Abstract
We have used cDNA microarray analysis to examine changes in gene expression during embryo development in soybean (Glycine max) and to compare gene expression profiles of two soybean cultivars that differ in seed isoflavonoid content. The analysis identified 5,910 genes that were differentially expressed in both soybean cultivars grown at two different locations for two consecutive years in one of the five different stages of embryo development. An ANOVA analysis with P value < 0.05 and < 0.01 indicated that gene expression changes due to environmental factors are greater than those due to cultivar differences. Most changes in gene expression occurred at the stages when the embryos were at 30 or 70 d after pollination. A significantly larger fraction of genes (48.5%) was expressed throughout the development and showed little or no change in expression. Transcript accumulation for genes related to the biosynthesis of storage components in soybean embryos showed several unique temporal expressions. Expression patterns of several genes involved in isoflavonoid biosynthesis, such as Phenylalanine Ammonia-Lyase, Chalcone Synthase (CHS) 7, CHS8, and Isoflavone Synthase2, were higher at 70 d after pollination in both the cultivars. Thus, expression of these genes coincides with the onset of accumulation of isoflavonoids in the embryos. A comparative analysis of genes involved in isoflavonoid biosynthesis in RCAT Angora (high seed isoflavonoid cultivar) and Harovinton (low seed isoflavonoid cultivar) revealed that CHS7 and CHS8 were expressed at significantly greater level in RCAT Angora than in Harovinton. Our study provides a detailed transcriptome profiling of soybean embryos during development and indicates that differences in the level of seed isoflavonoids between these two cultivars could be as a result of differential expression of CHS7 and CHS8 during late stages of seed development.
Soybean (Glycine max) L. Merr. is the world's most widely grown grain legume. It combines in one crop both the major supply of vegetable oil and protein, with a variety of uses in human food and animal feeds. Soybean also contains eight essential amino acids that are crucial for human nutrition and are not made naturally in the body (Carpenter et al., 2002).
Soybean seed is virtually devoid of endosperm and comprises a well-developed embryo and a surrounding seed coat. During embryo development, the fertilized egg cell differentiates into a mature embryo containing cells with different roles. The entire embryogenesis can be divided into five stages: globular, heart, cotyledon, maturation, and dormancy (Walbot, 1978). Each stage consists of unique morphogenic, cellular, and physiological events that are determined by changes in gene expression patterns. A complex regulatory network activates commencement of maturation and accumulation of storage products in the seeds. The whole process includes transcription and physiological reprogramming reconciled by many different pathways (Wobus and Weber, 1999). During maturation, embryonic cells synthesize a substantial amount of proteins and secondary metabolites. For example, storage proteins, lectins, trypsin inhibitors (Orthoefer, 1978), and isoflavonoids (Dhaubhadel et al., 2003) are made and accumulated in developing embryos.
Soybean seed has a unique chemical composition that makes it a valuable industrial and agricultural commodity. It contains 40% of the seed dry weight as proteins. Soybean seeds are a rich source of isoflavonoids that are associated with many health benefits (Dixon and Ferreira, 2002). The aim to optimize the protein and isoflavonoid content, and thereby improve the overall nutritional quality of soybean seeds, has received a lot of attention recently. Genetic and environmental factors control the synthesis of these compounds and their accumulation in the mature seed. A complex regulation and synchronization of various biosynthetic pathways during different stages of embryo development is critical to obtain a definite composition of seed reserves. We are interested in understanding the transcriptional changes during embryo development that may control seed quality in soybean. A natural starting point for this work would be to undertake an in-depth study of transcriptome changes during the seed development in soybean. Microarray technology offers an opportunity to accomplish this (Schena et al., 1995) and has been adopted successfully to study the changes in gene expression during seed filling in rice (Oryza sativa; Zhu et al., 2003), maize (Zea mays; Lee et al., 2002), Arabidopsis (Arabidopsis thaliana; Ruuska et al., 2002; Hennig et al., 2004), and tomato (Lycopersicon esculentum; Alba et al., 2005).
In this study, we have examined the temporal changes in gene expression during embryo development in soybean when seeds undergo major changes in metabolism. Soybean cDNA microarray chips consisting of over 18,000 cDNAs were used to define patterns of gene expression during five different embryo developmental stages in two soybean cultivars. We describe here the global gene expression profile during embryo development in soybean and discuss the transcriptional networks that synchronize the response to developmental programs, leading to the production of various components that accumulate in seeds. We also portray a detailed comparison of gene expression between soybean cultivars that differ in the seed isoflavonoid content and discuss the potential regulation points for isoflavonoid synthesis in soybean seeds.
RESULTS
The 18 K-A microarray slides were obtained from Dr. Lila Vodkin, University of Illinois (Vodkin et al., 2004). The arrays were comprised of 18,462 single-spotted PCR products from cDNAs of the low redundancy Gm-c1021, Gm-c1083, and Gm-c1070 unigene cDNAs and 64 choice clones spotted multiple times on the slide. The unigene set represented on the array provides highly representative mRNAs expressed in roots of seedlings and adult plants, flower buds, flowers, pods, and various stages of immature embryos and seed coats. These microarray slides have platform GPL3015 in the Gene Expression Omnibus (GEO).
We focused our study on five different developmental stages of embryos from two different soybean cultivars (RCAT Angora and Harovinton) to examine the steady-state transcript abundance. These stages included the early maturation stage as represented by 30, 40, and 50 d after pollination (DAP) to late maturation stage (60 and 70 DAP), when seeds had attained their full size but the process of desiccation was not complete (Fig. 1). A decrease in chlorophyll content of the embryos was noticed at 50 DAP. Embryos were collected from plants grown at two different locations (London and Delhi, Ontario, Canada) for two consecutive years (2003 and 2004). A dye swap and four biological replications were conducted per gene for each developmental stage to minimize the technical and biological variations, respectively. A total of 32 analyses per choice clone were performed, because they were represented four times in each array. The data analysis was done using computer software (GeneSpring v7.3, Agilent Technologies), and normalization was performed by per spot, per chip, intensity-dependent LOWESS. Only those genes that showed intensity >10 were combined together and selected for further analysis. This method allowed increased reliability in expression analysis.
Figure 1.
Developmental stages of soybean embryos during the study period. Representative embryos from early, mid, and late maturity (before complete desiccation) are shown. Numbers at the bottom indicate embryo developmental stages in DAP.
Effect of Environment and Cultivar Difference on Gene Expression
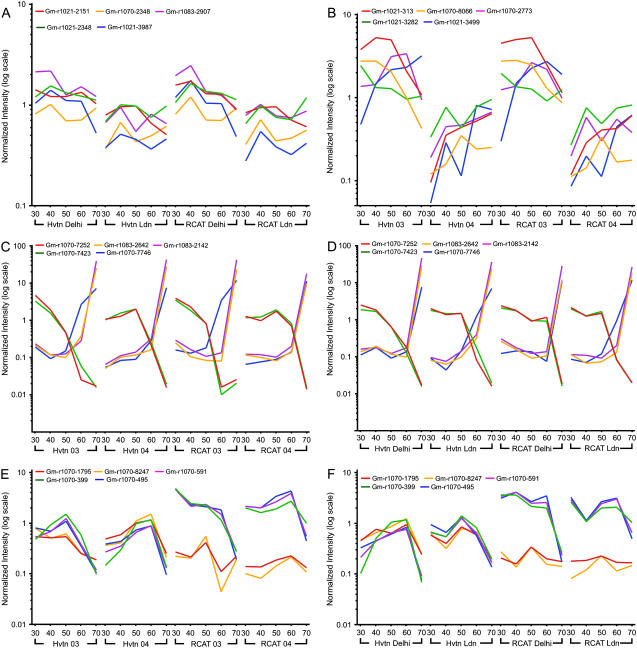
One of the major objectives of this research was to delineate the temporal patterns of gene expression during embryo development in soybean and interpret the results in the context of our existing knowledge of seed development and storage reserve accumulation. We identified several developmentally regulated genes by hybridizing the microarrays with mRNA samples from five distinct stages of seed development. A total of 11,480 genes showed intensities >10 at any time point during the development. These genes were chosen for further analysis by examining their hybridization intensities using a single channel method. An ANOVA test with P value < 0.05 and < 0.01 with different parameters was performed to calculate the number of genes that show a significant difference in their expression. An estimation of the effect of environmental factors on gene expression is summarized in Table I and Figure 2. A total of five representative genes (P < 0.01) were chosen from each parameter to demonstrate the common pattern of gene expression during the development. The results show that cultivar differences have very little effect compared to either the location or the growing season effects. The general patterns of gene expression were very similar in RCAT Angora and Harovinton for plants that were grown at the same location in different years, or in the same year at different locations, during different developmental stages (Fig. 2, A and B). A hierarchical clustering analysis using one-way ANOVA also grouped the cultivars together for particular developmental stages irrespective of location or the year (data not shown), confirming the greater effect of environmental factors on gene expression. A large difference in the temperature and precipitation was not observed between the locations during two growing seasons (Supplemental Table S1). However, the soil composition of the two locations was very different. Delhi soil had a significantly higher amount of sand compared to London (Table II). When ANOVA analysis was performed on developmental stages, genes were selected that displayed consistent developmental changes in their expression profiles. Thus, for this set of genes, only small differences were observed between the different locations and years for both RCAT Angora and Harovinton. The representative genes in Figure 2, C and D for both the cultivars showed a similar pattern of expression during embryo development irrespective of environmental conditions. The ANOVA analysis on cultivar was the most restrictive filter and resulted in the selection of only five genes. These genes have the highest probability of differential expression between the cultivars at all five developmental time points that were sampled (Fig. 2, E and F). Other cultivar-specific gene expression profiles that are developmentally specific were identified by further analyses (below).
