Abstract
Thioredoxins (TRXs) are small ubiquitous oxidoreductases involved in disulfide bond reduction of a large panel of target proteins. The most complex cluster in the family of plant TRXs is formed by h-type TRXs. In Arabidopsis (Arabidopsis thaliana), nine members of this subgroup were described, which are less well known than their plastidial counterparts. The functional study of type-h TRXs is difficult because of the high number of isoforms and their similar biochemical characteristics, thus raising the question whether they have specific or redundant functions. Type-h TRXs are involved in seed germination and self incompatibility in pollen-pistil interaction. Their function as antioxidants has recently been proposed, but further work is needed to clarify this function in plants. In this study, we describe two new h-type TRXs from pea (Pisum sativum; stated PsTRXh1 and PsTRXh2). By functional complementation of a yeast (Saccharomyces cerevisiae) trx1Δ trx2Δ double mutant, we demonstrate that PsTRXh1 is involved in the redox-imbalance control, possibly through its interaction with peroxiredoxins. In contrast, PsTRXh2 provokes a phenotype of hypersensitivity to hydrogen peroxide in the yeast mutant. Furthermore, we show differential gene expression and protein accumulation of the two isoforms, PsTRXh1 protein being abundantly detected in vascular tissue and flowers, whereas PsTRXh2 gene expression was hardly detectable. By comparison with previous data of additional PsTRXh isoforms, our results indicate specific functions for the pea h-type TRXs so far described.
Thioredoxins (TRXs) are small proteins (12–14 kD) with a characteristic folding and a conserved redox-active site WCG/PPC. The thiols of the Cys residues at the active site act as powerful reducing agents, able to disrupt disulphide bridges of target proteins. The large number of putative TRX targets identified during the last years indicates that TRXs are involved in an increasing number of processes in plants (Buchanan and Balmer, 2005).
TRXs are present in all types of organisms from bacteria to mammals, but plants exhibit the most complex TRX multigenic family, as shown by the analysis performed in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) genomes. The Arabidopsis genome encodes more than 25 different TRXs and other TRX-like proteins with multiple TRXs or non-TRX extra domains, being nuclear encoded and grouped by sequence similarity and intron positions (Meyer et al., 2002). In plants, the most complex subgroup is formed by the h-type TRXs, which contain nine isoforms in Arabidopsis (Meyer et al., 2005) and five isoforms in Populus trichocarpa (Gelhaye et al., 2004b). Oxidized h-type TRXs are reduced by NADPH-mediated NADP-TRX reductase, both proteins making up the so-called NADP-TRX reductase-TRX-system (Arner and Holmgren, 2000). Because of the little knowledge of h-type TRXs, contrary to the well-described chloroplastic TRXs (f and m), this group of TRXs has been intensively studied during the last years. Some of their functions have been established and include storage protein mobilization during seed germination (Kobrehel et al., 1992; Besse et al., 1996; Wong et al., 2002; Marx et al., 2003) and control of self-incompatibility processes in pollen-pistil recognition (Bower et al., 1996; Cabrillac et al., 2001). In addition, they have been related to Met metabolism (Mouaheb et al., 1998) and the response against biotic and abiotic stress (Mouaheb et al., 1998; Reichheld et al., 2002; Serrato and Cejudo, 2003; Laloi et al., 2004), this latter function not being well known yet. Type-h TRXs are generally assumed to be cytosolic proteins because of their lack of transit peptide (Florencio et al., 1988), but other subcellular locations have been proposed (Gelhaye et al., 2004a). They have also been described as an abundant protein in rice phloem sap (Ishiwatari et al., 1995; Schobert et al., 1998) where they are able to move between the sieve tube and nearby cells via plasmodesmata (Ishiwatari et al., 1998). In addition, they have been found to accumulate in the nucleus of seed cells (Serrato et al., 2001; Serrato and Cejudo, 2003).
In view of such diverse data, a great effort is still needed to clarify their role in the cell, the identification of multiple h-type TRX genes raising questions related to the specificity of their physiological roles. The first attempt to address these questions was the heterologous complementation of the yeast (Saccharomyces cerevisiae) mutant EMY63 (Müller, 1991) lacking both cytosolic TRX1 and TRX2, which revealed functional specificity associated with some of the Arabidopsis h-type TRXs (Mouaheb et al., 1998). Later, in recent years, several studies have been directed toward the search for TRX target proteins (Verdoucq et al., 1999; Maeda et al., 2004; Yamazaki et al., 2004). As a result, many proteins have been identified as putative TRX targets, thereby opening new fields of investigation on h-type TRXs.
In pea (Pisum sativum), five isoforms of the TRX family have been described. Three of them, f, m1, and m2, are chloroplastic (Jaramillo et al., 1997; Pagano et al., 2000), two h-type TRXs being cytosolic. These last cytosolic isoforms, PsTRXh3 and PsTRXh4, have recently been well characterized and related to diverse processes during seed germination (Montrichard et al., 2003). In this work, we address the gene, protein, and functional characterization of two additional h-type TRXs from pea, termed PsTRXh1 and PsTRXh2, with the aim to determine whether their role is specific or redundant with other isoforms. We first show that both h1 and h2 isoforms are true TRXs in vivo through their ability to complement some phenotypes of a yeast mutant devoid of endogenous cytosolic TRXs. Interestingly, we demonstrate that they confer divergent behavior to yeast trx1Δ trx2Δ cells toward oxidant molecules. Second, we show that their gene expression and protein accumulation patterns are also very different in pea tissues, especially in seedlings subjected to oxidative stress where their behavior confirms the results observed by yeast functional complementation. Finally, we discuss the possible physiological functions of PsTRXh1 and PsTRXh2 and propose that the role of the isoforms of the h-type cluster is not redundant in pea.
