Abstract
In tobacco (Nicotiana tabacum), an elicitor- and pathogen-induced 9-lipoxygenase (LOX) gene, NtLOX1, is essential for full resistance to pathogens, notably to an incompatible race of Phytophthora parasitica var. nicotianae (Ppn race 0). In this work, we aimed to identify those oxylipins induced during attempted infection by Ppn race 0 and down-regulated in NtLOX1 antisense plants. Here we show that colneleic and colnelenic acids, which significantly inhibit germination of Ppn zoospores, are produced in roots of wild-type plants inoculated with Ppn, but are down-regulated in NtLOX1 antisense plants. A search for a tobacco gene encoding the enzyme involved in the formation of these divinyl ether (DVE) fatty acids resulted in the cloning and characterization of a DVE synthase (DES) clone (NtDES1). NtDES1 is a 9-DES, specifically converting fatty acid 9-hydroperoxides into DVE fatty acids. NtDES1 has the potential to act in combination with NtLOX1 because, in the presence of the two enzymes, linoleic and linolenic acids were converted in vitro into colneleic and colnelenic acids, respectively. In addition, the pattern of NtDES1 gene expression was quite similar to that of NtLOX1. Their transcripts were undetected in healthy tissues from different plant organs, and accumulated locally and transiently after elicitation and fungal infection, but not after wounding. Visualization of NtDES1-yellow fluorescent protein and NtLOX1-cyan fluorescent protein fusion proteins in tobacco leaves indicated that both localize in the cytosol and are excluded from plastids, consistent with the presumed location of the 9-LOX pathway in plants and the lack of transit peptides for NtLOX1 and NtDES1, respectively. Our data suggest that, in tobacco, NtDES1 and NtLOX1 act together and form DVEs in response to pathogen attack and that this class of oxylipins modulates in vivo the outcome of the tobacco-Ppn race 0 interaction.
The oxylipin pathway in plants generates lipid-derived molecules through the oxygenation and subsequent enzymatic conversion of polyunsaturated fatty acids (PUFAs). The biosynthesis of most oxylipins is started by lipoxygenases (LOXs), although other PUFA oxygenases initiating distinct branches of the oxylipin pathway are encountered in plants (Hamberg et al., 1999; Blée, 2002). Plant LOXs mainly use linoleic acid (LA) and α-linolenic acid (LnA) as substrates and form, depending on their positional specificity, either 9- or 13-fatty acid hydroperoxides from which an array of additional oxylipins are synthesized through the so-called 9-LOX and 13-LOX pathways (Feussner and Wasternack, 2002). At least six different enzymes, as well as LOX itself, can use fatty acid hydroperoxides as substrates in plants. Besides LOX, three of them—namely, genes encoding allene oxide synthase (AOS), hydroperoxide lyase (HPL), and, more recently, divinyl ether (DVE) synthase (DES)—were cloned (Howe and Schilmiller, 2002). AOSs are the first dedicated enzymes of the biosynthetic pathway leading from 13-hydroperoxy-octadecatrienoic acid (13-HPOT) to jasmonates, that is, the plant growth regulator jasmonic acid (JA), its methyl ester methyl jasmonate, and some of its biosynthetic precursors like 12-oxo-phytodienoic acid (oPDA). HPLs cleave 9- or 13-hydroperoxides of C18-PUFAs to short-chain ω-oxo acids and aldehydes. DESs catalyze the conversion of LOX-derived fatty acid hydroperoxides into DVE fatty acids (Grechkin, 2002). Two DES cDNAs were cloned from tomato (Lycopersicon esculentum; Itoh and Howe, 2001) and potato (Solanum tuberosum; Stumpe et al., 2001); these enzymes exclusively convert 9-hydroperoxides and form the DVEs colneleic acid (CA) and colnelenic acid (CnA). Although their sequences are not published yet, two additional enzymes were described in Allium sp. and Ranunculus sp. that preferentially form the 13-DVE etheroleic and etherolenic acids (Grechkin et al., 1995; Hamberg, 1998). All AOS, HPL, and DES sequences isolated to date show significant similarity to each other and deduced proteins group within the CYP74 family, a new family of atypical P450s; the DESs whose cDNA was cloned so far belong to the CYP74D subfamily (Howe and Schilmiller, 2002). Members of the CYP74 family are unusual P450 monooxygenases in that they lack oxygenase activity and can use fatty acid hydroperoxides both as the oxygen donor and the substrate in contrast to classical P450 monooxygenases (Song et al., 1993; Itoh and Howe, 2001). This is reflected in recurrent amino acid changes in otherwise conserved domains of P450s (Kalb and Loper, 1988; Chapple, 1998). Other branches of the oxylipin pathway, leading to epoxy hydroxy fatty acids and hydroxy derivatives, are less well characterized at the molecular level (Feussner and Wasternack, 2002).
Oxylipins are involved in many different aspects of plant development (Porta and Rocha-Sosa, 2002) and plant responses to stress, notably pathogen attack (Blée, 1998, 2002; Feussner and Wasternack, 2002). Compounds formed through both the 13-LOX and 9-LOX pathways participate in plant defense against pests and pathogens. Illustrative of this, the few reported mutant or transgenic plants affected in synthesis, signaling, or perception of selected oxylipins show altered responses to pathogens (Shah, 2005). Underlying these phenotypes, both a signaling role and a direct antimicrobial effect are suggested for several oxylipins belonging to all categories tested so far (Prost et al., 2005; Shah, 2005).
The role of the 9-LOX pathway in plant defense is mainly emphasized in Solanaceous plants. Thus, 9-LOX gene expression and 9-LOX activity are strongly induced in tobacco and potato in response to two oomycete pathogens of these plants, Phytophthora parasitica var. nicotianae (Ppn) and Phytophthora infestans, respectively, and to oomycete-derived elicitors (Fournier et al., 1993; Véronési et al., 1996; Fidantsef and Bostock, 1998; Kolomiets et al., 2000; Göbel et al., 2001). Similarly, expression of the 9-DES gene StDES is increased in potato in response to elicitors and pathogen attack (Stumpe et al., 2001). Consistent with these observations, several 9-LOX-derived oxylipins, including 9-hydroperoxides, 9-hydroxides, CA and CnA, trihydroxy-oxylipins derived from 9-hydroperoxy LA (9-HPOD) and 9-hydroperoxy LnA (9-HPOT), accumulate in tobacco or potato in response to elicitor treatment or pathogen ingress (Rustérucci et al., 1999; Weber et al., 1999; Göbel et al., 2001, 2002). 9-LOX-silenced potato plants show decreased levels of 9-LOX-derived oxylipins (among which Ca, CnA, 9-hydroxy-octadecadienoic acid [9-HOD] and 9-hydroxy-octadecatrienoic acid) in leaves inoculated with the incompatible pathogen Pseudomonas syringae pv maculicola (Göbel et al., 2003). It was proposed that 9-hydroperoxides be endowed with cell death-promoting activity (Rustérucci et al., 1999; Knight et al., 2001), whereas CA and CnA act in vitro as antimicrobials against a range of pathogenic bacteria, fungi, and oomycetes (Prost et al., 2005). More specifically, LA- and LnA-derived DVEs inhibit mycelial growth and especially spore germination of several Phytophthorae, among which is Ppn (Weber et al., 1999; Prost et al., 2005). Other 9-LOX-derived compounds, such as 9-HPOD/Ts, 9-HOD/Ts, 9-oxo-octadecadienoic acid (9-KOD), and 9-oxo-octadecatrienoic acid (9-KOT) are antifungal and antioomycete in vitro; 9-KOT and 9-HPOD also exhibit some antibacterial activity (Prost et al., 2005). Even if open questions remain on the efficacy and actual occurrence of direct pathogen control via antimicrobial oxylipins in planta, these data suggest that 9-LOX-derived oxylipins might indeed be important contributors to the outcome of given plant-microbe interactions, notably in Solanaceous plants.