Table I.
Number of differentially expressed genes in developing soybean embryos
An ANOVA test was performed on 11,480 genes with different parameters and P values.
| Parameters | P < 0.05 | P < 0.01 |
|---|---|---|
| Developmental stage | 6,295 | 4,488 |
| Location | 526 | 88 |
| Year | 99 | 19 |
| Cultivar | 5 | 5 |
Figure 2.
Selected genes showing the greatest level of change in expression based on location, year, developmental stages, and cultivars. An ANOVA analysis with P value < 0.01 was performed on 11,480 genes that showed intensity >10 at least once during the development in soybean cultivars RCAT Angora (RCAT) and Harovinton (Hvtn). The analysis was performed using different parameters. A, Location. B, Year. C, Embryo developmental stage expressed in year. D, Embryo developmental stage expressed in location. E, Cultivars expressed in year. F, Cultivars expressed in location. Shown above are five genes from each analysis with the most significant P value that display various expression profiles when the RCAT and Hvtn were grown at London (Ldn) and Delhi or in year 2003 (03) and 2004 (04). Identity of genes with blast hit and cDNA source are: A, DNA polymerase λ (Gm-r1083-2907), pectinesterase (Gm-r1083-3987), hypothetical protein (Gm-r1070-9004), oligouridylate binding protein (Gm-r1021-2348), Asp aminotransferase (Gm-r1021-2151); B, hypothetical protein (Gm-r1070-8066, Gm-r1070-2773, Gm-r1021-3499), fiber protein Fb15 (Gm-r1021-3282), and ribosomal protein L28 (Gm-r1021-313); C and D, imbibition protein (Gm-r1083-2642), dehydrogenase (Gm-r1083-2142), splicing factor PRP8 homolog (Gm-r1070-7746), polyprotein (Gm-r1070-7423), annexin (Gm-r1070-7252); E and F, NPR (Gm-r1070-495), BBI (Gm-r1070-399), ARF 17 (Gm-r1070-8247), and hypothetical protein (Gm-r1070-591, Gm-r1070-1795).
Table II.
Soil characteristics of London and Delhi, Ontario
| Sand | Silt | Clay | |
|---|---|---|---|
| London | 35.60% | 48.70% | 15.70% |
| Delhi | 87.00% | 8.00% | 5.00% |
Cluster Analysis of 5,910 Differentially Expressed cDNAs in Developing Soybean Embryos
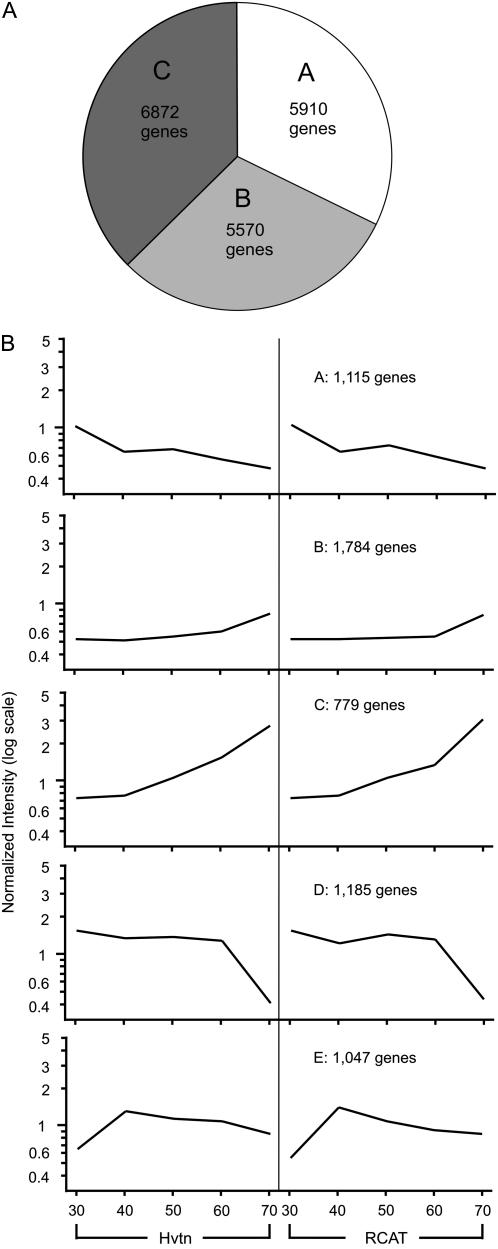
Our analysis grouped the total number of genes on the array into three categories based on the intensity and differential expression (Fig. 3A). Using single channel intensities for each cultivar, data was normalized per chip to the 50th percentile and then per gene to the median of the measurement for that gene, with a cutoff value >0.5 and <2.0 in all developmental stages for both cultivars. Only the genes that passed the filtering criteria were considered as genes that changed in expression based upon the experimental conditions. A total of 5,910 genes changed in their expression in all the biological replicates in at least one embryo developmental stage used in the study. Only those genes with intensities >10 were included in this group. A group of 5,570 genes showed intensities >10; however, their expression did not change significantly during the development. Another group of 6,872 genes changed in their expression with time but possessed very low intensity. A complete list of genes belonging to each of three groups is deposited in GEO (see “Materials and Methods”). To acquire an overall picture of gene expression changes, we clustered 5,910 differentially expressed genes by a k-means analysis. This separated the differentially expressed genes into five sets according to their profiles irrespective of locations or growing seasons. The general hypothesis of k-means cluster analysis is that genes involved in a similar function or common metabolic pathway will have similar profile of expression and thus likely to be grouped into the same group. The analysis was carried out for RCAT Angora and Harovinton separately. These cultivars do not possess a close common lineage that is evident from their pedigree information (Buzzell et al., 1991; G. Ablett, personal communication). Shown in Figure 3B is the average pattern of the genes that are included in the specific group. Each group consisted of several diverse genes with some functional correlations. Group A included genes whose transcripts accumulated to moderate level at 30 DAP and then remained lower after that during the development. Many cell wall-related genes, receptor kinases, Leu-rich repeat family proteins, homologs of genes encoding polyubiquitin, Suc synthase, and vacuolar protein sorting were included in group A.
Figure 3.
Cluster analysis of differentially expressed genes in soybean embryos. A, Grouping of 18 K genes into three categories. Category A included 5,910 genes that changed in their level at least once during the embryo development and had intensity >10. Category B included 5,570 genes that showed intensity >10 but level of gene expression did not change during embryo development, and category C included 6,872 genes that were differentially regulated but showed the intensity <10. B, Cluster analysis of 5,910 differentially regulated genes in RCAT Angora and Harovinton. The genes were classified using the k-means technique into five groups. Genes belonging to each group were averaged together and presented. The y axis is the normalized level of expression as a function of developmental stages (30, 40, 50, 60, or 70 DAP) in RCAT Angora (RCAT) and Harovinton (Hvtn).
Genes in groups B and C showed similar expression profiles in that they contain genes that were up-regulated at the later stages of embryo development. However, a considerable increase in the level of gene expression was noticed for the genes included in group C at 70 DAP. Genes encoding minor allergen, homolog to zinc binding protein, oxalyl-CoA decarboxylase, pathogen inducible trypsin inhibitor-like protein, and calmodulin are included in group B, while group C included genes such as a lipid transfer protein precursor, late embryogenesis abundant proteins, desiccation protective protein, catalase 4, Pro-rich protein, plasma membrane Ca2+ ATPase, and many transcription factor genes such as TATA box binding protein, APETALA2 domain-containing protein, and WRKY family transcription factors. This group included many genes that are required for seed maturity or are reported to accumulate during embryo maturation. For example, maturity related protein, many Cyt P450, senescence related proteins, receptor kinases, and ethylene responsive proteins were all within this group. Most of the genes participating in isoflavonoid biosynthesis are also clustered in group C.
