel tool for Arabidopsis transcriptome analysis. Plant Physiol 137 588–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Gomez-Mena C, Llorente F, Koornneef M, Salinas J, Martínez-Zapater JM (2005) Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol 139 1304–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyes DC, Zayed AM, Ascenzi R, McCaskill AJ, Hoffman NE, Davis KR, Görlach J (2001) Growth stage-based phenotypic analysis of Arabidopsis: a model for high throughput functional genomics in plants. Plant Cell 13 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin J-F, Wu S-H, Swidzinski J, Ishizaki K, et al (2005) Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J 42 567–585 [DOI] [PubMed] [Google Scholar]
- Calenge F, Saliba-Colombani V, Mahieu S, Loudet O, Daniel-Vedele F, Krapp A (2006) Natural variation for carbohydrate content in Arabidopsis: interaction with complex traits dissected by quantitative genetics. Plant Physiol 141 1630–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D, Fowler S, Fiehn O, Thomashow MF (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci USA 101 15243–15248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe ML, Serizet C, Thareau V, Aubourg S, Rouze P, Hilson P, Beynon J, Weisbeek P, van Hummelen P, Reymond P, et al (2003) CATMA: a complete Arabidopsis GST database. Nucleic Acids Res 31 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Randall PJ (1995) Characterization of a phosphate-accumulator mutant of Arabidopsis thaliana. Plant Physiol 107 207–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Purdy S, Christ A, Morot-Gaudry J-F, Wingler A, Masclaux-Daubresse C (2005) Characterization of new markers to determine the extent and variability of leaf senescence in Arabidopsis thaliana: a metabolic profiling approach. Plant Physiol 138 898–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz C, Saliba-Colombani V, Loudet O, Belluomo P, Moreau L, Daniel-Vedele F, Morot-Gaudry J-F, Masclaux-Daubresse C (2006) Leaf yellowing and anthocyanin accumulation are two genetically independent strategies in response to nitrogen limitation in Arabidopsis thaliana. Plant Cell Physiol 47 74–83 [DOI] [PubMed] [Google Scholar]
- Eberhard S, Drapier D, Wollman F-A (2002) Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J 31 149–160 [DOI] [PubMed] [Google Scholar]
- Ensminger I, Busch F, Huner NPA (2006) Photostasis and cold acclimation: sensing low temperature through photosynthesis. Physiol Plant 126 28–44 [Google Scholar]
- Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14 1675–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI (2005) Control of plant development and gene expression by sugar signalling. Curr Opin Plant Biol 8 93–102 [DOI] [PubMed] [Google Scholar]
- Hellmann H, Funck D, Rentsch D, Frommer WB (2000) Hypersensitivity of an Arabidopsis sugar signaling mutant toward exogenous Pro application. Plant Physiol 122 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himelblau E, Amasino RM (2001) Nutrients mobilized from leaves of Arabidopsis thaliana during leaf senescence. J Plant Physiol 158 1317–1323 [Google Scholar]
- Hurry V, Strand Å, Furbank R, Stitt M (2000) The role of inorganic phosphate in the development of freezing tolerance and the acclimatization of photosynthesis to low temperature is revealed by the pho mutants of Arabidopsis thaliana. Plant J 24 383–396 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, West MAL, van Leeuwen H, Kim K, Doerge RW, Michelmore RW, St Clair DA (2006) Genomic survey of gene expression diversity in Arabidopsis thaliana. Genetics 172 1179–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey S, Wingler A (2005) Natural variation in the regulation of leaf senescence and relation to other traits in Arabidopsis. Plant Cell Environ 28 223–231 [Google Scholar]
- Logemann J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163 16–20 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F (2002) Bay-0 × Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet 104 1173–1184 [DOI] [PubMed] [Google Scholar]
- Loudet O, Chaillou S, Merigout P, Talbotec J, Daniel-Vedele F (2003) Quantitative trait loci analysis of nitrogen use efficiency in Arabidopsis. Plant Physiol 131 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luquez VMC, Sasal Y, Medrano M, Martín MI, Mujica M, Guiamét JJ (2006) Quantitative trait loci analysis of leaf and plant longevity in Arabidopsis thaliana. J Exp Bot 57 1363–1372 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andrés C, Aubourg S, Bellaoui M, Bitton F, Bruyère C, Caboche M, Debast C, Gualberto J, Hoffmann B (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux C, Valadier M-H, Brugière N, Morot-Gaudry JF, Hirel B (2000) Characterization of the sink/source transition in tobacco (Nicotiana tabacum L.) shoots in relation to nitrogen management and leaf senescence. Planta 211 510–518 [DOI] [PubMed] [Google Scholar]