RESULTS
PsTRXh1 and PsTRXh2 Are TRX Genes Belonging to the h-Type Group
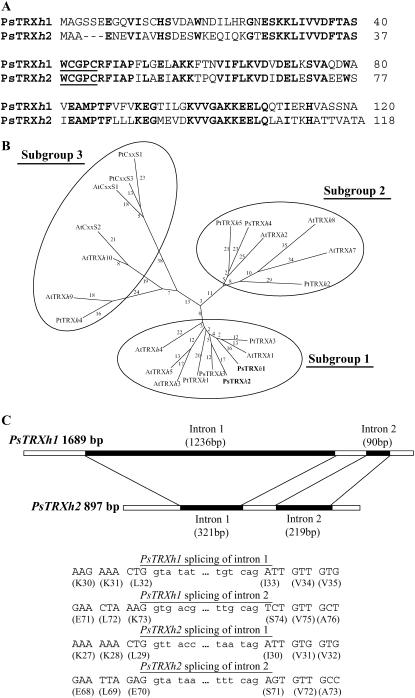
At the time we started this study, only chloroplastic TRXs had been described in pea, none of the cytosolic isoforms being discovered yet. Using an approach based on reverse transcription (RT)-PCR and RACE, we amplified from leaf total RNA two full-length cDNAs, termed PsTRXh1 (GenBank accession no. AJ310990) and PsTRXh2 (GenBank accession no. AJ319808). Both cDNAs contain a unique open reading frame (ORF) that encodes for polypeptides of 120 (PsTRXh1) and 118 residues (PsTRXh2; Fig. 1A). The deduced proteins exhibit similar molecular mass and present 60% identity at the amino acid level as well as the structural and functional amino acid features found in other h-type TRXs, including the canonical active site WCGPC (Fig. 1A). In addition, three-dimensional models for both deduced sequences, based on resolved h-type TRXs (Coudevylle et al., 2005), fit well to the classical TRX folding (data not shown). Compared to other TRXs from pea, we found a high similarity of both PsTRXh1 and PsTRXh2 with PsTRXh3 (64% and 72% identity, respectively); however, they both share less than 41% identity with PsTRXh4, the latest cytosolic isoform isolated up to now (Montrichard et al., 2003). Contrary to chloroplastic TRXs, PsTRXh1 and PsTRXh2 do not contain any transit peptide. A comparative phylogeny analysis of h-type TRXs from pea, Arabidopsis, and Populus confirmed that PsTRXh1 and PsTRXh2 belong, together with PsTRXh3, to h-type TRX subgroup 1 (also including AtTRXh1-3-4-5), while PsTRXh4 is positioned in subgroup 2 next to Arabidopsis h2-7-8 isoforms, probably because of the N-terminal extension that characterizes such TRXs (Fig. 1B).
Figure 1.
PsTRXh1 and PsTRXh2 deduced amino acid sequences, phylogenetic analysis, and gene structure. A, Alignment of PsTRXh1 (AJ310990) and PsTRXh2 (AJ319808) deduced amino acid sequences. The conserved amino acids are in bold and the active site underlined. B, DARWIN Phylogenetic tree of h-type TRXs from pea (Ps), P. trichocarpa (Pt), and Arabidopsis (At). Accession numbers are: PtTRXh1 (AF483625); PtTRXh2 (AF483266); PtTRXh3 (BU822062); PtTRXh4 (BU835000); PtTRXh5 (BU869308); PtCxxS1 (CA823821); PsCxxS3 (BU874060); AtTRXh1 (P29448); AtTRXh2 (S58123); AtTRXh3 (S58118); AtTRXh4 (S58119); AtTRXh5 (S58120); AtTRXh7 (AAD39316); AtTRXh8 (AAG52561); AtTRXh9 (AAG51342); AtTRXh10 (At3g56420); AtCxxS1 (AF144390); AtCxxS2 (ATU35639); PsTRXh1 (AJ310990); PsTRXh2 (AJ319808); PsTRXh3 (AY170650); and PsTRXh4 (AY170651). C, Gene structure of both pea h-type TRXs and determination of intron splicing sites by comparing cDNA and genomic sequences.
The genomic sequences of PsTRXh1 and PsTRXh2 genes were 1,689 bp and 897 bp long, each containing two introns of 1,236 and 90 bp in the PsTRXh1 gene and of 321 and 219 bp in the PsTRXh2 gene, respectively (Fig. 1C). Both intron positions were conserved within the h-type TRX cluster (Sahrawy et al., 1996). We could not find any significant similarity between either PsTRXh1 and PsTRXh2 intronic sequences or in 5′- and 3′-untranslated regions. According to both number and position of introns, both genes encode most probably h-type TRXs.
PsTRXh1 and PsTRXh2 Are True TRXs with Different Features in Vitro
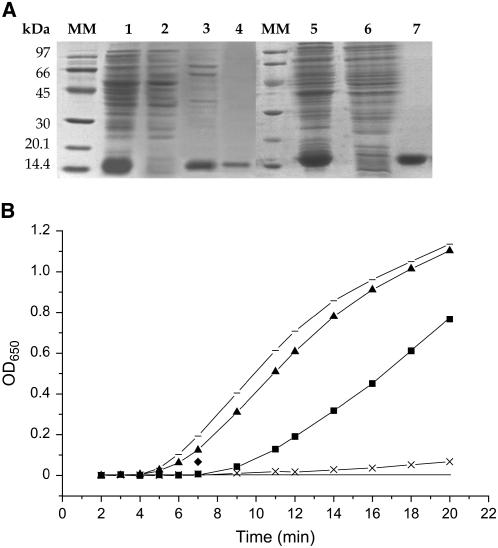
To determine whether PsTRXh1 and PsTRXh2 isoforms possess TRX activity in vitro, their corresponding ORFs were expressed in Escherichia coli to produce recombinant proteins. Both proteins accumulated at a high level in the soluble fraction and were purified to homogeneity (Fig. 2A). Interestingly, both proteins presented different thermo stability degrees, PsTRXh1 being resistant to higher temperatures (up to 75°C) than PsTRXh2 (45°C). In addition, PsTRXh1 was highly soluble during the purification process, while PsTRXh2 tended to form homodimeric structures (data not shown). TRX activity was determined in vitro by measuring the capacity of the recombinant proteins to reduce disulfide-bridged insulin, using as controls the well-characterized chloroplastic PsTRXf and PsTRXm1 (Pagano et al., 2000). Figure 2B shows that recombinant PsTRXh1 and PsTRXh2 are both able to reduce insulin, with isoform h1 showing higher efficiency, similar to that of the chloroplastic TRXm1. This result indicates that both PsTRXh1 and PsTRXh2 show efficient TRX activity in vitro.
Figure 2.
Purification and TRX activity of pea PsTRX h1 and h2 isoforms. A, SDS-PAGE analysis of protein samples from different steps of PsTRXh1 and PsTRXh2 purification procedure. Crude extract of E. coli transformed with pET-3d-TRXh1 (lane 1); resuspended pellet after warming up to 75°C (lane 2); supernatant after warming up to 75°C (lane 3); purification of PsTRXh1 by Sephadex G-50 and DEAE cellulose gel filtration (lane 4); crude extract of E. coli transformed with pET-3d-TRXh2 (lane 5); supernatant after warming up to 60°C (lane 6); and purification of PsTRXh2 by Sephadex G-50 and DEAE cellulose gel filtration (lane 7). Molecular weight markers (MM) are indicated in kilodaltons. B, Insulin-disulphide-reductase activity of pea TRXs. The rate of insulin reduction was followed by measuring the turbidity at 650 nm in reaction assays containing 5 μm of each TRX: PsTRXh1 (▴), PsTRXh2 (▪), PsTRXm1 (horizontal line), and PsTRXf (x). The negative control (no TRX) is represented by a line without a symbol.