Unlike the gene expression patterns for the genes belonging to group B and C, group D included genes that did not show major change in the level of expression from 30 to 60 DAP followed by a dramatic decrease in the level of transcript accumulation at 70 DAP. Examples include several chlorophyll a/b-binding proteins, β-conglycinin, Gly-rich protein, Bowman-Birk type protease inhibitor, P34 allergen protein, Rubisco small subunit, and brassinosteroid up-regulated protein 1 precursor.
Group E illustrates the expression patterns characteristic for a collection of genes that increased gradually in their transcript accumulation from 30 to 40 DAP followed by a slight increase or decrease in expression or reached a plateau. Examples include genes encoding lipooxygenase, WRKY family transcription factor, xyloglucan endotransglucosylase/hydrolase, seed calcium dependent protein kinase, and putative ATP-binding cassette transporters.
Expression Profiles of Genes Involved in Storage Product and Isoflavonoid Biosynthesis
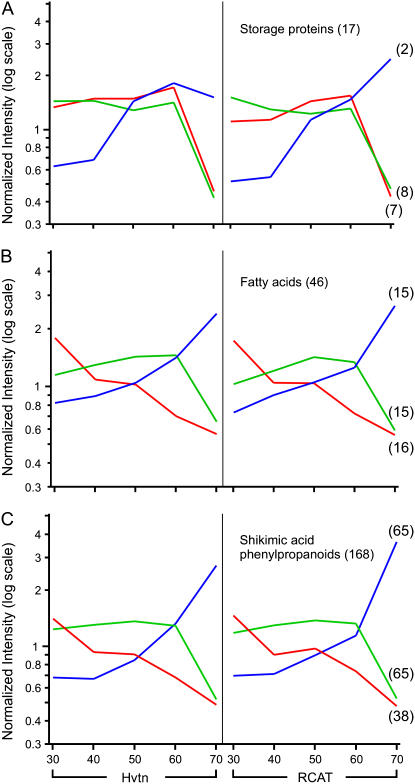
The major storage products of soybean seed are proteins and triacylglycerols. Significant amounts of isoflavonoids also accumulate in the seed during development (Dhaubhadel et al., 2003). Of the 5,910 genes described above, expression profiles of a total of 17 genes (0.3%) encoding soybean seed storage proteins were analyzed. All the storage protein cDNAs produced signals with very high intensity compared to other cDNAs on the array. The abundant glycinin (11S) and con-glycinin (7S) storage proteins consist of several unique subunits in soybean seeds. Our analysis classified the 17 genes into three major categories. A representation of each category is shown in Figure 4A. The first category with seven genes included three genes of α-subunit of β-conglycinin and four genes of glycinin with subunits G3, G4, and G5 (Fig. 4A, in red). The expression level of these genes was low at the beginning (30 DAP) and reached the peak activity within 40 to 60 DAP before declining toward maturation (70 DAP). The second category consisted of eight genes with an expression pattern very similar to the previous group but the level remained lower for the entire duration of development, followed by a sharp decline 60 DAP (Fig. 4A, in green). This group included seven α-subunit of β-conglycinin and a glycinin subunit G1. The two β-conglycinin genes with α and α′ subunits belonging to the third category showed a distinct profile of gene expression compared to other storage proteins. These genes were expressed at a lower level at 30 DAP, and the level of expression increased with time, reaching its maximum toward maturity (Fig. 4A, in blue).
Figure 4.
Expression profiles of genes encoding soybean seed storage compounds during embryo development. A group of: A, 17 seed storage protein genes; B, 46 fatty acid related genes; and C, 168 shikimic acid/phenylpropanoid pathway genes were analyzed by 3k-mean cluster analysis. The expression level of the genes in each cluster was averaged and presented by color-coded lines for RCAT Angora (RCAT) and Harovinton (Hvtn) during the embryo development (30, 40, 50, 60, or 70 DAP). The number in parentheses indicates total number of genes under each category.
The array used in this study allowed simultaneous analysis of 46 genes involved in fatty acid metabolism that showed intensity >10. These genes could be characterized by three main patterns of expression. The patterns of the first group followed a slow decline in expression from the early embryo development to late maturity stage (Fig. 4B, in red). Some of the representative genes in this category include: acyl carrier proteins (ACPs), Ω-3-fatty acid desaturase, β-ketoacyl-ACP synthetase, enoyl-ACP reductase, etc. The second group showed an increase in gene expression from 30 to 60 DAP followed by a sharp decrease in the expression. This group also includes many ACPs, two Ω-6-desaturase FAD 2 to 1, δ-12-fatty acid desaturase, and fatty acid elongase (Fig. 4B, in green). The third group showed a very different profile of gene expression. The expression of genes belonging to this group initially increased slowly and then more rapidly after 60 DAP. ω-6-desaturase FAD 2 to 1 and FAD2-2, acetyl CoA carboxylase, and many putative AIM1 proteins are included in this group (Fig. 4B, in blue).
We also studied the expression patterns of genes that are involved in shikimic acid and phenylpropanoid biosynthesis, because these routes lead to the isoflavonoid pathway. Out of 430 genes that are potentially involved in those pathways and were included on the array, only 168 (39%) showed the intensity >10. These genes were classified into three main groups according to their expression profiles (Fig. 4C). The first group started with high expression at 30 DAP, decreased at 40 DAP, and increased slightly at 50 DAP then decreased later on the development (Fig. 4C, in red). Some of the representative genes in this group are: Phe ammonia-lyase (PAL) 2, NAD(P)H-dependent 6-deoxychalcone synthase/reductase, and prephenate dehydrase. The second group was characterized by a set of genes with maximum expression at 60 DAP followed by a sharp decline in the expression level with time during the later stages of development (Fig. 4C, in green). For example, the expression of isoflavone reductase, 4-coumarate:CoA ligase isoenzyme 2, cinnamic acid 4-hydroxylase, and chalcone isomerase (CHI). The last group included genes with a distinctly different pattern, with maximum expression later in development (Fig. 4C, in blue). This group included PAL1, chalcone synthase (CHS) 7, CHS8, isoflavone synthase (IFS) 1, IFS2, and UDP-Glc:flavonoid glucosyltransferase. The complete list of genes for all three seed storage products belonging to each group is available in Supplemental Table S2.
Transcriptional Regulation of Isoflavonoid Biosynthesis in RCAT Angora and Harovinton
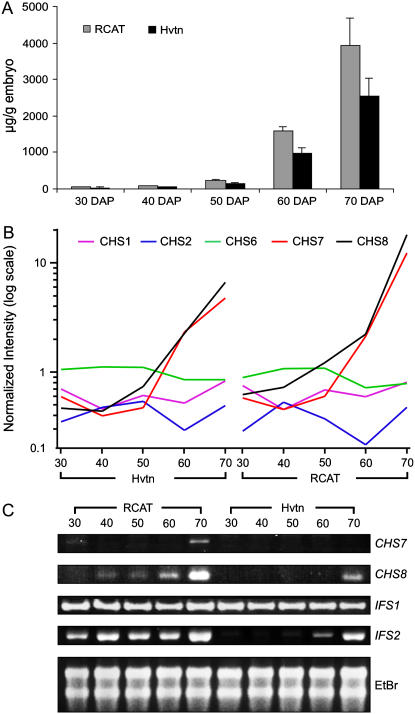
We have previously shown that RCAT Angora accumulates a higher level of seed isoflavonoids compared to Harovinton (Dhaubhadel et al., 2003). Here, we have measured the total isoflavonoid content in RCAT Angora and Harovinton during embryo development and used the same embryo development stages for microarray analysis to measure the differences in gene expression between the two cultivars that could possibly contribute to the differential isoflavonoid accumulation in the seed. Isoflavonoids were extracted from developing embryos grown in the year 2004 at the London and Delhi locations, hydrolyzed, and separated using HPLC. The peaks corresponding to isoflavonoids were identified and measured using authentic standards. The results indicate that RCAT Angora accumulates a significantly higher level of isoflavonoids compared to Harovinton in almost all the stages of embryo development (Fig. 5A). The level of isoflavonoid accumulation increased rapidly 60 DAP in both the soybean cultivars.
Figure 5.