In tobacco, expression of NtLOX1, a pathogen- and elicitor-induced 9-LOX gene, was previously demonstrated to be essential for resistance. On one hand, resistance of tobacco to an avirulent strain of Ppn is impaired when NtLOX1 expression is inhibited in transgenic plants by an antisense strategy (Rancé et al., 1998). On the other hand, NtLOX1-overexpressing lines show strongly decreased susceptibility to a virulent Ppn strain (Mène-Saffrané et al., 2003). Furthermore, especially in the dark, 9-LOX activity and products are necessary for cell death to proceed in the nonhost resistance response of tobacco to cryptogein, an elicitor produced by P. cryptogea (Rustérucci et al., 1999; Montillet et al., 2005). To further investigate the role of the oxylipin pathway in the tobacco-Ppn interaction, we first aimed to identify which oxylipins are down-regulated in NtLOX1 antisense plants. A search for tobacco genes encoding 9-hydroperoxide-converting enzymes was therefore undertaken. We report on the cloning and biochemical characterization of an elicitor- and pathogen-induced DES from tobacco. The role of the cloned enzyme in the formation of DVEs in response to pathogen attack is discussed in parallel with the role of NtLOX1.
RESULTS
Oxylipin Profiling of Wild-Type and NtLOX1 Antisense Plants Inoculated with Ppn Race 0
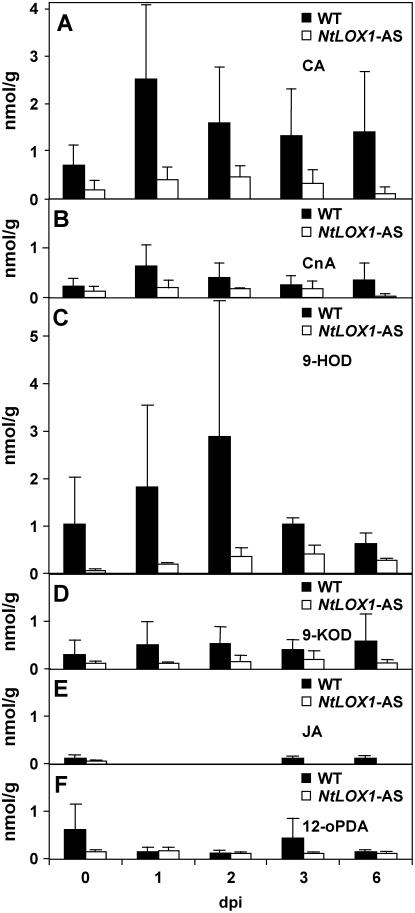
Previous work had shown that NtLOX1, a pathogen- and elicitor-induced 9-LOX gene, is essential for full resistance against the avirulent Ppn race 0. Because the resistance of tobacco to this pathogen is impaired when NtLOX1 expression is inhibited in transgenic plants expressing an antisense NtLOX1 construct (Rancé et al., 1998), we first aimed to identify those oxylipins that may be involved in this process. Thus, we performed oxylipin profiling of roots of wild-type and NtLOX1 antisense-infected plants as a function of time after inoculation with Ppn. The oxylipins whose amounts were increased during infection and that differed between wild-type and NtLOX1 antisense plants are shown in Figure 1, A to D. Despite variations in absolute amounts of oxylipins between independent experiments, it appears that the levels of CA (Fig. 1A) and CnA (Fig. 1B) were on average about 3-fold increased already at 1 d postinoculation (dpi) with Ppn race 0 in wild-type plants (black bars), and progressively decreased thereafter. Similar patterns were detected for 9-HOD (Fig. 1C), which peaked 2 dpi, and, to a much lesser extent, for 9-KOD (Fig. 1D). Of the above oxylipins, CA was the most abundant. Other oxylipins, most notably oPDA and JA derived from the AOS branch that metabolizes 13-hydroperoxides, remained at very low levels and did not vary much (Fig. 1, E and F). The NtLOX1 hydroperoxide products were not detected in the same samples and thus may not accumulate (data not shown). As compared to wild-type plants, the levels of CA, CnA, 9-HOD, and 9-KOD remained low in the NtLOX1 antisense plants under the same conditions (Fig. 1, A–D; white bars). From these data, we conclude that, at early stages of infection, the products of the DES branch of the 9-LOX pathway (CA and CnA) are major oxylipins involved in the plant defense response against Ppn because they accumulated faster than the product of the reductase branch 9-HOD.
Figure 1.
Quantification of oxylipins in root tissues inoculated with Ppn race 0 zoospores. CA (A) and CnA (B) accumulation is detectable already 1 dpi in wild-type plants (black bars), peaking at 2 dpi and decreasing thereafter. A similar pattern is shown by 9-HOD (C) and 9-KOD (D). Average content of the same compounds is essentially stable in roots of NtLOX1 antisense plant line 125-2-1 (white bars). E and F, JA and 12-oPDA content, respectively. The results are means of either three (wild type) or two independent experiments, with five pooled biological replicates and one measure per time point and trial.
Isolation of a NtDES1 cDNA
Based on the results described in the previous section, we aimed next to identify the involved DES. For this purpose, several pairs of degenerated primers were designed to target conserved sequence stretches in hydroperoxide-converting enzymes: AOS from flax (Linum usitatissimum), guayule (Parthenium argentatum), and Arabidopsis (Arabidopsis thaliana; Song et al., 1993; Pan et al., 1995; Laudert et al., 2000), HPL from pepper (Capsicum annuum; Matsui et al., 1996), and DES from tomato and potato (Itoh and Howe, 2001; Stumpe et al., 2001). Using DNA isolated from an elicitor-treated tobacco cell cDNA library (Véronési et al., 1995) as template, a 1,131-bp fragment (DESB3) was amplified, cloned, and sequenced. DESB3 showed moderate, although significant, sequence similarity to AOS and HPL genes and was highly similar to the tomato and potato DES sequences (Itoh and Howe, 2001; Stumpe et al., 2001). Southern-blot analysis of tobacco DNA with 32P-labeled DESB3 revealed DESB3 specifically hybridized to a small number of genomic sequences, most likely two (data not shown). DESB3 was used as a probe to screen the above-mentioned tobacco cDNA library. Several positive clones were isolated and found to be independent cDNA copies of the same transcript. DES1A, the longest of these clones, was sequenced and shown to contain a 1,434-bp open reading frame encompassing the DESB3 sequence, a 38-bp 5′-untranslated region, and a 169-bp 3′-untranslated region plus a poly(A)+ tail (Supplemental Fig. S1). 5′-RACE analysis confirmed the ATG in position 39 as the first in-frame start codon and showed that 16 bp only are lacking at the 5′-end in the cloned sequence compared to poly(A)+ RNA. The deduced amino acid sequence proved clearly related to other known hydroperoxide-converting enzymes, showing 44% to 48% identity to AOS sequences, 37% to 50% identity to HPL sequences, and 86.6% and 87.5% identity to tomato DES and potato DES, respectively (Itoh and Howe, 2001; Stumpe et al., 2001); the identity at the nucleotide level with the two latter genes reached 78.8% and 74.1%, respectively. The gene corresponding to DES1A was named NtDES1. Cross hybridization of DES1A with other CYP74 family members, including a partial DNA clone amplified from tobacco genomic DNA with a different pair of degenerated primers and virtually identical to the Nicotiana attenuata AOS clone (Ziegler et al., 2001), was investigated. DES1A did not cross hybridize to a guayule AOS clone, a pepper HPL clone, or the above-mentioned partial DNA from a putative tobacco AOS. Detailed comparison of the deduced NtDES1 primary sequence with P450 sequences showed conserved domains and characteristic substitutions, as observed for other known CYP74 enzymes (Supplemental Fig. S1).