PsTRXh1 and PsTRXh2 Complement Some Aspects of the trx1Δ trx2Δ Yeast Mutant in Vivo
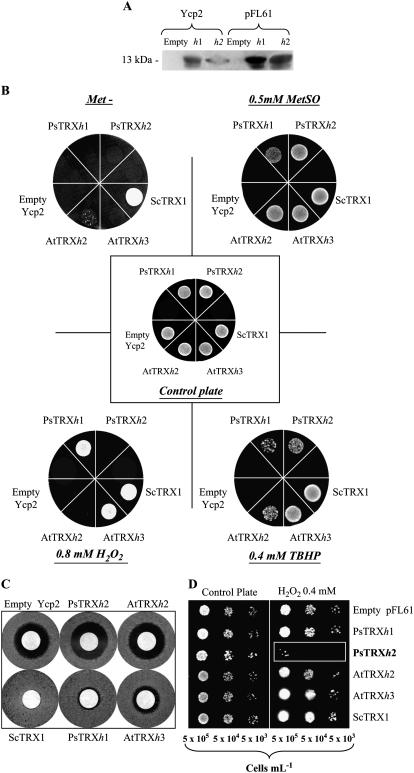
To know whether PsTRXh1 and PsTRXh2 are also functional TRXs in vivo, we tested their ability to complement the trx1Δ trx2Δ yeast mutant. In yeast, the simultaneous disruption of the two TRX-encoding genes, ScTRX1 and ScTRX2, leads to several growth defects, including organic sulfur auxotrophy and sensitivity to hydrogen peroxide (H2O2; Müller, 1991). Previous reports have already demonstrated the ability of some plant TRXs to complement specific aspects of such yeast mutant phenotype (Mouaheb et al., 1998; Issakidis-Bourguet et al., 1999; Vignols et al., 2003, 2005). PsTRXh1 and PsTRXh2 ORFs were then cloned into the low-copy centromeric Ycp2 (Mouaheb et al., 1998) and the high-copy pFL61 (Minet et al., 1992) vectors and successfully produced PsTRXh1 and PsTRXh2 proteins at corresponding low and high levels in the yeast trx1Δ trx2Δ mutant EMY63 (Fig. 3A). Because of their organic sulfur auxotrophy, trx1Δ trx2Δ yeast cells require Met for growth. We thus analyzed first the ability of PsTRXh proteins to restore mutant cell growth on a medium devoid of an organic sulfur source (without Met). None of the two pea TRXs produced from the Ycp2 vector efficiently restored yeast sulfate assimilation in trx1Δ trx2Δ yeast cells (Fig. 3B, above left), contrary to the Arabidopsis AtTRXh2 and the yeast ScTRX1, which were used as positive controls according to a previous work (Verdoucq et al., 1999). A faint complementation of Met auxotrophy was only observed when PsTRXh1 was produced at a high level from the pFL61 vector (data not shown). This result suggests that both PsTRXh1 and PsTRXh2 are unable to reduce efficiently the phosphoadenosine-5-phosphosulfate reductase, a protein involved in Met biosynthesis through sulfate assimilation that requires reduction, mostly by endogenous ScTRX1, for efficient activity in wild-type cells (Vignols et al., 2005). But interestingly, when Met was replaced by Met sulfoxide (an oxidized form of Met) in the assay, trx1Δ trx2Δ cells producing Ycp2-PsTRXh2 were able to grow similarly to positive controls (Fig. 3B, above right). A similar result was obtained for Ycp2-PsTRXh1 though at a lower extent. This result demonstrates that both pea h1 and h2 isoforms were able to replace the disrupted endogenous ScTRX1 and ScTRX2 to ensure reduction of the yeast Met sulfoxide reductase in vivo, an enzyme involved in oxidative damage repair by reversing Met oxidation (Moskovitz et al., 1997).
Figure 3.
Functional complementation of trx1Δ trx2Δ yeast cells by heterologous PsTRX h1 and h2 isoforms. A, Immunodetection of PsTRXh1 and PsTRXh2 produced in trx1Δ trx2Δ cells from Ycp2 and pFL61 vectors. Total protein samples (30 μg) were subjected to SDS-PAGE and electrotransferred. Blots were probed with anti-PsTRXh1 and anti-PsTRXh2 antibodies used at a dilution of 1:40,000 and 1:10,000, respectively. B, Growth of trx1Δ trx2Δ cells expressing heterologous PsTRXh proteins under different culture conditions. Yeast mutant (2.5 × 104 cells) carrying different Ycp2 constructs was spotted as a 50-μL drop on B media, lacking organic sulfur source (Cherest and Surdin-Kerjan et al., 1992): growth without Met (above left plate),or with 0.5 mm Met sulfoxide replacing Met (above right plate). Yeast cells were also grown in yeast nitrogen base-Gal media supplemented with all requirements (central control plate) or supplemented with oxidant molecules: 0.4 mm TBHP (below right plate) or 0.8 mm H2O2 (below left plate). The Arabidopsis AtTRXh2 (At5g39950) and AtTRXh3 (At5g42980), the yeast ScTRX1 (YLR043c), and the empty vector were used as controls (Verdoucq et al., 1999). C, Halo-inhibition assay showing the resistance of trx1Δ trx2Δ mutant cells expressing plant TRXs from Ycp2 vector in the presence of 0.8 mm H2O2. The inhibition measurement was performed according to the halo diameter after 72 h at 30°C. The 100% inhibition was arbitrarily attributed to Ycp2 empty vector. Inhibition determination for TRX-containing constructs was as follows: PsTRXh2, 120%; AtTRXh2, 102%; ScTRX1, 0%; PsTRXh1, 22%; and AtTRXh3, 40%. D, Hypersensitivity of trx1Δ trx2Δ cells toward H2O2 conferred by constitutive overproduction of PsTRXh2. trx1Δ trx2Δ cells carrying pFL61 constructs and expressing the various TRXs tested were grown to a density of 107 cells/mL. Seven microliters of serial dilutions were plated on solid medium without (control plate) or containing 0.4 mm H2O2.
Because trx1Δ trx2Δ yeast cells are also sensitive to oxidative stress, we analyzed the growth of trx1Δ trx2Δ mutant cells expressing heterologous PsTRXs on medium containing oxidant molecules. We found that PsTRXh2 conferred to trx1Δ trx2Δ cells the ability to grow in the presence of 0.4 mm terc-butyl-hydroperoxide (TBHP) at a lower extent than the positive controls AtTRXh3 and ScTRX1 but at a higher extent than AtTRXh2 (Fig. 3B, below right). Mutant cells expressing PsTRXh1 grew poorly. On the other hand, trx1Δ trx2Δ cells producing PsTRXh1 presented a rapid growth, similar to mutant cells producing ScTRX1 or AtTRXh3, on a medium containing 0.8 mm H2O2 (Fig. 3B, below left), whereas cells expressing PsTRXh2 did not grow in such oxidant growth conditions.
Interestingly, while confirming these results by halo-inhibition disc assays, we noticed that yeast cells expressing PsTRXh2 presented a higher sensitivity toward H2O2 than those transformed by the empty vector Ycp2, showing an excess of sensitivity (20%) in comparison to the negative control (Fig. 3C). To verify this hypersensitive effect of PsTRXh2, we repeated yeast mutant complementation assays to measure sensitivity to 0.4 mm H2O2 using pFL61 as shuttle vector. Figure 3D shows that all yeast mutant cells expressing both plant and yeast TRX from the phosphoglycerate kinase promoter, including those carrying the empty pFL61 vector, could grow in the presence of the oxidant molecules except those expressing PsTRXh2. This result confirmed that PsTRXh2 confers hypersensitivity to H2O2 to yeast cells in vivo. Taken together, our results demonstrate that both PsTRXh1 and PsTRXh2 are functional TRXs in vivo but confer different behavior to trx1Δ trx2Δ yeast cells, probably by interacting with different yeast targets.