Analysis of isoflavonoid content and IFS and CHS gene expression in RCAT Angora and Harovinton. A, Total embryo isoflavonoid content in RCAT Angora (RCAT) and Harovinton (Hvtn) during embryo development as determined by HPLC analysis. Data are mean values from three independent experiments. B, Comparison of temporal expression patterns of CHS genes in RCAT Angora (RCAT) and Harovinton (Hvtn) developing embryos at 30, 40, 50, 60, or 70 DAP obtained from microarray analysis. C, RT-PCR analysis of IFS1, IFS2, CHS7, and CHS8 transcripts with gene-specific primers. Template for RT-PCR was total RNA isolated from developing embryos (30–70 DAP) from RCAT Angora (RCAT) and Harovinton (Hvtn). Shown as control is rRNA visualized by staining with ethidium bromide (EtBr).
For the comparison of genes involved in isoflavonoid biosynthesis between RCAT Angora and Harovinton, a total of 430 genes that are involved either in shikimic acid pathway or in phenylpropanoid pathway were chosen. Results from different years and locations but for identical developmental time points were pooled together, and the mean normalized ratio between RCAT Angora and Harovinton for each gene was calculated. Of 430 genes, only 168 genes had intensity >10, and 26 genes showed a 1.5-fold difference in expression between two cultivars at one of the five embryo developmental stages under study (Table III). In fact, 19 genes were up-regulated, and seven genes were down-regulated in RCAT Angora compared to Harovinton. Only four genes revealed a 2-fold or greater change in expression, of which one was down-regulated. The expression of CHS7 and CHS8 genes was greater in RCAT Angora compared to Harovinton at 70 DAP (Fig. 5B; Table III). This difference was reproducible, statistically significant, and occurred at the stage when the embryo starts accumulating noticeably higher levels of isoflavonoids. The IFS2 (1.68-fold) and a putative dihydroflavonol reductase (2.19-fold) were also found to be expressed at higher levels in RCAT Angora versus Harovinton as were many upstream genes involved in the phenylpropanoid pathway, such as PAL, CHS, CHI, and certain Cyt P450 genes. The differences in isoflavonoid pathway gene expression between the two cultivars were even greater when separate analysis was performed for years and locations (data not shown).
Table III.
List of phenylpropanoid related genes with 1.5- or greater fold difference between RCAT Angora and Harovinton
| Source cDNA | RCAT/Hvtn (DAP)
|
GenBank No. | Annotation | ||||
|---|---|---|---|---|---|---|---|
| 30 | 40 | 50 | 60 | 70 | |||
| Gm-r1021-1507 | 1.23 | 1.61 | 1.55 | 1.03 | 2.81 | AI441937 | CHS8 |
| Gmr1021-698 | 1.46 | 1.27 | 1.38 | 0.96 | 2.62 | AI437793 | CHS7 |
| Gm-r1083-3853 | 1.25 | 1.30 | 1.18 | 0.94 | 1.86 | BE020072 | Phenyl-Ala ammonia-lyase (soybean) |
| Gm-r1070-2682 | 0.88 | 0.98 | 1.10 | 0.73 | 1.73 | AW460211 | Glutathione S-transferase 24 (soybean) |
| Gm-r1070-496 | 1.22 | 1.24 | 1.11 | 1.10 | 1.69 | AW397276 | Glutathione S-transferase 10 (soybean) |
| Gm-r1083-1082 | 1.16 | 1.08 | 0.98 | 1.08 | 1.65 | AI930620 | Putative cinnamyl alcohol dehydrogenase |
| Gm-r1083-3171 | 1.25 | 1.07 | 1.20 | 1.13 | 1.64 | AW704573 | Similar to chalcone-flavonone isomerase |
| Gm-r1083-179 | 0.81 | 0.89 | 0.72 | 0.73 | 1.61 | AW102036 | Putative Cyt P450 (Populus × canescens) |
| Gm-r1083-3468 | 1.03 | 0.79 | 0.81 | 0.76 | 1.58 | AW832016 | Cyt P450 (CYP72C; Madagascar periwinkle) |
| Gm-r1070-9192 | 0.83 | 0.93 | 0.72 | 0.82 | 1.55 | AW567634 | Putative UDP rhamnose-anthocyanidin-3-glucoside rhamnosyltransferase (Arabidopsis) |
| Gm-r1083-3218 | 1.46 | 1.31 | 1.69 | 1.20 | 1.22 | AW830241 | IFS2 (Beta vulgaris) |
| Gm-r1070-97 | 0.69 | 0.69 | 0.79 | 0.63 | 1.17 | AI735901 | Glutathione peroxidase (Malus × domestica) |
| Gm-r1070-1370 | 1.55 | 1.03 | 1.18 | 1.32 | 1.16 | AI794960 | Malonyl-CoA:ACP transacylase (Perilla frutescens) |
| Gm-r1083-646 | 1.65 | 0.80 | 1.05 | 0.72 | 1.10 | AW278307 | Isoflavone reductase homolog 1 (soybean) |
| Gm-r1070-17 | 0.87 | 0.63 | 0.75 | 0.72 | 1.07 | AI735873 | Cyt P450 (Arabidopsis) |
| Gm-r1070-8607 | 1.01 | 1.03 | 1.36 | 1.62 | 1.03 | AW507837 | CYP83D1p (soybean) |
| Gm-r1070-8518 | 0.46 | 0.59 | 0.98 | 1.21 | 1.02 | AW507888 | Putative CCR4-associated factor 1 (Arabidopsis) |
| Gm-r1070-323 | 1.57 | 0.92 | 1.17 | 1.22 | 1.01 | AW396976 | Putative malonyl-CoA:ACP transacylase (Arabidopsis) |
| Gm-r1083-4281 | 1.20 | 1.09 | 1.18 | 1.51 | 1.01 | BE020747 | UDP-glycose:flavonoid glycosyltransferase (Vigna mungo) |
| Gm-r1070-4384 | 0.83 | 1.13 | 0.67 | 0.81 | 0.96 | AW395890 | CHS2 CHS (Antirrhinum majus) |
| Gm-r1021-462 | 1.14 | 1.11 | 1.52 | 2.19 | 0.95 | AI437762 | Putative dihydroflavonol reductase (Ipomoea trifida) |
| Gm-r1070-5044 | 0.60 | 0.89 | 1.04 | 0.82 | 0.93 | AW458737 | 4-Coumarate-CoA ligase-like protein (Arabidopsis) |
| Gm-r1070-4479 | 0.65 | 1.21 | 0.95 | 0.77 | 0.86 | AW423274 | Laccase (diphenol oxidase; Arabidopsis) |
| Gm-b10BB-19 | 1.15 | 0.99 | 0.98 | 1.53 | 0.83 | BE805102 | Cyt P450 71D8 |
| Gm-r1070-4089 | 0.76 | 0.84 | 0.77 | 0.67 | 0.80 | AW099347 | Chalcone reductase (Sesbania rostrata) |
| Gm-r1070-3671 | 0.64 | 0.86 | 0.82 | 0.69 | 0.79 | AI966752 | Probable glutathione peroxidase (Arabidopsis) |
Verification of microarray results with reverse transcription (RT)-PCR analysis using gene-specific primers confirmed that IFS1 transcripts were present at similar levels in the developing embryos from early embryo development until maturity, while IFS2 transcript levels increased in both the cultivars during embryo development. Both CHS7 and CHS8 genes were expressed at a higher level in RCAT Angora compared to Harovinton at 70 DAP (Fig. 5C).
Comparative Analysis of RCAT Angora and Harovinton Developing Embryos
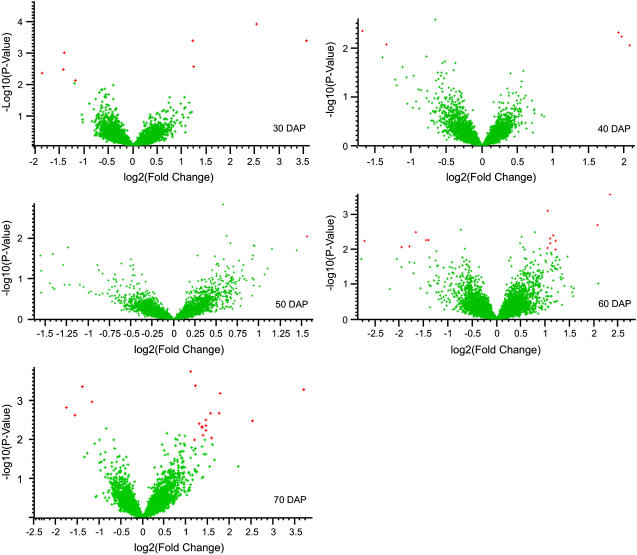
To identify other differentially expressed genes between RCAT Angora and Harovinton developing embryos, we prepared a list of genes that showed 2-fold or greater differences in expression between the cultivars at any particular stage of development (Table IV). To supplement the fold-change analysis, we performed a t test with P value of 0.01 by comparing a specific developmental stage between the two cultivars. The relationship between the P value from the t test and fold difference are represented by volcano plot for different stages of embryo development (Fig. 6).