- Masclaux-Daubresse C, Carrayol E, Valadier M-H (2005) The two nitrogen mobilisation- and senescence-associated GS1 and GDH genes are controlled by C and N metabolites. Planta 221 580–588 [DOI] [PubMed] [Google Scholar]
- Nielsen TH, Krapp A, Röper-Schwarz U, Stitt M (1998) The sugar-mediated regulation of genes encoding the small subunit of Rubisco and the regulatory subunit of ADP Glc pyrophosphorylase is modified by phosphate and nitrogen. Plant Cell Environ 21 443–454 [Google Scholar]
- Paul MJ, Pellny TK (2003) Carbon metabolite feedback regulation of leaf photosynthesis and development. J Exp Bot 54 539–547 [DOI] [PubMed] [Google Scholar]
- Poirier Y, Thoma S, Somerville C, Schiefelbein J (1991) A mutant of Arabidopsis deficient in xylem loading of phosphate. Plant Physiol 97 1087–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourtau N, Jennings R, Pelzer E, Pallas J, Wingler A (2006) Effect of sugar-induced senescence on gene expression and implications for the regulation of senescence in Arabidopsis. Planta 224 556–568 [DOI] [PubMed] [Google Scholar]
- Pourtau N, Marès M, Purdy S, Quentin N, Ruël A, Wingler A (2004) Interactions of abscisic acid and sugar signalling in the regulation of leaf senescence. Planta 219 765–772 [DOI] [PubMed] [Google Scholar]
- Prasad TK, Anderson MD, Martin BA, Stewart CR (1994) Evidence for chilling-induced oxidative stress in maize seedlings and a regulatory role for hydrogen peroxide. Plant Cell 6 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen H (1957) A modified ninhydrin colorimetric analysis for amino acids. Arch Biochem Biophys 67 10–15 [DOI] [PubMed] [Google Scholar]
- Stitt M, Lilley RMC, Gerhardt R, Heldt HW (1989) Metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol 174 518–552 [Google Scholar]
- Strand Å, Foyer CH, Gustafsson P, Gardeström P, Hurry V (2003) Altering flux through the Suc biosynthesis pathway in transgenic Arabidopsis thaliana modifies photosynthetic acclimation at low temperatures and the development of freezing tolerance. Plant Cell Environ 26 523–535 [Google Scholar]
- Strand Å, Hurry V, Gustafsson P, Gardeström P (1997) Development of Arabidopsis thaliana leaves at low temperatures releases the suppression of photosynthesis and photosynthetic gene expression despite the accumulation of soluble carbohydrates. Plant J 12 605–614 [DOI] [PubMed] [Google Scholar]
- Strand Å, Hurry V, Henkes S, Huner N, Gustafsson P, Gardeström P, Stitt M (1999) Acclimation of Arabidopsis leaves developing at low temperatures: increasing cytoplasmic volume accompanies increased activities of enzymes of the Calvin cycle an in the Suc-biosynthesis pathway. Plant Physiol 119 1387–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensson JT, Crosatti C, Campoli C, Bassi R, Michele Stanca A, Close TJ, Cativelli L (2006) Transcriptome analysis of cold acclimation in barley albina and xantha mutants. Plant Physiol 141 257–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thareau V, Dehais P, Serizet C, Hilson P, Rouze P, Aubourg S (2003) Automatic design of gene-specific sequence tags for genome-wide functional studies. Bioinformatics 19 2191–2198 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Thimm O, Redestig H, Blaesing OE, Palacios-Rojas N, Selbig J, Hannemann J, Piques CM, Steinhauser D, et al (2005) Extension of the visualization tool MapMan to allow statistical analysis of arrays, display of corresponding genes, and comparison with known responses. Plant Physiol 138 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Graaff E, Schwacke R, Schneider A, Desimone M, Flügge U-I, Kunze R (2006) Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol 141 776–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma DS, Zhang C-S (1999) Regulation of proline and arginine biosynthesis in plants. In BK Singh, ed, Plant Amino Acids. Biochemistry and Biotechnology. Marcel Dekker, Inc, New York, pp 248–265
- Wingler A, Brownhill E, Pourtau N (2005) Mechanisms of the light-dependent induction of cell death in tobacco plants with delayed senescence. J Exp Bot 56 2897–2905 [DOI] [PubMed] [Google Scholar]
- Wingler A, Lea PJ, Quick WP, Leegood RC (2000) Photorespiration—metabolic pathways and their role in stress protection. Philos Trans R Soc Lond B Biol Sci 355 1517–1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A, Marès M, Pourtau N (2004) Spatial patterns and metabolic regulation of photosynthetic parameters during leaf senescence. New Phytol 161 781–789 [DOI] [PubMed] [Google Scholar]
- Wingler A, Purdy S, MacLean JA, Pourtau N (2006) The role of sugars in integrating environmental signals during the regulation of leaf senescence. J Exp Bot 57 391–399 [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentgraf U, Jobst J, Kolb D, Rentsch D (2004) Senescence-related gene expression profiles of rosette leaves of Arabidopsis thaliana: leaf age versus plant age. Plant Biol 6 178–183 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.