PsTRXh1, PsTRXh2 mRNAs, and Derived Proteins Are Differently Accumulated in Pea Organs
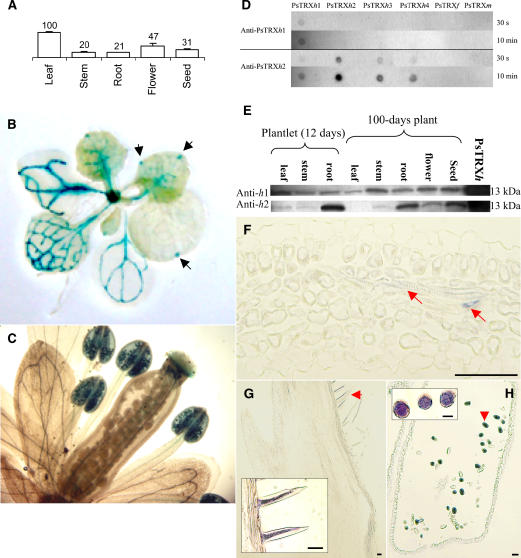
The above results suggest that in addition to a true functional TRX activity in vivo, PsTRXh1 and PsTRXh2 may display a divergent function in pea tissues in light of their different behavior in yeast cells under oxidative stress. To characterize the pattern of expression of PsTRXh1 and PsTRXh2 genes, we first performed a relative quantification of their corresponding mRNAs using real-time PCR on different tissues excised from 100-d-old pea plants. PsTRXh1 transcripts were detected in all the organs tested, showing a higher accumulation in leaves and flowers (Fig. 4A). In contrast, PsTRXh2 mRNA was almost undetectable, presenting a much weaker accumulation than PsTRXh1 mRNA, with mean values lower than 1% as compared with the PsTRXh1 level of mRNA (data not shown). A higher level of PsTRXh2 gene expression could be detected only in plantlets younger than 12 d but always remained at a maximum expression level of 18% and 16% in respect to the values of PsTRXh1 transcripts in leaves and roots, respectively (data not shown). These results were confirmed in transgenic Arabidopsis plants expressing the β-glucuronidase (GUS; UidA) reporter gene under the control of the promoter of either PsTRXh1 or PsTRXh2 genes. In such plants, we found that PsTRXh1 promoter produced strong UidA expression in vascular veins of the whole plant, except in the root, with expression being also strong in hydathodes (Fig. 4B, arrows). In addition, GUS activity under PsTRXh1 promoter was detected in pollen grains and stigmatic papillae in flowers (Fig. 4C). In contrast, we were unable to detect UidA expression under the control of PsTRXh2 promoter in transgenic seedling and mature plants grown under standard growth conditions (data not shown).
Figure 4.
PsTRXh1 and PsTRXh2 gene expression and protein accumulation in pea tissues. A, PsTRXh1gene expression determined by real-time PCR. Total RNA was obtained from dissected tissues of mature plants (100-d-old). Final values are the result of three independent experiments and were normalized by the 2ΔΔCT method using the 18S rRNA as standard (Livak and Schmittgen, 2001). Expression level of 100% was arbitrarily attributed to the maximum value obtained with leaves. B and C, Histochemical localization of GUS activity in transgenic lines of Arabidopsis plants expressing the UidA reporter gene under the control of PsTRXh1 promoter (PrPsTRXh1∷GUS). Signal was detected in vascular tissues from the green organs and in the hydathode structure in leaves (B, black arrows). In flowers, the signal was detected in the pollen grains and stigma papillae (C). D, Cross reaction of various pea TRX (10 ng each) with anti-PsTRXh1 and anti-PsTRXh2 antibodies, as indicated. Exposure times are indicated on the right. E, Western-blot analysis of PsTRXh1 and h2 isoforms in pea tissues. Protein extracts (20 μg) from dissected tissues were loaded. F to H, Immunolocalization of pea TRXh1 protein. Sections of 10 μm were incubated with either the anti-PsTRXh1 antibody or the preimmune serum (data not shown). Signals were detected in vascular tissue from leaves (F), stigma papillae (G), and pollen grains (H). Bars = 100 μm.
We next analyzed the accumulation of PsTRXh1 and PsTRXh2 proteins in pea tissues and whether their accumulation correlated their corresponding gene expression. For this purpose, recombinant PsTRXh1 and PsTRXh2 proteins were used to obtain antibodies that were subsequently checked for their specificity. Figure 4D shows that the anti-PsTRXh1 antibody is specific to its antigen, recognizing only the PsTRXh1 isoform. No recognition signal was obtained with other PsTRXh proteins despite long-time exposure or when preimmune serum was used. This information allowed us to search for the presence of the PsTRXh1 isoform in pea tissues. Western-blot analysis of protein extracts from different tissues probed with the anti-PsTRXh1 antibody detected a band, close to 13 kD, which was more abundant in leaves of 12-d-old plantlets (Fig. 4E). As the plant became mature (100-d-old plants), the level of PsTRXh1 decreased in leaves while increasing in stem. A high accumulation of PsTRXh1 protein was detected in flowers and dry seeds (Fig. 4E). A more detailed analysis of PsTRXh1 localization using immunohistochemistry revealed that PsTRXh1 accumulates in vascular tissues of the whole plant, as shown in leaves (Fig. 4F). A high signal was also detected in floral tissues, the stigmatic papillae (Fig. 4G), and pollen grains (Fig. 4H). These results corroborate the data obtained in transgenic plants carrying the PromPsTRXh1∷UidA construct (Fig. 4, B and C).
Regarding PsTRXh2, the antibody raised against this isoform recognized its antigen but also detected all the h-type isoforms tested (Fig. 4D). Such cross detections were obtained regardless of shortened time exposure and adapted antibody concentrations. When western blots were probed with this antibody, a strong signal was detected in roots from both 12-d-old plantlets and mature plants and also in flowers and seeds (Fig. 4E). This pattern of protein accumulation was not consistent with PsTRXh2 mRNA accumulation. It is likely that these levels mostly correspond to cross recognition of other abundant isoforms such as PsTRXh3, for which similar patterns were previously reported (Montrichard et al., 2003). Therefore, PsTRXh2 accumulation could not be further investigated.