Identification of NtDES1 as a 9-DES
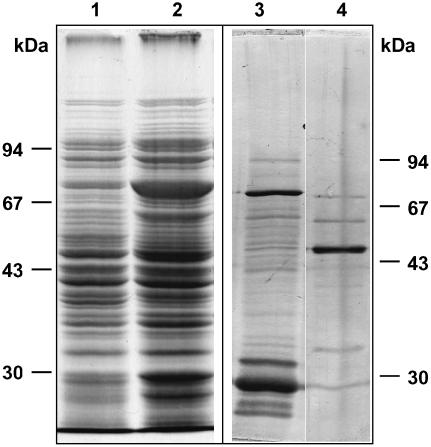
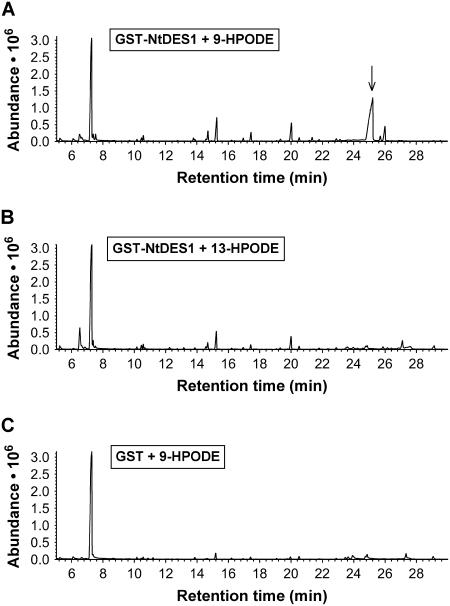
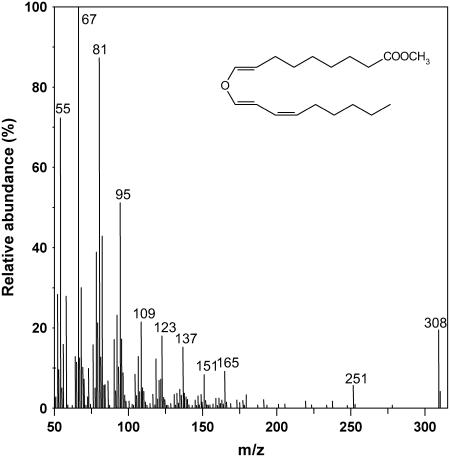
To study the enzymatic activity of the protein encoded by NtDES1 cDNA, a glutathione S-transferase (GST)-NtDES1 fusion protein was expressed in Escherichia coli. SDS-PAGE analysis of solubilized proteins showed that bacterial cultures harboring the pGEX-NtDES1 recombinant vector accumulated an additional, prominent protein of the expected size (around 80 kD) upon isopropyl-β-d-thiogalactopyranoside (IPTG) induction (Fig. 2, lane 2) as compared to control extracts from noninduced bacteria (Fig. 2, lane 1) or from induced bacteria bearing the pGEX-5X-3 empty vector. The fusion protein was purified by affinity chromatography on glutathione-Sepharose 4B (Fig. 2, lane 3). Protease digestion of the GST-NtDES1 fusion directly on the affinity column yielded a 54-kD protein (Fig. 2, lane 4). Hydroperoxide-converting activity of crude extracts and purified GST-NtDES1 or GST-free NtDES1 proteins was assayed with pure 9-HPOD, 9-HPOT, 13-hydroperoxy-octadecadienoic acid (13-HPOD), or 13-hydroperoxy-octadecatrienoic acid (13-HPOT). All protein solutions containing either NtDES1 or the fusion protein, but not those prepared from bacteria expressing the pGEX-5X-3 empty vector or from noninduced bacteria were able to metabolize 9-hydroperoxides as indicated by a decrease in A234 nm, whereas 13-hydroperoxides appeared to be very poor substrates (data not shown). Thus, the rate of hydroperoxide-degrading activity in the presence of the purified NtDES1 protein was about 180 nmol s−1 mg−1 protein when 9-hydroperoxides were used as substrates, whereas undetectable with 13-hydroperoxides under the same conditions. The ability of NtDES1 to catalyze the breakdown of fatty acid hydroperoxides is consistent with homology to CYP74 enzymes. Preference for 9-LOX-derived hydroperoxides together with high sequence similarity with potato and tomato 9-DES suggested that NtDES1 encodes a tobacco 9-DES. This was further confirmed by identification of the major oxylipin generated from 9-HPOD by GST-NtDES1. Following incubation of 9-HPOD with protein extracts from E. coli expressing either pGEX-NtDES1 or the empty vector, the reaction products were extracted and analyzed by gas chromatography (GC)-mass spectrometry (MS) as methyl ester derivatives. Comparison of the GC profiles revealed a major peak eluting at about 25 min when pGEX-NtDES1 bacterial extracts were used (Fig. 3A), which was not present if 13-HPOD was given as a substrate or in the mock experiment (Fig. 3, B and C). The mass spectrum associated with this peak, showing prominent ions at mass-to-charge ratio = 308 (M+; 20%), 251 (7%), 165 (9%), 151 (8%), 137 (15%), 123 (19%), 109 (22%), 95 (51%), 81 (88%), and 67 (100%; Fig. 4), is in agreement with previous reports on the methyl CA spectrum (Galliard and Phillips, 1972; Itoh and Howe, 2001). Thus, NtDES1 is able to form CA from 9-HPOD.
Figure 2.
Purification of NtDES1 expressed in E. coli. Soluble protein extracts correspond to 180 μL of E. coli culture from control (lane 1) or IPTG-induced (lane 2) bacterial cells carrying pGEX-NtDES1. The GST-NtDES1 protein bound to the affinity column was either eluted with 10 mm reduced glutathione (lane 3, 875 ng protein) or digested with factor Xa, leading to the release of NtDES1 in the incubation buffer (lane 4, 210 ng protein). After separation on 10% polyacrylamide gel, proteins were stained with Coomassie Blue (lanes 1 and 2) or silver nitrate (lanes 3 and 4). Position of protein standards are indicated for each gel.
Figure 3.
Hydroperoxide-degrading activity of NtDES1. A, Recombinant NtDES1 converts 9-HPOD into a major product with a GC retention time of about 25 min, later identified as CA (arrow). B, NtDES1 cannot convert efficiently 13-HPODs. C, GST alone (negative control) cannot degrade 9-HPOD.
Figure 4.
Mass spectrum of the major product generated by recombinant NtDES1 from 9-HPOD. 9-HPOD was incubated with a protein extract from E. coli expressing GST-NtDES1. Reaction products were extracted and analyzed by GC-MS as methyl ester derivatives. The mass spectrum of the major product is shown and corresponds to methyl CA (inserted formula).
NtDES1 and NtLOX1 Can Cooperate in Vitro to Form 9-DVEs
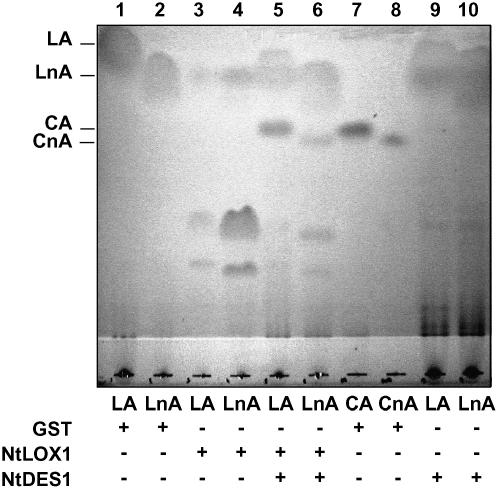
In tobacco, 9-hydroperoxides are expected to be formed by NtLOX1, an elicitor- and pathogen-inducible 9-LOX (Fournier et al., 1993; Véronési et al., 1996). Therefore, the ability of NtDES1 to utilize hydroperoxides formed through NtLOX1 action was investigated in a qualitative thin-layer chromatography (TLC) assay. The LOX substrates, LA and LnA, were incubated with crude bacterial extracts, prepared from E. coli strains expressing either the GST-NtDES1 fusion protein or NtLOX1, alone or in combination. NtLOX1 massively degraded the two fatty acids (Fig. 5, lanes 3 and 4) in contrast to the control (lanes 1 and 2 and 9 and 10). The combination of the two enzymes (lanes 5 and 6) yielded compounds comigrating with CA and CnA (lanes 7 and 8) derived either from C18:2 or C18:3 fatty acid substrates, respectively. This showed that NtLOX1 and NtDES1 are able to cooperate to form DVE fatty acids from PUFAs, at least in vitro. Although the retained TLC assay was qualitative, it might be significant that NtLOX1 and NtDES1 used in combination (lanes 5 and 6) apparently depleted fatty acid substrates to a lesser extent than NtLOX1 alone (lanes 3 and 4). This could reflect the previously reported inhibitory activity of DVE fatty acids toward 9-LOX (Corey et al., 1987; Itoh and Howe, 2001).
Figure 5.
In vitro-coupled activity of recombinant NtLOX1 and NtDES1. Soluble protein extracts prepared from E. coli transformed with pGEX-5X-3 (GST) as control, pT7-NtLOX1 (LOX), or pGEX-NtDES1 (DES) were incubated alone or in combination with LA or LnA. Reaction products were extracted and separated by TLC and compared to CA and CnA standards. Oxylipins and fatty acids were stained with iodine.