Table IV.
List of genes that are differentially expresseda in RCAT Angora and Harovinton (Hvtn) during embryo development expressed as RCAT/Hvtn
| Source cDNA | RCAT/Hvtn (DAP)
|
Genbank No. | Annotation | ||||
|---|---|---|---|---|---|---|---|
| 30 | 40 | 50 | 60 | 70 | |||
| Genes overexpressed in RCAT Angora | |||||||
| Gm-r1070-399 | 12.84 | 3.71 | 1.58 | 2.21 | 4.71 | AW397761 | BBI (Glycine soja) |
| Gm-r1070-591 | 9.12 | 4.13 | 2.54 | 4.19 | 3.03 | AW395691 | Unknown protein (Arabidopsis) |
| Gm-r1070-495 | 6.16 | 3.78 | 2.87 | 5.25 | 2.99 | AW395780 | NPR (Vigna radiata) |
| Gm-r1070-7835 | 2.56 | 1.34 | 1.39 | 1.07 | 0.77 | AW471705 | Pro-pol-dUTPase polyprotein (Mus musculus) |
| Gm-r1070-8548 | 2.41 | 1.34 | 1.55 | 1.69 | 1.61 | AW508008 | Histone deacetylase family protein (Arabidopsis) |
| Gm-r1070-687 | 1.80 | 1.64 | 1.59 | 2.57 | 1.49 | AW397511 | Putative ADPATP carrier (Arabidopsis) |
| Gm-r1083-91 | 1.33 | 0.92 | 1.23 | 2.31 | 0.63 | AW101976 | Dicyanin (tomato) |
| Gm-r1021-951 | 1.31 | 0.94 | 1.27 | 2.26 | 0.70 | AI443187 | Polyubiquitin |
| Gm-r1021-2958 | 1.21 | 0.98 | 1.38 | 2.21 | 1.10 | AI495397 | β 1 Tubulin (maize) |
| Gm-r1083-1766 | 1.39 | 0.99 | 1.43 | 2.26 | 0.70 | AW133197 | Mcp20 (Matricaria chamomilla) |
| Gm-r1021-1736 | 1.24 | 0.94 | 1.25 | 2.13 | 0.67 | AI461256 | Ubiquitin (tomato) |
| Gm-r1070-4767 | 1.21 | 1.64 | 1.51 | 2.13 | 1.23 | AW458996 | Putative ZF-HD homeobox protein |
| Gm-r1070-8715 | 1.30 | 1.42 | 1.40 | 1.76 | 12.26 | AW508321 | Putative protein (Arabidopsis) |
| Gm-r1070-8811 | 1.35 | 1.53 | 1.67 | 2.02 | 6.35 | AW508210 | Unknown (Arabidopsis) |
| Gm-r1070-1797 | 0.47 | 0.53 | 0.55 | 0.59 | 5.29 | AI941073 | β-Glucosidase (Arabidopsis) |
| Gm-r1070-1770 | 0.74 | 0.47 | 0.60 | 0.70 | 2.83 | AI930811 | ATP-dependent Clp protease proteolytic subunit (ClpR3) putative (Arabidopsis) |
| Gm-r1083-2390 | 0.93 | 1.18 | 0.89 | 1.02 | 3.24 | AW704319 | Putative tropinone reductase (Arabidopsis) |
| Gm-r1070-7506 | 1.07 | 1.30 | 1.25 | 1.28 | 2.92 | AW433146 | Putative immunophilin (Hordeum vulgare) |
| Gm-r1021-1507 | 1.23 | 1.61 | 1.55 | 1.03 | 2.81 | AI441937 | CHS8 (soybean) |
| Gm-r1021-957 | 0.99 | 0.99 | 0.94 | 0.89 | 2.65 | AI443665 | Putative tropinone reductase (Arabidopsis) |
| Gm-r1021-698 | 1.46 | 1.27 | 1.38 | 0.96 | 2.62 | AI437793 | CHS7 (soybean) |
| Gm-r1083-556 | 1.32 | 1.47 | 1.40 | 1.03 | 2.49 | AW278434 | Disease resistance response protein (Arabidopsis) |
| Gm-r1021-955 | 1.24 | 1.06 | 1.21 | 1.64 | 2.50 | AI443646 | Unnamed protein (Arabidopsis) |
| Gm-r1021-1051 | 1.12 | 1.04 | 1.34 | 1.65 | 2.53 | AI443281 | Unknown protein (Arabidopsis) |
| Gm-r1021-633 | 1.25 | 1.04 | 1.27 | 1.43 | 2.23 | AI438104 | ADP-ribosylation factor (Vigna unguiculata) |
| Gm-r1070-7823 | 0.70 | 1.23 | 0.78 | 1.07 | 2.17 | AW472392 | Hypothetical protein, imported (Arabidopsis) |
| Gm-r1070-760 | 1.04 | 1.17 | 1.13 | 1.45 | 2.28 | AW100830 | Putative lipase/acylhydrolase (Arabidopsis) |
| Gm-r1021-3678 | 1.05 | 1.31 | 1.37 | 1.30 | 2.14 | AI496545 | Protein F28K19.28, imported (Arabidopsis) |
| Genes overexpressed in Harovinton | |||||||
| Gm-r1070-1795 | 0.35 | 0.32 | 0.39 | 0.30 | 0.76 | AI941182 | Putative protein (Arabidopsis) |
| Gm-b10BB-57 | 0.41 | 0.37 | 0.55 | 0.42 | 0.83 | AF207687 | Gly soja apyrase GS50 mRNA |
| Gm-r1070-8247 | 0.30 | 0.31 | 0.34 | 0.16 | 0.71 | AW508839 | ARF 17 (Arabidopsis) |
| Gm-r1070-5473 | 1.00 | 0.95 | 0.87 | 0.41 | 1.30 | AW459595 | Putative acyl CoA thioesterase (Arabidopsis) |
| Gm-r1070-7210 | 0.79 | 0.76 | 0.71 | 0.38 | 1.13 | AW278706 | Ribosomal protein S14 (Lupinus luteus) |
| Gm-r1070-5532 | 0.64 | 0.92 | 0.71 | 0.36 | 1.22 | AW509288 | Unnamed protein (M. musculus) |
| Gm-r1021-1039 | 0.81 | 0.77 | 0.60 | 0.31 | 0.92 | AI441825 | Pyrophosphate-dependent phosphofructo-1-kinase-like protein (Arabidopsis) |
| Gm-r1070-8357 | 0.80 | 0.77 | 1.21 | 0.82 | 0.44 | AW509456 | bZIP transcription factor ATB2 (soybean) |
| Gm-r1070-8922 | 0.80 | 0.81 | 1.09 | 0.55 | 0.38 | AW567823 | Unknown protein F2P9.17, imported (Arabidopsis) |
| Gm-r1083-3569 | 1.00 | 0.98 | 0.81 | 1.10 | 0.36 | AW832203 | Anthranilate N-hydroxycinamoyl/bensoiltransferase-like protein (Cicer arietinum) |
| Gm-r1083-13 | 1.02 | 1.16 | 1.11 | 0.51 | 0.28 | AI736505 | Galactinol synthase (soybean) |
2-fold difference.
Figure 6.
Comparison of level of gene expression between RCAT Angora and Harovinton. The relation between the 2-fold difference and statistical significance using t test are presented by volcano plots in RCAT Angora (RCAT) and Harovinton (Hvtn) at 30, 40, 50, 60, or 70 DAP. For each time point, the genes with the greatest significant difference (RCAT/Hvtn ratio) are shown in red.
The genes selected by this analysis show a significant difference between the two cultivars that are independent of location or year. Most of the differences observed between the two cultivars were at 30 DAP or near maturity. Three genes that were up-regulated in RCAT Angora at most of the developmental stages under study were a NADPH-protochlorophyllide oxidoreductase (NPR), a Bowman-Birk type protease isoinhibitor C (BBI), and a homolog to hypothetical protein from Arabidopsis. The maximum difference in normalized intensity of BBI between RCAT Angora and Harovinton was 12.8-fold at 30 DAP. The gene encoding NPR was expressed 6.1-fold higher in RCAT Angora at 30 DAP, and the minimum fold difference was 2.9 at 50 and 70 DAP. The transcripts for an auxin response factor (ARF) 17 and an unknown protein were accumulated to a significantly higher level in Harovinton compared to RCAT Angora. The normalized intensity of ARF17 was 6.3-fold higher at 60 DAP and 3-fold higher at other stages of development in Harovinton than in RCAT Angora.