PsTRXh1 and PsTRXh2 Genes Are Differentially Expressed in Response to Oxidative Stress in Pea Tissues
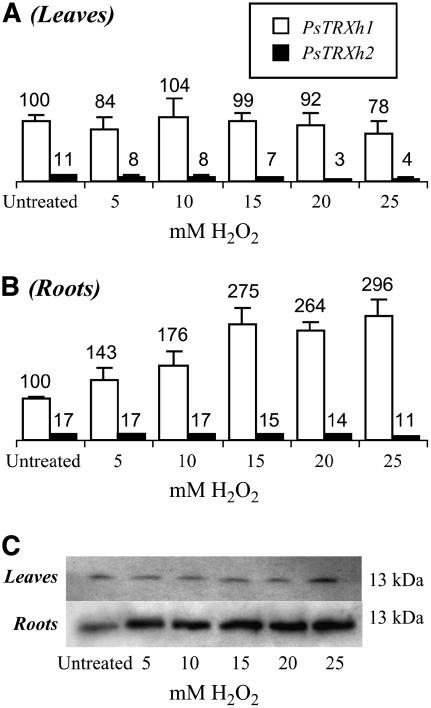
Heterologous complementation of the yeast EMY63 mutant with pea TRXs h1 and h2 isoforms suggested that they could be specifically involved in different detoxification pathways toward oxidant molecules. To test whether PsTRXh1 and PsTRXh2 accumulation patterns in planta were also influenced by oxidative stress, we performed a series of experiments in which 12-d-old pea plantlets were treated with H2O2. The expression of PsTRXh1 and PsTRXh2 genes was analyzed by real-time PCR on leaves and roots collected from control or stressed plants, and, when possible, the patterns of protein accumulation were also analyzed. When pea plantlets were grown in the presence of increasing concentrations of H2O2, the amount of PsTRXh1 transcripts remained unchanged in leaves (Fig. 5A), whereas it strongly increased in roots (Fig. 5B) as compared to untreated plants. This effect on the amount of transcripts correlated with the protein level as shown by western-blot analysis performed on the same plantlets using the anti-PsTRXh1 antibody, which showed the higher accumulation of PsTRXh1 in roots from plantlets treated with H2O2, whereas no significant effect was observed on leaves (Fig. 5C).
Figure 5.
PsTRXh1 and PsTRXh2 gene expression in pea tissues under oxidative stress. A and B, Relative PsTRXh1 (white bars) and PsTRXh2 (black bars) gene expression was measured by real-time PCR on pea leaves (A) and roots (B) treated with increasing concentrations of H2O2. Final values on top of bars are the result of three independent experiments and were normalized by the 2ΔΔCT method using the 18S rRNA as standard (Livak and Schmittgen, 2001). C, Immunodetection of PsTRXh1 in pea tissues under oxidative stress. Protein extracts from untreated and stressed leaves (50 μg) and roots (30 μg) were loaded in a Tris-tricine gel and transferred onto nitrocellulose membrane. PsTRXh1 was detected using the anti-PsTRXh1 antibody. Concentrations of the hydroperoxide in millimolars are indicated at the bottom.
Regarding PsTRXh2 gene, its messengers were scarcely detectable, as previously mentioned (Fig. 5A, black bars). As it was observed in previous analyses (Fig. 4D), the cross detection of other PsTRX isoforms by anti-PsTRXh2 antibody did not allow the further analysis of PsTRXh2 accumulation in stressed tissues.
DISCUSSION
In plants, h-type TRXs constitute a complex group of TRXs. Whereas nine TRXs of this type have been described in Arabidopsis among more than 40 TRX-related sequences (Meyer et al., 2005), five members have been reported in Populus sp. (Gelhaye et al., 2004b) and three in wheat (Triticum aestivum; Cazalis et al., 2006). In pea, a legume of agronomical interest, only two isoforms, PsTRXh3 and PsTRXh4, have been characterized, both being related to seed development and germination (Montrichard et al., 2003). The first task of this work was to extend our knowledge of the TRX family in this plant. We describe here two new h-type isoforms, PsTRXh1 and PsTRXh2, and show their different pattern of expression. The second aim of this work was to unravel the biological function of h1 and h2 isoforms by comparison with the two previously described PsTRXh3 and PsTRXh4 isoforms. Based on physiological and functional approaches, we provide evidence that h1 and h2 isoforms have different function in the redox imbalances.
The phylogenic analysis confirmed that both PsTRXh1 and PsTRXh2 isoforms belong to the h-type subgroup 1 of TRXs, as their counterpart PsTRXh3 (Montrichard et al., 2003; Gelhaye et al., 2004b). The absence of target peptide in the amino acid sequence of both deduced proteins suggests that they are most probably cytosolic. For PsTRXh1, we demonstrate its presence in vascular veins in agreement with previous reports describing h-type TRXs as abundant proteins in the phloem sap (Ishiwatari et al., 1995). Both isoforms are functional TRXs in vitro as shown by the insulin reduction assay, though PsTRXh1 showed higher activity. However, PsTRXh1 and PsTRXh2 showed differences regarding thermostability and dimerization capacity.
Though PsTRXh1 and PsTRXh2 are grouped with PsTRXh3 in the same phylogenic subgroup, they differ from the other pea cytosolic TRX characterized up to now by their pattern of accumulation in plant tissues characterized up to now. PsTRXh3 and PsTRXh4 have been described as abundant seed proteins (Montrichard et al., 2003), PsTRXh3 decreasing slowly after germination and PsTRXh4 disappearing after radicle protrusion. The corresponding genes were mostly expressed in cotyledons and axes during seed imbibition, whereas expression in plantlets was detected at higher level in roots (Montrichard et al., 2003). The expression analysis of PsTRXh1 and PsTRXh2 genes reveals a differential expression in pea tissues. In contrast to PsTRXh3 and PsTRXh4 genes, PsTRXh1 gene expression was detected in all the organs analyzed from young and mature plants, whereas PsTRXh1 protein accumulated preferentially in vascular tissues. However, PsTRXh2 gene was expressed at a very low level, which suggests a low accumulation of the corresponding protein, as shown for some of the h-type TRXs of Arabidopsis (Reichheld et al., 2002). Due to lack of specificity, the anti-PsTRXh2 polyclonal antibody produced in this study could not be used to determine the physiological conditions in which PsTRXh2 accumulation may be modified. Interestingly, and contrary to previous data (Montrichard et al., 2003), we found that both PsTRXh1 and PsTRXh2 mRNAs are present in dry seeds and that at least the PsTRXh1 protein accumulated in this organ. Whether h1, h3, and h4 share common function and targets in seeds remains to be investigated. However, the fact that the four isoforms exhibit divergent patterns and levels of accumulation in pea tissues suggests that their functions are, at least in part, not redundant.