Coordinated Expression of NtDES1 and NtLOX1 during Pathogenesis
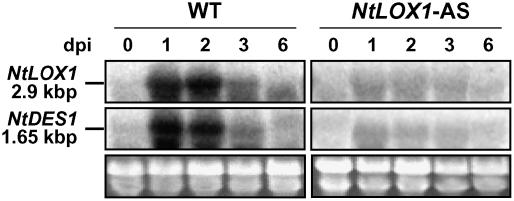
Expression of NtDES1 in tobacco was preliminarily investigated in defense- and pathogenesis-related conditions. A time-course study of NtDES1 expression in elicitor-treated tobacco cells showed that NtDES1 transcript accumulation was already detectable 2 h after addition of the elicitor (30 μg/mL) and was maximal after 4 h, whereas it was undetected in control samples (Supplemental Fig. S2A). The same elicitor preparation, as well as CBEL, a glycoprotein elicitor from Ppn (Villalba-Mateos et al., 1997), also efficiently induced accumulation of NtDES1 transcript when infiltrated in tobacco leaf tissues, this effect being limited to the infiltrated area (data not shown). These data prompted us to investigate NtDES1 expression in healthy tobacco plants, as well as in response to wounding and pathogen attack. NtDES1 transcripts could not be detected in leaves, stems, or roots from healthy tobacco either in northern-blot or reverse transcription-PCR experiments, nor in wounded leaves over several extended time-course experiments with very close time points (data not shown). In contrast, when tobacco plants were challenged with Ppn in a stem inoculation assay, NtDES1 transcript already accumulated 12 h after inoculation as compared to control samples from mock-inoculated stems. NtDES1 transcripts were not detected in the leaf closest to the infected area (Supplemental Fig. S2B). NtLOX1 expression was previously found to be only locally induced as well, both in promoter-β-glucuronidase (B. Verdaguer, J. Fournier, and M.T. Esquerré-Tugayé, unpublished data) and northern experiments (Rancé et al., 1998). We therefore went further to the analysis of NtDES1 and NtLOX1 transcript accumulation during interaction with the pathogen in conditions simulating real-life infection processes; that is, in zoospore-inoculated roots. RNA was extracted from plant samples that were also analyzed for oxylipins in Figure 1. The results showed that both transcripts are already detectable 1 dpi of wild-type plants (Fig. 6). A peak in expression was evident by 2 dpi for both genes and transcripts were no longer detectable 6 dpi. Unexpectedly, the NtDES1 expression pattern was influenced by the silencing of NtLOX1 because in antisense plants challenged with Ppn race 0 no such induction as in wild-type roots and stems was visible (Fig. 6; Supplemental Fig. S2B).
Figure 6.
NtDES1 and NtLOX1 transcriptional profile upon infection. Northern analysis was performed on 20 μg total RNA isolated from roots inoculated with zoospores of Ppn race 0 at the indicated time points. NtLOX1 (2.9 kb; top) and NtDES1 (1.65 kb; middle) transcripts were detected with radiolabeled probes. Bottom image shows ethidium bromide staining of the gel. One representative image out of two independent trials is reported.
Localization of NtDES1 in Tobacco Cells
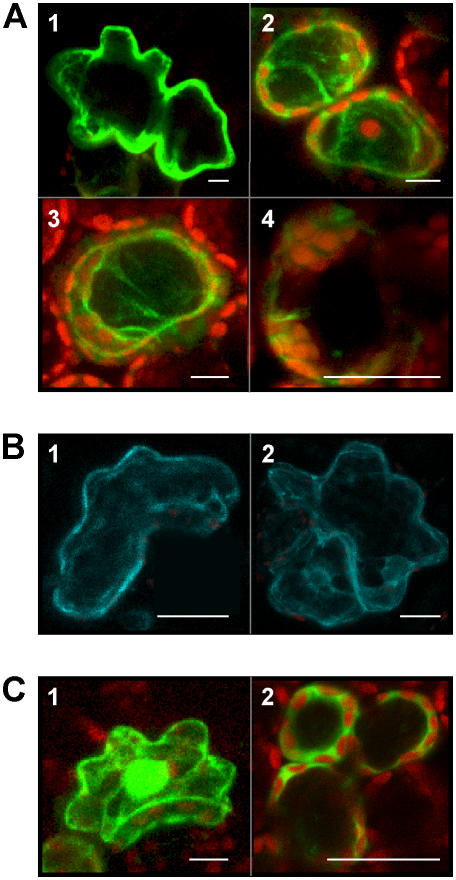
Similar to other DES sequences and in contrast to related CYP74 enzymes, NtDES1 does not contain the chloroplast-targeting N-terminal transit peptide. Expression of a NtDES1-yellow fluorescent protein (YFP) fusion protein in transiently transformed tobacco leaves showed cytosolic localization of a 9-hydroperoxide-metabolizing CYP74, NtDES1 (Fig. 7A). As compared to control leaves overexpressing YFP alone in which fluorescence mostly concentrated in the nucleus (Fig. 7C), the pGreenII-0229-NtDES1-YFP construct labeled the cytoplasm very brightly, whereas the fluorescent fusion protein was excluded from the nuclei and chloroplasts (Fig. 7A). We analyzed the location of NtLOX1-cyan fluorescent protein (CFP) for comparison. It localized in the cytoplasmic compartment as well, as shown in Figure 7B, fluorescence being excluded from nuclei and cytoplasmic organelles.
Figure 7.
Intracellular localization of NtDES1 and NtLOX1. A, Tobacco leaves transformed to transiently express the NtDES1-YFP fusion protein are labeled in the cytoplasm. Epidermal cell, total depth 5.54 μm, resolution 0.36 × 0.36 × 2.77 μm (1); palisade cells, total depth 43.3 μm, resolution 0.36 × 0.36 × 1.80 μm (2 and 3); palisade cell, total depth 26.7 μm, resolution 0.16 × 0.16 × 7.3 μm (4). B, Epidermal cells overexpressing the NtLOX1-CFP fusion protein also show a fluorescent cytoplasm (total depth 14.91 μm, 0.21 × 0.21 × 4.97 μm; 1 and 2). C, Control leaves transformed with the vector pGreenII0029-2 × 35S-TL-YFP have fluorescent cytoplasm and nucleus; epidermal cell, total depth 17 μm, 0.36 × 0.36 × 1.71 μm (1); mesophyll cells, total depth 36 μm, 0.11 × 0.11 × 1.6 μm (2). Pictures were taken 2 to 4 d after Agrobacterium-mediated transformation. Bars in figures represent 10 μm.
DISCUSSION
In tobacco, expression of NtLOX1, a pathogen- and elicitor-induced 9-LOX gene, is essential for full resistance to several pathogens. Notably, resistance of wild-type tobacco line 46-8 to Ppn race 0 is impaired when NtLOX1 expression is inhibited in transgenic plants by an antisense strategy (Rancé et al., 1998). However, given several metabolic routes into which LOX-issued hydroperoxides can be channeled, it was not clear which oxylipins produced by NtLOX1 were to be held responsible for such a phenotype. Therefore, we first identified those oxylipins that are induced in resistant tobacco root tissues challenged with Ppn race 0 zoospores and down-regulated in NtLOX1 antisense plants. We could show that the induction of 9-LOX-derived DVE fatty acids (especially CA) and 9-HOD was lowered at least 3-fold in the NtLOX1 antisense line 125-2-1 (Fig. 1). Thus, we aimed to clone the enzymes involved besides NtLOX1 and a search for tobacco genes encoding 9-hydroperoxide-converting enzymes potentially acting downstream of NtLOX1 was undertaken, concentrating at first on the CA and CnA biosynthetic protein. A PCR-based search for CYP74-related sequences in a cDNA library from elicitor-treated tobacco cells resulted in the isolation of a cDNA named DES1A. The corresponding NtDES1 gene encodes a polypeptide of 478 amino acids whose identity was strongly suggested by high sequence similarity to the recently reported DESs from potato and tomato (Itoh and Howe, 2001; Stumpe et al., 2001). As in other CYP74 enzymes lacking oxygenase activity, the O2-binding pocket is modified in NtDES1. The conserved Thr residue found in this region in P450 monooxygenases is replaced by Ala (Ala-290) as in tomato and potato DES (Itoh and Howe, 2001; Stumpe et al., 2001). Thus, the core consensus sequence in the modified O2-binding site appears to be AGLNA in Solanaceous plant DESs instead of GGxx(I/V/L) as in other CYP74 enzymes. The nature of NtDES1 was further confirmed by activity measurements showing its ability to form CA from 9-HPOD. Because NtDES1 accepted only 9-LOX-derived fatty acid hydroperoxides as substrates in vitro, the enzyme was identified as a 9-DES (Figs. 2–4; Supplemental Fig. S1).