A β-glucosidase was expressed 5.2-fold higher in RCAT Angora compared to Harovinton at 70 DAP, while no major change in the level of expression for this gene in both the cultivars was observed at earlier stages of embryo development. Interestingly, a gene encoding apyrase GS50 was expressed 2.5-fold higher at 30, 40, and 60 DAP, and the level dropped down to 1.5-fold at 50 and 70 DAP in Harovinton as compared to RCAT Angora. Many genes with unknown functions were also differentially expressed in RCAT Angora and Harovinton.
Finally, to determine whether the differential gene expression observed between Harovinton and RCAT Angora was due to the difference in their genome structure and copy number, we performed microarray hybridization of an 18 K-A chip with the probes derived from the genomic DNA from each of the cultivars. The result indicated that the differences in transcript accumulation for the genes described above are not due to major differences in gene copy number with the genomes of each of the cultivars.
DISCUSSION
One of the major challenges of plant developmental biology is to identify the genes involved in seed development and determine their functions. During seed development, various amino acids and metabolites are transported into the developing embryo and distributed to different biosynthetic pathways for the synthesis of major seed storage compounds. Transcriptome analysis is an important step toward gaining an understanding of the complexity and coordination of the various pathways. Here, we present a comprehensive analysis of the soybean transcriptome at five stages of embryo development. Our analysis identified 11,480 genes that are expressed in developing embryos of soybean. To select constitutively expressed genes that do not change, we filtered for genes that possessed the normalized intensity >0.5 and <2.0 in both the cultivars at both the locations and years. A total of 5,570 genes showed consistent expression level throughout development in our study. Thus, almost 50% of the genes that were detected as expressed genes did not change in their expression level during seed development. Using solution hybridization, Goldberg et al. (1981) have shown that many mRNAs present in the maturation stage in soybean embryos are also present in the cotyledon stage embryo, suggesting that many of these messages are present in the embryos throughout development. We identified 5,910 genes that changed in their expression at least once during the development. Grouping of these genes into different categories according to their expression profiles led to clusters of genes that change in a similar fashion and possibly share certain functional characteristics (Fig. 3). Of 5,910 genes that showed differential expression, 760 genes (12.8%) were annotated as unknown. Some of the unknown genes were highly expressed at certain stages of development and may possibly play a major role in seed development and metabolism. Therefore, this new information on expression profiling can guide potential work in functional genomics and may offer the foundation for reverse genetic methods to identify the function of these highly expressed genes during embryo development in soybean. The expression profiles of the vast majority of the genes follow the same pattern in RCAT Angora and Harovinton. The similarity in expression suggests that there are minor cultivar-specific differences in soybean, and thus the results may be broadly applicable. Two genes that displayed greater hybridization signals in RCAT Angora were BBI and NPR (Fig. 2, E and F). BBI is a sulfur-rich protein that inhibits trypsin and chymotrypsin proteases (Wilson, 1997). NPR catalyzes the first light-dependent reaction in chlorophyll biosynthesis, and its expression is related to chlorophyll synthesis in green tissues (Kuroda et al., 1995). Because both the cultivars were grown under the same conditions and possess a similar maturity profile, it is not clear why RCAT Angora embryos accumulate higher levels of these transcripts compared to Harovinton.
A major change in gene regulation was observed at 30 and 70 DAP as compared to other stages of development. This was particularly true for the three main storage products of the soybean seeds (Fig. 4). The storage proteins of soybean seeds are comprised of two multimeric globulins, namely 7S and 11S globulins. The 7S globulin consists of β-conglycinin subunits and the 11S consists of glycinin proteins (Hill and Breidenbach, 1974; Thanh and Shibasaki, 1976). From a total of 17 transcripts for seed storage proteins, only two showed a varied pattern compared to transcripts for other seed storage proteins. The expression pattern of these two storage protein transcripts matched with their protein profile (Hajduch et al., 2005). A similar pattern of temporal expression of storage protein genes has been found in Arabidopsis during seed filling (Ruuska et al., 2002). The other 15 storage protein transcripts were expressed at a high level throughout the earlier stages of embryo development with a sharp decline toward maturity, despite that the storage proteins themselves seem to accumulate throughout seed development (Hajduch et al., 2005). This pattern of storage protein gene expression was also observed during rice grain filling (Duan and Sun, 2005). It appears that there is a developmental stage specific to posttranscriptional regulation of storage protein gene expression in soybean embryos. Identification of regulatory factors that contribute to the control of storage protein gene expression will allow us to dissect the mechanism of storage protein synthesis in seed.
Regulation of fatty acid composition is one of the challenging areas in any oilseed breeding program, and this has gained a lot of attention in soybean breeding. Oils low in polyunsaturated fatty acids and high in 18:1 have increased stability and possibly nutritional benefits (Liu and White, 1992; Yadav, 1996). The seed-specific expression of microsomal Ω-6 desaturases FAD2-1 and FAD2-2 play a role in desaturation of 18:1 in soybean seeds (Heppard et al., 1996). These genes are up-regulated when embryos approach maturity phase. Most of the ACPs and fatty acid elongases were expressed at a higher level during early maturity stage and subsequently declined in expression at 70 DAP. In Arabidopsis, fatty acid associated genes are expressed at a higher level during seed maturity (Ruuska et al., 2002).
One of the major emphases of this study was to analyze expression patterns of genes involved in isoflavonoid synthesis and to correlate the patterns with seed isoflavonoid accumulation. A rate-limiting enzyme for isoflavonoid synthesis is IFS. This Cyt P450 enzyme converts naringenin and liquiritigenin to their corresponding isoflavones and defines a branch point in the synthesis of these natural products (Steele et al., 1999; Jung et al., 2000). Among two IFS genes, IFS1 and IFS2, IFS2 increased in expression during embryo development and showed greatest intensity near embryo maturity at 70 DAP (Supplemental Table S3). These results are concordant with a previous study of IFS1 and IFS2 gene expression that employed gene-specific primers and RT-PCR methods (Dhaubhadel et al., 2003). The expression patterns of IFS, CHS7, and CHS8 correlate well with seed isoflavonoid accumulation, indicating there is a close relationship between expression of these genes and metabolite accumulation in the seed. Because RCAT Angora and Harovinton differ in the level of seed isoflavonoids, it was hypothesized that some genes in the biosynthetic pathway may differ in their expression level. Indeed, CHS7 and CHS8 genes were expressed at a significantly higher level in RCAT Angora compared to Harovinton (Table III; Fig. 5B). These two members of the CHS multigene family belong to the same clad and share a high degree of sequence identity (Matsumura et al., 2005). A tissue-specific expression of CHS7 and CHS8 in the seed coat has been observed, and an increase in transcript accumulation was correlated with pigmented seed coat color (Tuteja et al., 2004). Our results suggest that CHS7 and CHS8 genes have diversified in their tissue-specific expression patterns. Despite the difference in isoflavonoid accumulation in the embryos of RCAT Angora and Harovinton (Fig. 5A), no significant difference in the level of IFS gene expression was observed between the two cultivars.
Past studies that have explored the control of isoflavonoid accumulation in plant tissues may help us to interpret our data. For example, expression of a chimeric R and C1 transcription factor, which increases anthocyanin levels in maize tissues (Bruce et al., 2000), induced the accumulation of isoflavones in transgenic soybean seeds compared to their wild type (Yu et al., 2003). In contrast, the introduction of IFS genes into a nonlegume background resulted in the production of isoflavones in Arabidopsis (Liu et al., 2002), tobacco (Nicotiana tabacum; Jung et al., 2000), and rice (Sreevidya et al., 2006). The introduction of IFS expression possibly diverts the naringenin substrate to isoflavonoid synthesis and adds a new branch in phenylpropanoid pathway in nonlegumes. However, increased isoflavonoid synthesis in legumes does not necessarily result from higher levels of IFS expression. An increase in the level of CHS expression at the protein level and transcripts for PAL, CHR, dihydroflavonol reductase, cinnamic acid 4-hydroxylase, and CHI genes were also demonstrated in the chimeric R and C1 seeds versus wild type. Our results are congruent with this finding. Thus, genes upstream of IFS were up-regulated in RCAT Angora versus Harovinton embryos, resulting in greater isoflavonoid accumulation in RCAT Angora.
For isoflavonoid biosynthesis, chalcone is a critical metabolite that is produced by CHS/CHR and is either used in isoflavonoid synthesis or diverted to the other branch of the pathway. It is possible that the increase in CHS expression enhances production of chalcone that may be diverted toward isoflavonoid synthesis in RCAT Angora and not affect the rest of the phenylpropanoid pathway. In contrast, no significant increase in isoflavone level was observed when CHS was expressed in soybean seed under the control of seed-specific promoter (Yu and McGonigle, 2005). It appears that CHS7 and CHS8 genes are crucial for isoflavonoid synthesis and that enhanced expression of one or both of these genes during development is specifically associated with higher seed isoflavonoid content at maturity.