To analyze further the function of h-type TRXs in pea, we tested the ability of PsTRXh1 and PsTRXh2 to complement the trx1Δ trx2Δ yeast mutant. Until now, this mutant has been used to study the functional specificity of the TRXs from Arabidopsis (Mouaheb et al., 1998; Issakidis-Bourguet et al., 2001; Vignols et al., 2003), identify TRX targets (Verdoucq et al., 1999), and find determinants for the specificity into TRX architecture (Bréhélin et al., 2000). The first major conclusion that may be drawn from our results is that contrary to AtTRXh2 from Arabidopsis (Mouaheb et al., 1998), neither PsTRXh1 nor PsTRXh2 seem to be involved in sulfate assimilation. Both pea h1 and h2 isoforms were unable to restore trx1Δ trx2Δ yeast cell growth in the absence of Met, indicating their incapacity to reduce the phosphoadenosine-5-phosphosulfate reductase, an enzyme that requires reduction by TRX for correct sulfate assimilation (Russel et al., 1990; Draculic et al., 2000). This result suggests that another pea isoform may be involved in sulfate assimilation. Interestingly, PsTRXh4 and AtTRXh2 are members of subgroup 2 of the h-type TRXs cluster (Gelhaye et al., 2004b), suggesting that the pea h4 isoform, or a yet unknown TRXh, could fulfill this role in yeast and therefore in pea cells.
The second contribution of this study was to reveal a potential function of PsTRXh1 and h2 isoforms in physiological responses to oxidative stress. We provide evidence that both h1 and h2 pea TRXs are able to restore the inability of trx1Δ trx2Δ mutant cells to use Met sulfoxide as a source of sulfur, the h2 isoform being much more efficient. The inability of the trx yeast mutant to use Met sulfoxide as the only sulfur source is due to a defect of the Met sulfoxide reductase (MSR) activity (Müller, 1991), an enzyme that normally requires reduction by TRX to be active. MSR is an antioxidant enzyme that exerts its protection against oxidative stress by maintaining a low level of oxidized Met, either as protein-bound or as free amino acid form (Moskovitz et al., 1997). Therefore, we propose that PsTRXh2 and, to a lower extent, PsTRXh1 are involved in the reduction of MSR enzyme in pea cells, which is consistent with the isolation of plant MSRs as putative TRX targets (Buchanan and Balmer, 2005).
Finally, our study shows that the function of h1 and h2 isoforms is not restricted to an antioxidant response through the reduction of MRS enzymes but may be extended to the reduction of other antioxidant enzymes such as peroxiredoxins (PRXs). PsTRXh1 and PsTRXh2 conferred differential response of trx1Δ trx2Δ mutant cells toward hydroperoxides. PsTRXh1 conferred high resistance to H2O2 and low to TBHP, whereas PsTRXh2 conferred hypersensitivity to H2O2 and resistance to TBHP. Previous studies have shown that trx1Δ trx2Δ yeast mutant complementation of H2O2 sensitivity involves TRX interaction with the yeast AHP1 type-II PRX (cTPxII; Verdoucq et al., 1999), suggesting that this yeast protein could be preferentially the target of PsTRXh1. Regarding PsTRXh2, its hypersensitizing effect in trx1Δ trx2Δ yeast mutant toward H2O2 was directly correlated with the type of vector used for heterologous complementation, the growth of mutant yeast cells being greatly affected when using high-copy vector pFL61. The effect of PsTRXh2 in trx1Δ trx2Δ yeast mutant is very similar to that of AtTRXm3 (Issakidis-Bourguet et al., 2001) despite the fact that both pea and Arabidopsis isoforms are phylogenetically distant (Meyer et al., 2005). It is too preliminary to know the exact function of these isoforms, but our data indicate that PsTRXh2 and PsTRXh1 have different roles most probably by capturing specific targets that are differentially involved in oxidative stress response. This is corroborated by the effect of TBHP on the trx1Δ trx2Δ yeast mutant, which showed better growth when complemented with PsTRXh2 than with PsTRXh1. In yeast, the two types of TRX-dependent peroxidases (cTPxI/TSA1 and cTPxII/AHP1) exhibit opposite preferences for H2O2 or alkyl hydroperoxide reduction (Jeong et al., 1999). In contrast to cTPxI protein, cTPxII is not abundant and is highly inducible by peroxides. Both corresponding mutants are sensitive to TBHP but with a higher hypersensitivity to TBHP and higher resistance to H2O2 for cTPxII in fermentative and respiratory conditions (Munhoz and Netto, 2004). We suggest that both PsTRXh1 and h2 isoforms could interact specifically with these different members of the PRX family involved in detoxification.
Several data suggest that at least the PsTRXh1 isoform could be involved in reactive oxygen species (ROS) detoxification in pea tissues through PRX reduction. Plant PRXs act as a common antioxidant (Dietz et al., 2002; Dietz, 2003), the TRXs being identified as one of their natural reductants (Chae et al., 1994; Kwon et al., 1994). PRXII isoforms have been detected in Arabidopsis tissues where TRXs are also present (Bréhélin et al., 2003), and PRXs have been identified as putative TRX targets (for review, see Buchanan and Balmer, 2005). In addition, direct interaction between Arabidopsis PRXs and one cytosolic h-type TRX has been demonstrated (Yamazaki et al., 2004). Interestingly, PsTRXh1 is induced by oxidative stress. This is consistent with the ability of the h1 isoform to confer resistance to oxidative stress to TRX-deficient yeast cells and with a function as PRX reductant required to maintain the redox imbalances in pea tissues. The presence of PsTRXh1 in vascular veins could be related with phloem antioxidant systems (Walz et al., 2002), including those related to TRXs, as PRXs or glutaredoxins (Szederkenyi et al., 1997; Rouhier et al., 2001). Moreover, it could contribute to long-distance redox signaling between different parts of the plant as it has been recently suggested (Balmer et al., 2006).
Additionally, we show that PsTRXh1 is present in pollen and stigma papillae. In a recent work, relatively large amounts of ROS, principally H2O2, have been detected in stigma papillae from several species across a range of different angiosperm groups. This effect has been related to possible functions such as pollen-pistil interaction, redox signaling, or resistance to pathogen (McInnis et al., 2006). The presence of PsTRXh1 in these tissues may be an additional indication of its involvement in ROS metabolism. The PsTRXh1 gene is also expressed in hydathodes as has also been described for AtTRXh4 (Reichheld et al., 2002), but the function of these structures is not yet understood. Regarding the function of PsTRXh2 in planta, our results suggest that it might differ from that of PsTRXh1 due to its very low level of expression and its ability to provoke hypersensitivity of yeast trx1Δ trx2Δ mutant cells to H2O2.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Pea (Pisum sativum) L. cv Lincoln seeds were germinated on vermiculite in plastic trays and grown in a growth chamber for 10 to 15 d under 16-/8-h photoperiod at a light intensity of 200 μmol m−2 s−1, with a day/night temperature of 20°C/25°C. After 3 weeks, plants were transplanted into a soil:vermiculite (3:1, w/w) medium. At different times thereafter, plants were collected and dissected into different parts (flowers, stems, roots, leaves, and seeds) and then processed immediately or frozen until use. For stress treatments, roots of 12-d-old pea seedlings were placed in the oxidant solutions (0, 5, 10, 15, 20, and 25 mm of either H2O2 or TBHP) and maintained for 15 h in darkness. Roots and leaves were then washed in distilled water and either analyzed or frozen in liquid nitrogen and stored at −80°C until use.