The NtDES1 primary sequence lacks both a hydrophobic N terminus that cooperates in anchoring numerous eukaryotic P450s to the endoplasmic reticulum and the N-terminal sequence targeting some CYP74s to the plastid (Song et al., 1993; Laudert et al., 2000). Indeed, localization studies via YFP tagging indicated that NtDES1 is located to the cytosolic compartment and excluded from both the nucleus and other organelles. This is in agreement with the lack of an organelle-targeting sequence, although the absence of an obvious transit peptide does not always correlate with the cytosolic location as exemplified by barley (Hordeum vulgare) AOSs (Maucher et al., 2000). The cytosolic location of NtDES1 is consistent with its specificity toward 9-hydroperoxides that at least in Solanaceous plants are exclusively synthesized in the cytosol (Feussner and Wasternack, 2002). Indeed, NtLOX1, the only tobacco 9-LOX characterized so far, localized to the cytoplasm as well, as shown by confocal microscopy on transiently transformed leaves (Fig. 7).
Several lines of evidence, in agreement with the results recently reported for potato-pathogen interactions (Kolomiets et al., 2000; Göbel et al., 2001, 2002; Stumpe et al., 2001), further strengthen the hypothesis of a concerted action of NtDES1 and NtLOX1 leading to the synthesis of CA and CnA during the defense response of tobacco. First, the expression pattern of NtDES1 was coincident with that of NtLOX1 (Véronési et al., 1996; this work; Fig. 6; Supplemental Fig. S2). Second, the two enzymes cooperated in vitro to form the DVEs CA and CnA from their respective fatty acid precursors (Fig. 5). Third, CA and CnA, which are not normally found in healthy tissues, were produced in tobacco upon pathogen challenge such as infection of leaves by Tobacco mosaic virus (Weber et al., 1999) or of stem (L. Mène-Saffrané, unpublished data) and roots (this work; Fig. 1) by Ppn. To our knowledge, this is the first report on oxylipin synthesis in inoculated root tissue. The fact that the C18:2-derived DVE CA rather than CnA accumulated in infected tobacco roots might be due to the relatively higher abundance of LA than LnA in nonphotosynthesizing cells. The opposite is true in green tissues, where LnA is preferentially found in association with chloroplastic membranes (Somerville et al., 2000). Whether the 9-LOX-derived DVEs accumulate into the cytosol or diffuse either to other compartments or even to outside the roots remains to be elucidated.
Further studies provided additional data linking NtDES1 and NtLOX1, adding to our knowledge of the 9-LOX pathway in plants. Our results showed that transcripts of both enzymes are not detectable in healthy tobacco roots, stems, and leaves nor in untreated cells (i.e. strictly inducible). They also demonstrated that tissue responsiveness to induction is not root specific because it is present in cell culture and both in underground and above-ground infected tissues. They further indicated that NtDES1 inducibility is compromised in NtLOX1 antisense plants because induction of NtDES1 by pathogen attack was hampered unexpectedly both in roots and stems of silenced plants (Fig. 6; Supplemental Fig. S2B). The reasons underlying this unresponsiveness are unknown at the moment; the hypothesis that NtLOX1 products might at least partially regulate NtDES1 transcription awaits further testing. Expression of NtDES1 (and NtLOX1) was restricted to the elicited or inoculated area in plants, thereby indicating that these genes are not systemically induced in tobacco (Supplemental Fig. S2). NtDES1 was not induced by salicylic acid either, a signal molecule related to systemic induction of defense in various plants (data not shown). This suggested that the DES branch of the 9-LOX pathway participates in local rather than systemic defense response through production of antimicrobial CA and CnA at the site of pathogen attack. Most importantly, NtLOX1 antisense plants, which are compromised in their resistance to Ppn race 0 both at the stem and at the root level (Rancé et al., 1998), displayed reduced accumulation of DVEs upon inoculation with zoospores of the black shank agent (Fig. 1). The previously published oxylipin profile of potato leaves undergoing hypersensitive response to the incompatible pathogen P. syringae pv maculicola (Göbel et al., 2003) shows that, in the absence of POTLX-3 (a pathogen-inducible 9-LOX from vegetative tissues), 9-LOX-derived oxylipins are substantially less abundant than in wild-type potato leaves. POTLX-3-silenced potato plants, however, are not more susceptible than the wild type to this bacterial pathogen. This apparent discrepancy might be due to the fact that 13-LOX-derived oxylipins (such as JA, oPDA, 13-HOD, and 13-HOT) are also induced in the potato-Pseudomonas pathosystem, whereas during the tobacco-Ppn race 0 interaction they are not (Fig. 1, E and F). Furthermore, their induction by pathogens seems amplified in the absence of 9-LOX in potato, but not in tobacco and, indeed, their higher abundance compared to wild-type plants is suggested to compensate for the decreased production of 9-LOX-derived oxylipins in cell death promotion (Göbel et al., 2003). Interestingly, clear accumulation of 9-hydroperoxides is detectable in Pseudomonas-infected potato plants, but not in Ppn-challenged tobacco. Differences both at the phenotypical level in 9-LOX-silenced infected plants and between the oxylipin profiles of the two pathosystems might be linked to tissue-specific, besides plant- or pathogen-specific, features.
In conclusion, from this work and previous reports, it appears that the 9-LOX/9-DES pathway is highly induced in Solanaceous plants upon challenge with microbial pathogens, notably oomycetes. Previous studies show that CA and CnA are able to inhibit the germination of Ppn zoospores in vitro besides possessing general antimicrobial activity and are therefore good candidates to act as defense compounds in the early stages of this interaction (Prost et al., 2005). With this work, we provided a demonstration of a measurable contribution by 9-LOX-derived oxylipins to resistance in vivo and of their role as phytoalexins. In tobacco, a picture emerges linking the early induction of phospholipase A2 (Roy et al., 1995; Dhondt et al., 2000), 9-LOX (Véronési et al., 1996), and 9-DES activities (this article), and the subsequent production of CA and CnA, pointing to an important function of the 9-LOX pathway. The question of whether other branches of the same pathway contribute to the resistance of tobacco to Ppn should be envisaged. However, given the data of the literature, it is unlikely that either 9-HPL or 9-AOS are crucial players in the altered phenotype of NtLOX1 antisense plants. Indeed, no HPL from Solanaceous plants has been described so far that metabolizes 9-LOX products. As to the 9-AOS branch recently reported in potato roots (Itoh et al., 2002; Stumpe et al., 2006), it appears that the relevant products are far less abundant than DVEs, especially if 9-HPOD derived (Stumpe et al., 2006); furthermore, their potential as antimicrobial and signaling molecules has not been investigated yet. In any case, because 9-LOX-derived oxylipins other than DVEs, mainly 9-HOD, are formed in root tissues during the interaction of wild type 46-8, but not of NtLOX1 antisense plants with Ppn race 0, their regulation needs be addressed in the future to further discriminate the relative contribution of different classes of compounds to resistance in planta.
MATERIALS AND METHODS
Biological Material
Tobacco (Nicotiana tabacum) plant line 46-8 and the derived NtLOX1 antisense line 125-2-1 (Rancé et al., 1998) were grown on vermiculite in a growth chamber for root inoculations and in soil for stem inoculations. A cell suspension culture of tobacco cv Wisconsin 38 was grown as reported (Rickauer et al., 1997). Phytophthora parasitica var. nicotianae (Ppn) race 0, to which wild-type tobacco 46-8 is resistant, was grown on oatmeal agar at 25°C in the dark (Fournier et al., 1993). An elicitor fraction was obtained by autoclaving a cell wall preparation of Ppn (Rickauer et al., 1997).