The hypothesis that CHS7 and CHS8 expression may influence seed isoflavonoid content is supported by independent, quantitative trait loci (QTL) analyses that were performed to identify markers associated with isoflavone levels. These past studies have identified several QTL that lie in the same linkage group as CHS genes (Kassem et al., 2004; Primomo et al., 2005). For example, the QTLs for glycitein share the same linkage groups D1a and B1 as CHS7 and CHS8, respectively, supporting the proposal that these genes are key players for increased isoflavonoid production in seeds. The linkage groups A1 and K also share QTL for isoflavone aglycones and possess the CHS2 and CHS6 genes.
In conclusion, our results illustrate that transcriptional control during soybean embryo development is a highly coordinated process. We found much evidence demonstrating that the synthesis and transport of storage proteins, fatty acids, and isoflavonoids are transcriptionally regulated from the early developmental stage to maturity in soybean embryo. Our results show that environmental effects on the transcriptome of the developing seed are large and exceed cultivar-specific effects. The information obtained from this study provides a powerful tool for studying and understanding gene functions for many unidentified genes that may have crucial roles in regulating and coordinating the expression of nutrient partitioning genes during embryo development. This study also provides a detailed comparison of gene expression between two cultivars that differ in isoflavonoid content and points to CHS7 and CHS8 as genes that influence isoflavonoid biosynthetic flux in soybean seeds.
MATERIALS AND METHODS
Plant Materials and Tissue Preparation
Soybean (Glycine max) L. Merr. cv RCAT Angora (3150 CHU) and cv Harovinton (3100 CHU) were obtained from Dr. Istvan Rajcan (Department of Plant Agriculture, University of Guelph, Ontario) and Agriculture and Agri-Food Canada, Harrow, respectively. Both the cultivars belong to late maturity group I to early maturity group II. Soybean seeds were planted at two Agriculture and Agri-Food Canada experimental stations in southern Ontario, London and Delhi, in 2003 and 2004. Regular agronomic practices and planting dates were followed. The pods were tagged on the first day of pollination and harvested at 30, 40, 50, 60, and 70 DAP. The pods were collected randomly from five to seven plants, and embryos were excised from seeds, frozen in liquid nitrogen, and stored at −80°C.
RNA Isolation and RT-PCR Analysis
Total RNA was isolated from developing embryos following the procedure of Wang and Vodkin (1994). Total RNA was quantified using a spectrophotometer, and samples of total RNA (2 μg each) were electrophoretically separated in formaldehyde gels (1.5% w/v) and stained with ethidium bromide to ensure concentration and integrity. Samples of 400 μg total RNA were used to purify poly(A) RNA using MicroPoly(A) Purist TM kit (Ambion) according to the manufacturer's protocol with some modifications. The concentration and purity of poly(A) RNA was assessed spectrophotometrically. RT-PCR reactions for IFS1 and IFS2 were performed as described previously (Dhaubhadel et al., 2003). Gene-specific primer sequences for CHS7 and CHS8 were: CHS7, 5′-CCCTCCCATCCACTCTCTC-3′, 5′-CCCGCTAGCAAACAAGGTTAC-3′; CHS8, 5′-CCCCAAATAGCTCCCAGTACT-3′, 5′-GGCCATCCAGGGAGGTAA-3′. PCR conditions were as follows: 94°C for 1 min, 63°C for 30 s, and 72°C for 1 min 45 s (35 cycles) for CHS7. For CHS8, the PCR conditions were the same as for CHS7 except that annealing temperature was 58°C.
Experimental Design, Probe Labeling, Hybridization, and Data Analysis
Soybean cDNA microarray slides consisting of 18,432 cDNAs spotted onto amine-coated glass slides (18 K-A) were obtained from Dr. Lila Vodkin (University of Illinois, Urbana). A total of 42 microarray slides were hybridized, 40 from four separate biological samples using independent samples of mRNA for each experiment with dye swaps to minimize technical variation. Each experiment included mRNA samples from soybean cv RCAT Angora and cv Harovinton at five different embryo developmental stages (30, 40, 50, 60, and 70 DAP). An additional two slides were hybridized with labeled genomic DNA to compare the two cultivars at the genomic level.
Probe labeling was performed by using CyScribe First-Strand cDNA Labeling kit (Amersham BioSciences) according to the manufacturer's instruction. Samples of 1.5 μg mRNA were used in labeling reaction with CyDye-labeled dCTP. Purification of labeled cDNA and removal of unincorporated nucleotides were performed by using CyScribe GFX Purification kit (Amersham BioSciences) according to the manufacturer's instruction, except that the labeled cDNAs were eluted in two steps to a total elution volume of 80 μL. The incorporation of cyanine-3- and cyanine-5-labeled nucleotides into cDNA was determined spectrophotometrically by measuring the absorption at 550 nm and 650 nm, respectively.
Microarray slides were exposed to an additional cross-linking at 50 mJ cm−2 and then prehybridized for 45 min at 42°C in prehybridization buffer containing 5× SSC, 0.1% SDS, and 1% bovine serum albumin, followed by two washes in 0.1× SSC at room temperature. The slides were rinsed with sterile water and dried by centrifugation. Equal amounts of purified CyDye-labeled probes were combined together, dried under vacuum at 45°C (Speed Vac, Savant Instrument), and resuspended in a hybridization solution (40 μL total volume) containing 1.25 ng/μL poly(A) DNA, 50% (v/v) formamide, and 25% (v/v) hybridization buffer (Amersham BioSciences). The hybridization mix was denatured at 100°C for 2 min, cooled to room temperature, and applied to the prehybridized slide. The slide was covered with a 24- × 60-mm coverslip and placed in a hybridization chamber containing 10 μL water. The hybridization was carried out at 42°C for 20 h, the coverslip removed in 2× SSC, 0.1% SDS, followed by one posthybridization wash in 2× SSC, 0.1% SDS at 42°C for 5 min, two washes in 0.1× SSC, 0.1% SDS at 42°C for 2 min, and two room temperature washes in 0.1× SSC for 1 min. Slides were rinsed with sterile water and dried under nitrogen gas prior to laser scanning (Bio-Rad ChipReader with VersArray ChipReader v3.0, Bio-Rad). Spot intensities were quantified individually for background signals (Array Vision v6.0 software). Background subtracted intensities were imported into GeneSpring v7.3 (Agilent Technologies) and normalized by per spot, per chip, intensity-dependent LOWESS. A dye swap was performed in each experiment, and final normalized ratios were averaged from each location and year for a particular cultivar at a given time point. The differential gene expression between RCAT Angora and Harovinton at a particular stage of embryo development was monitored by taking the ratio of RCAT Angora and Harovinton from the two-color hybridization of each slide. The relative gene expression for embryo development as a function of time was calculated as an alternate approach to the data using a single channel input for each cultivar imported into GeneSpringGX, normalized by per chip to 50th percentile, and per gene to the median of the measurement of that gene. To further validate this approach, two-color hybridizations were performed on two time points of the same cultivar, RCAT Angora 30 DAP and 70 DAP. A comparative genomic DNA hybridization using genomic DNA from RCAT Angora and Harovinton was conducted as described in Gijzen et al. (2006). Data were background subtracted and genes with intensity <10 were removed from the list. Data was MIAME validated and deposited to the GEO (National Center for Biotechnology Information, http://www.ncbi.nih.gov) series GSE 4194; samples GSM94935 to GSM94976.
Technical and Biological Variation
To estimate technical variation, control experiments in which the same RNA was labeled with cyanine-3 and cyanine-5 were performed. The degree of biological and environmental variability in the embryos was assessed by growing them for two different years at two different locations.
Isoflavonoid Analysis
Embryo samples of identical developmental stages that were used in the microarray experiment were ground to a fine powder in liquid nitrogen and extracted with 50% acetonitrile in water followed by sonication for 30 min in an ice water bath. The samples were centrifuged for 25 min at 3,000g and the supernatant was collected. The extraction process was repeated two times with the pellet and the supernatant fractions were pooled together and filtered (Acrodisc, nylon, 0.45 μm). The malonyl- and acetyl-isoflavonoids were converted to their corresponding glucosides by hydrolyzing the filtered samples with 1.3% KOH at room temperature for 4 h followed by neutralization of the sample with 3% KH2PO4. The solvent was evaporated and samples were redissolved in 40% dimethyl sulfoxide prior to HPLC analysis. Isoflavonoids were separated by injecting 20 μL of the samples on a C18 column (Symmetry Column, Waters Corporation, 5 μm). A guard column of the identical packing material was connected before the analytical column. The samples were run at room temperature applying a mobile-phase gradient of 10% to 35% acetonitrile in 0.1% acetic acid over 45 min at a flow rate of 1 mL/min (Waters Limited). The total separation time was 63 min, which included a 4-min wash and 14-min equilibration. Isoflavonoid peaks were compared with the retention time and UV spectra of the aglycone and glucoside standards (LC Laboratories) and quantified using the Millenium32 Software (Waters Limited).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Table S1. Temperature and precipitation in London and Delhi in 2003 and 2004.