Nucleic Acid Extraction and PCR Techniques
DNA was extracted and purified from pea leaves using the Plant DNAzol Reagent kit (Invitrogen). The mRNA was obtained from 12-d-old green leaves using the mRNA Isolation kit (Roche). RT-PCR was performed using the Access RT-PCR system (Promega) according to the manufacturer's instructions. PCR-Walking was performed on a pea genomic DNA library as template following the methods described by Devic et al. (1997).
PsTRXh1 and PsTRXh2 Cloning
All primers used for cloning are described in Table I. Th2N/2C (for h1isoform) and ThN/2C (for h2 isoform) pairs of primers were first used in a RT-PCR on RNA extracted from pea leaf to amplify h1 and h2 partial cDNAs. Based on these sequences, new primers were designed to amplify full-length cDNAs by RACE. 3′-RACE experiments were performed using Th3F and Th4F primers for h1 and h2, respectively, whereas DNA-Walk experiments (Devic et al., 1997) were performed using Th3R/Th4RN (for h1) and Psw1/Psw2 (for h2) pairs of primers to amplify 5′-coding and 5′-UTR sequences. Genomic sequences flanking introns in both PsTRXh1 and PsTRXh2 genes were obtained using PeahN/PeahC and Peah2N/Peah2C pairs of primers.
Table I.
Primers used in this work
Ta, Annealing temperature.
| Primers | Restriction Sequencea | Sequence | Ta |
|---|---|---|---|
| °C | |||
| PeahN | NcoI | 5′-agaccatggcaggttcatcagaagagg-3′ | 60 |
| PeahC | BamHI | 5′-ttggatccctaagcattagatgaagccac-3′ | 60 |
| Peah2N | NcoI | 5′-agaccatggcagcagaaaatgaggtg-3′ | 60 |
| Peah2C | BamHI | 5′-ttggatcctcaagcagtagcaacagtt-3′ | 60 |
| ThN | 5′-atggctgcagaaggtgaagtgatc-3′ | 63 | |
| Th2N | 5′-gaggagggacaagtgatcgcttgccac-3′ | 63 | |
| Th2C | 5′-ctcctctttctttgcaccaacaactttatc-3′ | 63 | |
| Th4F | 5′-caaaaaactgattgtggtggattt-3′ | 50 | |
| Th3F | 5′-tgtgtttgtgaaagaaggaacg-3′ | 50 | |
| Th3R | 5′-gtgaaggatatcgttccatgc-3′ | 60 | |
| Th4RN | 5′-tttcttggattcattgcctct-3′ | 54 | |
| Psw1 | 5′-cttccatgattcatcggaatgaac-3′ | 54 | |
| Psw2 | 5′-gttcctttctggatctgttgttccttcc-3′ | 60 | |
| MluIhN | MluI | 5′-agacgcgtatggcaggttcatcagaa-3′ | 60 |
| MluIh2N | MluI | 5′-agacgcgtatggcagcagaaaatgag-3′ | 60 |
| NotIhN | NotI | 5′-ttgcggccgcatggcaggttcatca-3′ | 67 |
| NotIhC | NotI | 5′-ttgcggccgcctaagcattagatgaa-3′ | 67 |
| NotIh2N | NotI | 5′-ttgcggccgcatggcagcagaaaat-3′ | 65 |
| NotIh2C | NotI | 5′-ttgcggccgctcaagcagtagcaac-3′ | 65 |
| Ycp2-5′ | 5′-cctctatactttaacgtcaagg-3′ | 55 | |
| Ycp2-3′ | 5′-acgtgtacaccgggtcacgt-3′ | 55 | |
| FLDR | 5′-ctattattttagcgtaaaggatgg-3′ | 55 | |
| FLGA | 5′-ctcttttttacagatcatcaagg-3′ | 55 | |
| H1up | 5′-tcttcattgttgttgggatatcttg-3′ | 55 | |
| H1down | 5′-tcatcctccaccattatttaagacg-3′ | 55 | |
| H2up | 5′-tcatatcaagatcatcaataacatagtgaact-3′ | 55 | |
| H2down | 5′-ttgaagtaggccaataatgtgcc-3′ | 55 | |
| 18Sup | 5′-agtaagcgcgagtcatcagct-3′ | 55 | |
| 18Sdown | 5′-cattcaatcggtaggagcgac-3′ | 55 |
Restriction sequences are underlined.
Gene Expression Analysis by Real-Time PCR
Samples of total RNA (500 ng) extracted from stressed plants using the RNeasy Plant Mini kit (Qiagen) were reverse transcribed using the Multiscribe Reverse Transcriptase and random hexamer primers (Applied Biosystems). For quantification, the SYBR Green technology, an ABI Prism 7700 Sequence detector (Applied Biosystems), and the QuantiTect SYBR Green PCR kit (Qiagen) were used. Specific primers at the 3′-untranslated region were designed using the Primer Express (Applied Biosystems) software (H1up/down for PsTRXh1, H2up/down for PsTRXh2, and 18Sup/down for rRNA 18S; Table I). Relative quantification of gene expression was monitored after normalization by the 18S rRNA expression as internal control, as fold variation over a calibrator using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
Overexpression of PsTRXh1 and PsTRXh2 in Escherichia coli
To produce recombinant PsTRXh1 and PsTRXh2 in Escherichia coli, the corresponding ORFs were cloned into the expression plasmid pET-3d (Novagen) via NcoI and BamHI restriction sites. Induction of expression of recombinant proteins was performed according to Wangensteen et al. (2001) with modifications. For PsTRXh1 purification, the soluble fraction obtained after recombinant protein induction was heated to 70°C for 10 min. The supernatant was then fractionated with (NH4)2SO4 (40%–85%, w/v), and the final precipitate dissolved in TRX buffer (30 mm Tris-HCl, pH 7.9, 100 mm NaCl, 1.4 mm 2-mercaptoethanol) supplemented with 0.1 mm phenylmethanesulfonyl fluoride. The solution was filtered through a Sephadex G-50 column, and the fractions containing TRX were analyzed by SDS-PAGE (Laemmli, 1970) and further concentrated. Nucleic acids were eliminated through DEAE-cellulose column using a linear gradient (100–600 mm NaCl) in the TRX buffer. PsTRXh2 was produced as described for PsTRXh1, but with the following modifications: NaCl-free TRX buffer was used, no heating step was applied, and double Sephadex G-50 filtration step was required. Protein concentration was determined by the Bradford method (Bradford, 1976).
Determination of TRX Activity
The insulin-disulphide reduction assay was performed (Holmgren, 1979). Recombinant TRXs were used at final concentrations of 2.5 and 5 μm in potassium phosphate buffer 100 mm, pH 7.0, 2 mm EDTA, 0.13 mm bovine insulin, and 0.25 mm dithiothreitol as reductant.