Treatment of Plant Material
Stem inoculations by Ppn were performed on 8-week-old plants as previously described (Rancé et al., 1998), whereas root inoculations were carried out on 5- to 6-week-old plants by dipping their roots in a zoospore suspension (106 total zoospores in 3 mL/plant). Both for the purpose of RNA extraction and oxylipin analysis, root tissue of five to eight plants per time point was collected. Elicitor treatments were carried out on 7-d-old tobacco cell cultures to which the Ppn elicitor was added to the final concentration of 30 μg/mL (Rickauer et al., 1997). In wounding experiments, the veins and interveinal tissues of mature detached leaves were crushed with forceps.
cDNA Cloning
PCR was performed with pairs of degenerated primers targeted to CYP74 hydroperoxide-converting enzymes, using an aliquot of a cDNA library from elicitor-treated tobacco cells in λ-ZAPII as template (Véronési et al., 1995). A DNA fragment of 1,131 bp (DESB3) was amplified with the primer pair: 5′-SAAYATGCCICCKGGCCC-3′ (HPCEf, forward) and 5′-TTICCIGCRCAYTGYTTRTT-3′ (HPCEr, reverse), with the following amplification program: 5-min denaturation at 94°C, 35 cycles of 30 s at 94°C, 30 s at 50°C, 1.5 min at 72°C, final extension step of 5 min at 72°C. DESB3 was A/T cloned into pGEM-T and sequenced. After α32P random primer labeling, DESB3 was used to probe plaque lifts of about 106 clones of the above-mentioned tobacco cDNA library. After three rounds of screening, 20 positive clones were converted into phagemids by in vivo excision. One of these clones, named DES1A, was retained for complete sequencing of its 1,641-bp insert. 5′-RACE analysis (5′-full RACE core set; TaKaRa Biomedicals) was performed according to the manufacturer's instructions on about 5 μg total RNA from elicited tobacco cells harvested 4 h after elicitor treatment and on 2.5 μg of poly(A)+ RNA isolated from tobacco cells elicitor treated for 15 h. The sequence of the phosphorylated gene-specific primer used for the initial reverse transcription-PCR step was 5′-GCCACCAATTTGTAG-3′. After RNA ligation of the resulting first-strand cDNA leading to circularization or concatemer formation, the gene-specific oligonucleotides used for further amplification of the unknown 5′-region were: pair 1, 5′-TGTTTGGTTTCATAGCAAAG-3′ (forward) and 5′-CTTCACCTTGTTTGTAAAAG-3′ (reverse); pair 2, 5′-CCACCGTTGTCAAAATCAAC-3′ (forward) and 5′-CGTATCGATCTTTAATGGCG-3′ (reverse). Amplified fragments were cloned into pGEM-T and sequenced.
Database searches were performed with the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST). Multiple alignments were carried out using ClustalW (http://www.ch.embnet.org/software/ClustalW.html) and Boxshade (http://www.ch.embnet.org/software/BOX_form.html) software. The nucleotide sequence reported in this article was deposited in GenBank (accession no. AF070976).
Expression of Recombinant NtDES1 and NtLOX1 in Escherichia coli
Forward and reverse primers were designed to amplify DNA fragments covering the NtDES1 and NtLOX1 open reading frames from cDNA clones DES1A and TL-J2, respectively (Véronési et al., 1995). An EcoRI and an NdeI site were added at the 5′-end of the DES1A- and TL-J2-derived PCR products, respectively. The reverse primer was targeted to the T7 promoter in the pBluescript II vector in both PCR reactions. PCR-modified products were cloned into the pGEM-T vector. The DES1A-modified fragment was subcloned via EcoRI and XhoI digestion into the pGEX-5X-3 expression vector (Pharmacia) to generate a GST-NtDES1 fusion protein. The TL-J2-derived fragment was subcloned via NdeI and ScaI partial digestion into the pT7-7 expression vector (Novagen) to produce untagged NtLOX1. The resulting pGEX-NtDES1 and pT7-NtLOX1 constructs were entirely sequenced and transferred into Escherichia coli strain BL21. Expression of the recombinant proteins was induced by treating log-phase bacteria with 0.1 mm IPTG for pGEX-NtDES1 or 0.4 mm IPTG for pT7-NtLOX1. Soluble protein extracts to be used as a crude source of enzyme were prepared from induced cultures by spinning bacteria down by centrifugation and suspending them in phosphate-buffered saline (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.3, 50 μL/mL culture) or in 100 mm sodium phosphate buffer, pH 6.5 (100 μL/mL culture), then lysing by sonication on ice for three cycles of 30 s. Affinity purification of the GST-NtDES1 protein was performed in batch on glutathione-Sepharose 4B (Pharmacia) with proteins solubilized in 1× phosphate-buffered saline. The fusion protein fixed on the affinity matrix was either eluted from the matrix in reduced glutathione elution buffer (10 mm reduced glutathione in 50 mm Tris-HCl, pH 8.0) or directly digested with 50 μg/mL factor Xa (Boehringer Mannheim) in 50 mm Tris-HCl, 150 mm NaCl, 1 mm CaCl2, pH 7.5, for 2 to 4 h under gentle shaking, leading to the release of GST-free NtDES1 in the supernatant after centrifugation. Aliquots from all purification steps were electrophoresed under denaturing conditions (10% polyacrylamide gel) and stained either by the silver nitrate method or with Coomassie Blue.
Extraction of RNA and Northern Analysis
Total RNA was extracted from shock-frozen plant material by TRIzol (Invitrogen). Twenty micrograms of total RNA were subjected to denaturing electrophoresis in a 1.2% agarose gel in 2.2 m formaldehyde and 20 mm MOPS, 8 mm sodium acetate, 1 mm EDTA, pH 7.0, and in the presence of ethidium bromide to check for equal loading. After destaining, RNA was passively transferred onto nylon membranes (Hybond; Amersham) and fixed at 80°C for 1 h or by UV cross linking. Membranes were hybridized with α32P-labeled probes (Appligene oligolabeling random priming kit) in hybridization buffer (50 mm PIPES, 100 mm NaCl, 50 mm sodium phosphate buffer, pH 6.5, 1 mm EDTA, 5% [w/v] SDS) at 60°C after prehybridization in the same buffer, and washed in 5% (w/v) SDS, 1× SSC at 60°C.
Hydroperoxide-Converting Activity Assay
Decrease in A234 nm was monitored in a 400-μL reaction containing 50 μm pure fatty acid hydroperoxide (Cayman Chemicals) in 50 mm sodium phosphate buffer, pH 7.0, for 2 min following addition of 1 to 20 μL of GST-free NtDES1. Activity was calculated from the initial reaction rate and expressed as nkatal/mg protein. Proteins were quantified with the Bio-Rad protein assay.
Product Analysis and TLC Assay
Products of the in vitro enzymatic assay were analyzed as methyl ester derivatives by GC-MS. Briefly, 100 μL of total protein extracts (corresponding to 4 mL of IPTG-induced bacterial cultures, OD600 nm = 0.6) in borate buffer, 100 mm, pH 9.0, were incubated for 30 min at 30°C in the presence of 200 nmol of 9- or 13-HPOD in 2 mL of the same buffer. The reaction was stopped by acidification to pH 4.0 by 1 m citric acid, derivatized and analyzed by GC-MS by standard methods (Weber et al., 1997). For TLC assays (Caldelari and Farmer, 1998), soluble protein extracts from IPTG-induced recombinant strains of E. coli in phosphate buffer were used as source of enzymes. Enzymatic reactions proceeded for 10 min at 30°C in 10 mL (final volume) of 100 mm sodium phosphate buffer, pH 6.5, 360 μm LA or LnA (Larodan), and 10% glycerol. Reactions were initiated adding an aliquot (1 mL) of soluble proteins and stopped by acidification to pH 4.0 with 1 m citric acid, then extracted twice with diethyl ether. The organic layer was dried on anhydrous MgSO4 and concentrated under a stream of nitrogen down to approximately 50 μL. These extracts were submitted to TLC on silica plates with concentrating zone (Merck) and hexane:diethyl ether:formic acid (70:30:1 [v/v/v]) as the mobile phase. Unsaturated compounds were revealed with iodine vapor. CA and CnA standards were prepared as previously described (Weber et al., 1999).