Supplemental Table S2. List of genes involved in seed storage products in RCAT Angora and Harovinton developing embryos.
Supplemental Table S3. List of genes with 3.5 or greater fold change in expression during embryo development.
Supplementary Material
Acknowledgments
We thank Istvan Rajcan (University of Guelph) and Vaino Poysa (Agriculture and Agri-Food Canada, Harrow) for soybean seeds, Dr. Lila Vodkin (University of Illinois) for 18 K-A cDNA microarray slides, and Alex Molnar and Aldona Gaidauskas-Scott (Agriculture and Agri-Food Canada, London) for technical assistance.
This work was supported by Agriculture and Agri-Food Canada's Canadian Crop Genomics Initiative (grant to S.D.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Sangeeta Dhaubhadel (dhaubhadels@agr.gc.ca).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruce W, Folkerts O, Garnaat C, Crasta O, Roth B, Bowen B (2000) Expression profiling of the maize flavonoid pathway genes controlled by estradiol-inducible transcription factor CRC and P. Plant Cell 12 65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzell RI, Anderson TR, Hamill AS, Welacky TW (1991) Harovinton soybean. Can J Plant Sci 71 525–526 [Google Scholar]
- Carpenter J, Felsot A, Goode T, Hamming M, Onstad D, Sankula S (2002) Comparative Environmental Impacts of Biotechnology Derived and Traditional Soybean, Corn and Cotton Crops. Council for Agricultural Science and Technology, Ames, IA, pp 15–50
- Dhaubhadel S, McGarvey BD, Williams R, Gijzen M (2003) Isoflavonoid biosynthesis and accumulation in developing soybean seeds. Plant Mol Biol 53 733–743 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Ferreira D (2002) Genestein. Phytochemistry 60 205–211 [DOI] [PubMed] [Google Scholar]
- Duan M, Sun SSM (2005) Profiling the expression of genes controlling rice grain quality. Plant Mol Biol 59 165–178 [DOI] [PubMed] [Google Scholar]
- Gijzen M, Kuflu K, Moy P (2006) Gene amplification of the Hps locus in Glycine max. BMC Plant Biol 6 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg RB, Hoschek G, Tam SA, Ditta GS, Breidenbach RW (1981) Abundance, diversity, and regulation of mRNA sequence sets in soybean embryogenesis. Dev Biol 83 201–217 [DOI] [PubMed] [Google Scholar]
- Hajduch M, Ganapathy A, Stein JW, Thelen JJ (2005) A systematic proteomic study of seed filling in soybean: establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteomic database. Plant Physiol 137 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Gruissem W, Grossniklaus U, Köhler C (2004) Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol 135 1765–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppard EP, Kinney AJ, Stecca KL, Miao G-H (1996) Developmental and growth temperature regulation of two different microsomal omega-6 desaturase genes in soybeans. Plant Physiol 110 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Breidenbach RW (1974) Proteins of soybean seeds. II. Accumulation of the major protein components during seed development and maturation. Plant Physiol 53 747–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung W, Yu O, Lau SM, O'Keefe DP, Odell J, Fader G, McGonigle B (2000) Identification and expression of isoflavone synthase, the key enzyme for biosynthesis of isoflavones in legumes. Nat Biotechnol 18 208–212 [DOI] [PubMed] [Google Scholar]
- Kassem MA, Meksem K, Iqbal MJ, Njiti VN, Banj WJ, Winters TA, Wood A, Lightfoot DA (2004) Definition of soybean genomic regions that control seed phytoestrogen amounts. J Biomed Biotechnol 1 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Masuda T, Ohta H, Shioi Y, Takamiya K (1995) Light enhanced gene expression of NADPH-protochlorophyllide oxidoreductase in cucumber. Biochem Biophys Res Commun 210 310–316 [DOI] [PubMed] [Google Scholar]
- Lee J-M, Williams ME, Tingey SV, Rafalski JA (2002) DNA array profiling of gene expression changes during maize embryo development. Funct Integr Genomic 2 13–27 [DOI] [PubMed] [Google Scholar]
- Liu C-J, Blount JW, Steele CL, Dixon RA (2002) Bottlenecks for metabolic engineering of isoflavone glycoconjugates in Arabidopsis. Proc Natl Acad Sci USA 99 14578–14583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HR, White PJ (1992) Oxidative stability of soybean oils with altered fatty acid composition. J Am Oil Chem Soc 69 528–532 [Google Scholar]
- Matsumura H, Watanabe S, Harada K, Senda M, Akada S, Kawasaki S, Dubouzet EG, Minaka N, Takahashi R (2005) Molecular linkage mapping and phylogeny of the chalcone synthase multigene family in soybean. Theor Appl Genet 110 1203–1209 [DOI] [PubMed] [Google Scholar]
- Orthoefer FT (1978) Processing and utilization. In AG Norman, ed, Soybean Physiology, Agronomy, and Utilization. Academic Press, New York, pp 219–246
- Primomo VS, Poysa V, Ablett GR, Jackson C-J, Gijzen M, Rajcan I (2005) Mapping QTL for individual and total isoflavone content in soybean seeds. Crop Sci 45 2454–2464 [Google Scholar]
- Ruuska SA, Girke T, Benning C, Ohlrogge JB (2002) Contrapuntal networks of gene expression during Arabidopsis seed filling. Plant Cell 14 1191–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270 467–470 [DOI] [PubMed] [Google Scholar]
- Sreevidya VS, Srinivasa Rao C, Sullia SB, Ladha JK, Reddy PM (2006) Metabolic engineering of rice with soybean isoflavone synthase for promoting nodulation gene expression in rhizobia. J Exp Bot 57 1957–1969 [DOI] [PubMed] [Google Scholar]
- Steele CL, Gijzen M, Qutob D, Dixon RA (1999) Molecular characterization of the enzyme catalyzing the aryl migration reaction of isoflavonoid biosynthesis in soybean. Arch Biochem Biophys 367 146–150 [DOI] [PubMed] [Google Scholar]
- Thanh VH, Shibasaki K (1976) Heterogeneity of beta-conglycinin. Biochim Biophys Acta 181 404–409 [DOI] [PubMed] [Google Scholar]
- Tuteja JH, Clough SJ, Chan WC, Vodkin LO (2004) Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell 16 819–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vodkin LO, Khanna A, Shealy R, Clough SJ, Gonzalez DO, Philip R, Zabala G, Thibaud-Nissen F, Sidarous M, Stromvik MV, et al (2004) Microarray analysis for global expression constructed with a low redundancy set of 27,500 sequenced cDNAs representing an array of developmental stages and physiological conditions of the soybean plant. BMC Genomics 5 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walbot V (1978) Control mechanisms for plant embryogeny. In M Clutter, ed, Dormancy and Developmental Arrest. Academic Press, New York, pp 113–166
- Wang CS, Vodkin LO (1994) Extraction of RNA from tissues containing high levels of procyanidins that bind RNA. Plant Mol Biol 12 132–145 [Google Scholar]
- Wilson KA (1997) The protease inhibitors of seeds. In BA Larkins, IK Vasil, eds, Cellular and Molecular Biology of Plant Seed Development. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 331–374
- Wobus U, Weber H (1999) Seed maturation: genetic programmes and control signals. Curr Opin Plant Biol 2 33–38 [DOI] [PubMed] [Google Scholar]
- Yadav NS (1996) Genetic modification of soybean oil quality. In DPS Verma, RC Shoemaker, eds, Soybean Biotechnology. CAB International, Wallingford, UK, pp 165–188
- Yu O, McGonigle B (2005) Metabolic engineering of isoflavone biosynthesis. Adv Agron 86 147–190 [Google Scholar]
- Yu O, Shi J, Hession AO, Maxwell CA, McGonigle B, Odell CA (2003) Metabolic engineering to increase isoflavone biosynthesis in soybean seed. Phytochemistry 63 753–763 [DOI] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Chen W, Nicholas P, Chang H-S, Guimil S, Su W, Estes B, Zou G, Wang X (2003) Transcriptional control of nutrient partitioning during rice grain filling. Plant Biotechnol J 1 59–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.