Oxylipin Profiling
Oxylipin analysis was performed as described, with some modifications (Göbel et al., 2001). For each experiment, 1.5 g of frozen roots from 8-week-old tobacco plants (five to eight plants per time point) were ground in liquid nitrogen and extracted by adding 10 mL of extraction medium (n-hexane:2-propanol, 3:2 [v/v] with 0.0025% [w/v] butylated hydroxytoluene) and immediately homogenized with an Ultra Turrax under a stream of argon on ice for 30 s. The extract was centrifuged at 3,200g at 4°C for 10 min. The clear upper phase was dried under nitrogen stream, redissolved in 1 mL isopropanol:chloroform (1:2 [v/v]), and applied to an aminopropyl column (Supelclean LC-NH2 3 mL SPE tubes; Supelco). The subsequent fraction was eluted with 9 mL diethylether:acetic acid (98:2 [v/v]) and dried under nitrogen stream. Samples were dissolved in 80 μL methanol:water:acetic acid (75:25:0.1 [v/v/v]). Analysis of oxylipins was carried out on an Agilent 1100 HPLC system coupled to a diode array detector. At first, oxylipins were purified on reversed phase-HPLC on an ET250/2 Nucleosil 120 to 5 C18 column (2.1 × 250 mm, 5-μm particle size; Macherey-Nagel), with a solvent system methanol:water:acetic acid (85:15:0.1 [v/v/v]) and a flow rate of 0.18 mL min−1. For detection of hydroxy fatty acids, A234 nm indicating the conjugated diene system was recorded. DVEs and keto dienoic fatty acids were detected by monitoring A250 nm and A272 nm, respectively. For quantification of the hydroxy and keto fatty acids and for purification of the oPDA/JA- and CA/CnA-containing fraction, respectively, straight phase-HPLC was carried out on a Zorbax Rx-SIL column (2.1 × 150 mm, 5-μm particle size; Agilent) with a solvent system of n-hexane:2-propanol:trifluoroacetic acid (100:1:0.02 [v/v/v]), and a flow rate of 0.2 mL min−1. oPDA and JA were quantified by GC-MS as described (Göbel et al., 2002). For quantification of CA and CnA, a second reversed-phase-HPLC analysis was then performed using an ET 250/2 Nucleosil 120 to 5 C18 column (2.1 × 250 mm, 5-μm particle size; Macherey-Nagel) with a solvent system methanol:water:acetic acid (90:10:0.1 [v/v/v]), and a flow rate of 0.18 mL min−1. For quantification, 13-γ-HOT was added to each sample as internal standard and calibration curves (five-point measurements) were established. The experiment was repeated twice for NtLOX1 antisense plants and three times for wild-type plants.
Subcellular Localization of Fluorescent Protein-Tagged NtDES1 and NtLOX1
For fluorescent protein-tagging experiments, the open reading frames of NtDES1 and NtLOX1 were cloned into plant expression vectors pGreenII0229-2 × 35S-TL and pGreenII0029-2 × 35S-TL, respectively (Hellens et al., 2000), downstream of an enhanced cauliflower mosaic virus 35S promoter in fusion with the tobacco etch virus translational leader (Restrepo et al., 1990) using NcoI/NotI sites inserted by PCR at the 5′ and 3′ ends of the coding sequences. All amplicons were fully sequenced before further subcloning. YFP or CFP were then inserted via NotI downstream of NtDES1 and NtLOX1, respectively. Cultures of transformed Agrobacterium tumefaciens strain GV3101∷pMP90 were grown overnight at 28°C from individual colonies in 2 mL of YEB medium (0.5% [w/v] beef extract, 0.1% [w/v] yeast extract, 0.5% [w/v] peptone, 0.5% [w/v] saccharose, 2 mm MgSO4) containing rifampicin (20 μg mL−1) and kanamycin (50 μg mL−1). Four hundred microliters of cell culture were pelleted by centrifugation and resuspended in 2 mL of induction medium [60 mm K2HPO4, 33 mm KH2PO4, 7.5 mm (NH4)2SO4, 1 mm MgSO4, 0.2% (w/v) Glc, 0.5% (v/v) glycerol, 50 μm acetosyringone, 10 mm morpholine ethane sulfonic acid (MES), pH 5.6]. After overnight incubation at 28°C, cells were pelleted again, washed in Murashige and Skoog medium containing 10 mm MES, pH 5.6, and resuspended to A600 nm = 0.5 in Murashige and Skoog-MES with 150 μm acetosyringone. Young, fully expanded leaves were used for agroinfiltration; the C-terminal translational fusion proteins NtDES1-YFP and NtLOX1-CFP were visualized 3 d after on a Leica TCS SP2 confocal microscope, using a long-distance 40× water-immersion objective (HCX Apo 0.80). The Ar laser bands of 514 and 458 nm were used to excite YFP and CFP, respectively. The two signals were detected at the specific emission windows of 525 to 600 nm for YFP and 466 to 550 nm for CFP; the former channel was then false colored in green to maximize definition in RGB acquisition mode. Chlorophyll fluorescence was detected in red (633–771 nm) with both excitation wavelengths. PGreenII0229-2 × 35S-TL-YFP was used to transform control leaves under the same conditions.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AF070976.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence of NtDES1.
Supplemental Figure S2. NtDES1 transcript accumulation upon biotic stress.
Supplementary Material
Acknowledgments
We are grateful to K. Matsui and D. Werck-Reichhart for kindly providing the pepper HPL and the guayule RPP clones, respectively. We also thank D. Valentino, L. Plizzari, F. Carbonne, S. Freitag, and P. Sanchez for technical help and G. Tamietti for continuous support. A.F. ran all bioassays with transgenic plants and analyzed NtLOX1/NtDES1 expression and oxylipins in roots. F.C. performed all molecular biology work, studied NtDES1 localization and expression, and purified recombinant NtDES1 together with J.F. C.G. performed oxylipin profiling with A.F., confirmed oxylipins by GC-MS, and analyzed data. L.M.-S. confirmed NtDES1 activity and performed MS of CA and TLC test. J.F., M.T.E.-T., I.F., and F.C. planned and supervised the work, planned the concept of the article, and wrote the manuscript.
This work was supported by a predoctoral fellowship from the Ministero della Pubblica Istruzione Università e Ricerca and the Università Italo-Francese (travel grant to A.F.), by the Italian Consiglio Nazionale della Ricerche (grant no. 203.12.20/1 code 12.03.02, RAISA to F.C.), and by the Deutsche Forschungsgemeinschaft (grant to I.F.). L.M.-S. was supported by a predoctoral fellowship from the French Ministère de l'Education Nationale, de la Recherche et de la Technologie. M.-T.E-T, J.F., and L.M.-S. were supported by the Ministère de l'Education Nationale de l'Enseignement Supérieur et de la Recherche and the Centre National de la Recherche Scientifique (France).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Francesca Cardinale (francesca.cardinale@unito.it).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Blée E (1998) Phytooxylipins and plant defense reactions. Prog Lipid Res 37 33–72 [DOI] [PubMed] [Google Scholar]
- Blée E (2002) Impact of phyto-oxylipins in plant defense. Trends Plant Sci 7 315–322 [DOI] [PubMed] [Google Scholar]
- Caldelari D, Farmer EE (1998) A rapid assay for the coupled cell free generation of oxylipins. Phytochemistry 47 599–604 [Google Scholar]
- Chapple C (1998) Molecular-genetic analysis of plant cytochrome p450-dependent monooxygenases. Annu Rev Plant Physiol 43 311–343 [DOI] [PubMed] [Google Scholar]
- Corey EJ, Nagata R, Wright SW (1987) Biomimetic total synthesis of colneleic acid and its function as a lipoxygenase inhibitor. Tetrahedron Lett 28 4917–4920 [Google Scholar]
- Dhondt S, Geoffroy P, Stelmach BA, Legrand M, Heitz T (2000) Soluble phospholipase A2 activity is induced before oxylipin accumulation in tobacco mosaic virus-infected tobacco leaves and is contributed by patatin-like enzymes. Plant J 23 431–440 [DOI] [PubMed] [Google Scholar]
- Feussner I, Wasternack C (2002) The lipoxygenase pathway. Annu Rev Plant Biol 53 275–297 [DOI] [PubMed] [Google Scholar]
- Fidantsef AL, Bostock RM (1998) Characterization of potato tuber lipoxygenase cDNAs and lipoxygenase expression in potato tubers and leaves. Physiol Plant 102 257–271 [Google Scholar]
- Fournier J, Pouénat M-L, Rickauer M, Rabinovitch-Chable H, Rigaud M, Esquerré-Tugayé M-T (1993) Purification and characterization of elicitor-induced lipoxygenase in tobacco cells. Plant J 3 63–70 [Google Scholar]
- Galliard T, Phillips DR (1972) Enzymic conversion of linoleic acid into 9-(nona-1′,3′-dienoxy)non-8-enoic acid, a novel unsaturated ether derivative isolated from homogenates of Solanum tuberosum tubers. Biochem J 129 743–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göbel C, Feussner I, Hamberg M, Rosahl S (2002) Oxylipin profiling in pathogen-infected potato leaves. Biochim Biophys Acta 1584 55–64 [DOI] [PubMed] [Google Scholar]
- Göbel C, Feussner I, Rosahl S (2003) Lipid peroxidation during the hypersensitive response in potato in the absence of 9-lipoxygenases. J Biol Chem 278 52834–52840 [DOI] [PubMed] [Google Scholar]
- Göbel C, Feussner I, Schmidt A, Scheel D, Sanchez-Serrano J, Hamberg M, Rosahl S (2001) Oxylipin profiling reveals the preferential stimulation of the 9-lipoxygenase pathway in elicitor-treated potato cells. J Biol Chem 276 6267–6273 [DOI] [PubMed] [Google Scholar]
- Grechkin AN (2002) Hydroperoxide lyase and divinyl ether synthase. Prostaglandins Other Lipid Mediat 68-69 457–470 [DOI] [PubMed] [Google Scholar]
- Grechkin AN, Fazliev FN, Mukhtarova LS (1995) The lipoxygenase pathway in garlic (Allium sativum L.) bulbs: detection of the novel divinyl ether oxylipins. FEBS Lett 371 159–162 [DOI] [PubMed] [Google Scholar]
- Hamberg M (1998) A pathway for biosynthesis of divinyl ether fatty acids in green leaves. Lipids 33 1061–1071 [DOI] [PubMed] [Google Scholar]
- Hamberg M, Sanz A, Castresana C (1999) α-Oxidation of fatty acids in higher plants: identification of a pathogen-inducible oxygenase (PIOX) as an α-dioxygenase and biosynthesis of 2-hydroperoxylinolenic acid. J Biol Chem 274 24503–24513 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM (2000) pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol Biol 42 819–832 [DOI] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL (2002) Oxylipin metabolism in response to stress. Curr Opin Plant Biol 5 230–236 [DOI] [PubMed] [Google Scholar]
- Itoh A, Howe GA (2001) Molecular cloning of a divinyl ether synthase: identification as a CYP74 cytochrome P-450. J Biol Chem 276 3620–3627 [DOI] [PubMed] [Google Scholar]
- Itoh A, Schilmiller AL, McCaig BC, Howe GA (2002) Identification of a jasmonate-regulated allene oxide synthase that metabolizes 9-hydroperoxides of linoleic and linolenic acids. J Biol Chem 277 46051–46058 [DOI] [PubMed] [Google Scholar]
- Kalb VF, Loper JC (1988) Proteins from eight eukaryotic cytochrome P-450 families share a segmented region of sequence similarity. Proc Natl Acad Sci USA 85 7221–7225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight VI, Wang H, Lincoln JE, Lulai EC, Gilchrist DG, Bostock RM (2001) Hydroperoxides of fatty acids induce programmed cell death in tomato protoplasts. Physiol Mol Plant Pathol 59 277–286 [Google Scholar]
- Kolomiets MV, Chen H, Gladon RJ, Braun EJ, Hannapel DJ (2000) A leaf lipoxygenase of potato induced specifically by pathogen infection. Plant Physiol 124 1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudert D, Schaller F, Weiler EW (2000) Transgenic Nicotiana tabacum and Arabidopsis thaliana plants overexpressing allene oxide synthase. Planta 211 163–165 [DOI] [PubMed] [Google Scholar]
- Matsui K, Shibutani M, Hase T, Kajiwara T (1996) Bell pepper fruit fatty acid hydroperoxide lyase is a cytochrome P450 (CYP74B). FEBS Lett 394 21–24 [DOI] [PubMed] [Google Scholar]
- Maucher H, Hause B, Feussner I, Ziegler J, Wasternack C (2000) Allene oxide synthases of barley (Hordeum vulgare cv. Salome): tissue specific regulation in seedling development. Plant J 21 199–213 [DOI] [PubMed] [Google Scholar]
- Mène-Saffrané L, Esquerré-Tugayé M-T, Fournier J (2003) Constitutive expression of an inducible lipoxygenase in transgenic tobacco decreases susceptibility to Phytophthora parasitica var. nicotianae. Mol Breed 12 271–282 [Google Scholar]
- Montillet JL, Chamnongpol S, Rusterucci C, Dat J, van de Cotte B, Agnel JP, Battesti C, Inze D, Van Breusegem F, Triantaphylides C (2005) Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol 138 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Durst F, Werck-Reichhart D, Gardner HW, Camara B, Cornish K, Backhaus RA (1995) The major protein of guayule rubber particles is a cytochrome P450: characterization based on cDNA cloning and spectroscopic analysis of the solubilized enzyme and its reaction products. J Biol Chem 270 8487–8494 [DOI] [PubMed] [Google Scholar]
- Porta H, Rocha-Sosa M (2002) Plant lipoxygenases: physiological and molecular features. Plant Physiol 130 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prost I, Dhondt S, Rothe G, Vicente J, Rodriguez MJ, Kift N, Carbonne F, Griffiths G, Esquerre-Tugaye MT, Rosahl S, et al (2005) Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol 139 1902–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancé I, Fournier J, Esquerré-Tugayé M-T (1998) The incompatible interaction between Phytophthora parasitica var. nicotianae race 0 and tobacco is suppressed in transgenic plants expressing antisense lipoxygenase sequences. Proc Natl Acad Sci USA 95 6554–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickauer M, Brodschelm W, Bottin A, Véronési C, Grimal H, Esquerré-Tugayé M-T (1997) The jasmonate pathway is involved differentially in the regulation of different defence responses in tobacco cells. Planta 202 155–162 [Google Scholar]
- Roy S, Pouénat M-L, Caumont C, Cariven C, Prévost M-C, Esquerré-Tugayé M-T (1995) Phospholipase activity and phospholipid patterns in tobacco cells treated with fungal elicitor. Plant Sci 107 17–25 [Google Scholar]
- Rustérucci C, Montillet JL, Agnel JP, Battesti C, Alonso B, Knoll A, Bessoule JJ, Etienne P, Suty L, Blein JP, et al (1999) Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J Biol Chem 274 36446–36455 [DOI] [PubMed] [Google Scholar]
- Shah J (2005) Lipids, lipases, and lipid-modifying enzymes in plant disease resistance. Annu Rev Phytopathol 43 229–260 [DOI] [PubMed] [Google Scholar]
- Somerville C, Browse J, Jaworski JG, Ohlrogge J (2000) Lipids. In BB Buchanan, W Gruissem, R Jones, eds, Biochemistry and Molecular Biology of Plants. American Society of Plant Physiologists, Rockville, MD, pp 456–527
- Song WC, Funk CD, Brash AR (1993) Molecular cloning of an allene oxide synthase: a cytochrome P-450 specialized for the metabolism of fatty acid hydroperoxides. Proc Natl Acad Sci USA 90 8519–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpe M, Göbel C, Demchenko K, Hoffmann M, Klosgen RB, Pawlowski K, Feussner I (2006) Identification of an allene oxide synthase (CYP74C) that leads to formation of alpha-ketols from 9-hydroperoxides of linoleic and linolenic acid in below-ground organs of potato. Plant J 47 883–896 [DOI] [PubMed] [Google Scholar]
- Stumpe M, Kandzia R, Gobel C, Rosahl S, Feussner I (2001) A pathogen-inducible divinyl ether synthase (CYP74D) from elicitor-treated potato suspension cells. FEBS Lett 507 371–376 [DOI] [PubMed] [Google Scholar]
- Véronési C, Rickauer M, Fournier J, Marolda M, Esquerré-Tugayé M-T (1995) Nucleotide sequence of an elicitor-induced tobacco lipoxygenase cDNA (GenBank X84040). Plant Physiol 108 1342 [Google Scholar]
- Véronési C, Rickauer M, Fournier J, Pouénat M-L, Esquerré-Tugayé M-T (1996) Lipoxygenase gene expression in the tobacco-Phytophthora parasitica nicotianae interaction. Plant Physiol 112 997–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba-Mateos F, Rickauer M, Esquerré-Tugayé M-T (1997) Cloning and characterization of a cDNA encoding an elicitor of Phytophthora parasitica var. nicotianae that shows cellulose-binding and lectin-like activities. Mol Plant-Microbe Interact 10 1045–1053 [DOI] [PubMed] [Google Scholar]
- Weber H, Chetelat A, Caldelari D, Farmer EE (1999) Divinyl ether fatty acid synthesis in late blight-diseased potato leaves. Plant Cell 11 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H, Vick BA, Farmer EE (1997) Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proc Natl Acad Sci USA 94 10473–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler J, Keinanen M, Baldwin IT (2001) Herbivore-induced allene oxide synthase transcripts and jasmonic acid in Nicotiana attenuata. Phytochemistry 58 729–738 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.