Heterologous Complementation Analysis
Complementation experiments were performed with the yeast (Saccharomyces cerevisiae) strain EMY63 (Mata ade2-1 ade3-100 his3-11 leu2-3 lys2-801 trp1-1 ura3-1 trx1∷TRP1 trx2∷LEU2; Müller, 1991) according to previous reports (Mouaheb et al., 1998; Issakidis-Bourguet et al., 2001). Plant TRX expression in yeast cells was achieved using either the inducible Ycp2 shuttle vector (Cole et al., 1990) or the constitutive pFL61 vector (Minet et al., 1992). Each plant TRX was amplified from the corresponding full-length cDNA by PCR using specific pairs of oligonucleotides designed to introduce either MluI (5′)/BamHI (3′) or NotI (5′)/NotI (3′) sites necessary for cloning into Ycp2 and pFL61 vectors, respectively (MluIhN/PeahC and NotIhN/NotIhC to amplify and clone h1 ORF; MluIh2N/Peah2C and NotIh2N/NotIh2C for h2 cloning; Table I). ScTRX1 (YLR043c), AtTRXh2 (At5g39950), and AtTRXh3 (At5g42980) ORFs carried by both vectors were used as positive controls according to a previous work (Mouaheb et al., 1998), the empty vectors being used as negative controls. All constructs were introduced into the yeast EMY63 cells by lithium acetate method (Ito et al., 1983). Transformants were verified by PCR on EMY63 total genomic DNA using Ycp2-5′/Ycp2-3′ and FLDR/FLGA pairs of primers (Table I). Yeast complementation and halo-inhibition assays were performed according to Vignols et al. (2005) and Issakidis-Bourguet et al. (2001), respectively.
Western-Blot and Antibodies Cross-Reaction Analyses
Plant tissues were homogenized with liquid nitrogen and ground in ice-cold mortar with extraction buffer (50 mm Tris-HCl, pH 7.9, 0.2 mm EDTA, 0.5 mm phenylmethanesulfonyl fluoride, and 5 mm 2-mercaptoethanol). The homogenates were centrifuged, and the supernatant was processed or stored at −80°C. For western-blot analysis, protein samples were separated on SDS-PAGE 16.5% (w/v) polyacrylamide (Schagger and Vonjagow, 1987) and then electrotransferred onto polyvinylidene difluoride membranes. Immunodetection was performed using antibodies raised against pea TRXsh1 (1:40,000) and h2 (1:10,000), coupled with ECL western-blotting detection system (Amersham Biosciences). In cross-reaction assays, 2 μL (5 ng/μL) of recombinant proteins were dropped on polyvinylidene difluoride membranes and immunodetected using the anti-PsTRXh1 (1:10,000) and PsTRXh2 (1:2,500). Plastidial PsTRXf and PsTRXm (Pagano et al., 2000) were already available in the lab as pET constructs, and PsTRXh3 and PsTRXh4 recombinant proteins were kindly provided by Dr. F. Montrichard (Angers, France).
Immunohistochemistry
Small pieces of plant tissues were dissected and fixed by incubation in FAE (50% ethanol, 5% acetic acid, 3.7% formaldehyde) with occasional vacuum, dehydrated in a graded series of aqueous ethanol solution, and embedded in Paraffin M.P. (Panreac) as described in González et al. (1998). Sections (10-μm thick) were cut in a Leica RM 2025 microtome and placed on poly-l-Lys-coated microscope slides. After deparaffinizing in Histo-Clear (National Diagnostic) and rehydrating in decreasing concentrations of ethanol, sections were blocked for 3 h in Tris-buffered saline (TBS) buffer containing 1% (w/v) bovine serum albumin. Anti-PsTRXh1 antibody (diluted 1:4,000 in TBS) or preimmune serum were added to the samples and incubated overnight at 4°C. Unbound primary antibodies were removed by three washes of 10 min in TBS. Tissue sections were then incubated with alkaline phosphatase-conjugated goat anti-rabbit IgG for 2 h at 37°C. The reaction of alkaline phosphatase was developed with nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl-P.
Transgenic Plants and GUS Assays
PsTRXh1 and PsTRXh2 promoter regions were isolated from genomic DNA by PCR walking (Devic et al., 1997), using a reverse primer to introduce a unique BamHI site upstream from the ATG codon. Fragments of 1.03 kb (PsTRXh1) and 0.3 kb (PsTRXh2) were isolated and cloned into the binary vector pBI101.1 (Clontech) at the initiation codon of the promoterless GUS gene. The resulting plasmids were introduced into Agrobacterium tumefaciens (C58pMP90). Arabidopsis (Arabidopsis thaliana; ecotype Columbia) was transformed with Agrobacterium by the floral dip method (Clough and Bent, 1998). T1, T2, and T3 seedlings were selected in vitro on Murashige and Skoog medium supplemented with 1% (w/v) Suc, 0.8% (w/v) agar, and 30 μg/mL kanamycin under a 16-h-light/8-h-dark regime at 22°C. Plants were cultivated either in vitro on the same medium or in soil:vermiculite (3:1, w/w) medium and under the same conditions as described above. GUS histochemical staining was performed according to Jefferson et al. (1987).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers PsTRXf (CAA45098) and PsTRXm1 (CAA53900). The accession numbers for the rest of the sequences are in the Figure 1 legend.
Acknowledgments
We thank Dr. F. Montrichard for the gift of PsTRXh3 and PsTRXh4 recombinant proteins and Dr. R. Cooke for revising the manuscript.
This work was supported by the Dirección General de Investigación Científica y Técnica, Spain (grant nos. PB98–0474 and BF12002–00401), by the Junta de Andalucía, Spain (grant no. CVI 154), by CSIC (Acción Integrada grant no. HF2001–0136), and by the Spanish government (fellowship FPI98 to J.A.T.).
This paper is dedicated to the loved and esteemed memory of Professor Julio López Gorgé, who died on June 7, 2004, at the age of 69.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: José A. Traverso (jose.traverso@isv.cnrs-gif.fr).
References
- Arner ESJ, Holmgren A (2000) Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem 267 6102–6109 [DOI] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schürmann P, Hurkman WJ, Buchanan BB (2006) A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proc Natl Acad Sci USA 103 2988–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse I, Wong JH, Kobrehel K, Buchanan BB (1996) Thiocalsin: a thioredoxin-linked, substrate-specific protease dependent on calcium. Proc Natl Acad Sci USA 93 3169–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower MS, Matias DD, Fernandes-Carvalho E, Mazzurco M, Gu T, Rothstein SJ, Goring DR (1996) Two members of the thioredoxin-h family interact with the kinase domain of a Brassica S locus receptor kinase. Plant Cell 8 1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Bréhélin C, Meyer EH, de Souris JP, Bonnard G, Meyer Y (2003) Resemblance and dissemblance of Arabidopsis type II peroxiredoxins: similar sequences for divergent gene expression, protein localization, and activity. Plant Physiol 132 2045–2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bréhélin C, Mouaheb N, Verdoucq L, Lancelin JM, Meyer Y (2000) Characterization of determinants for the specificity of Arabidopsis thioredoxins h in yeast complementation. J Biol Chem 275 31641–31647 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56 187–220 [DOI] [PubMed] [Google Scholar]
- Cabrillac D, Cock JM, Dumas C, Gaude T (2001) The S locus receptor kinase is inhibited by thioredoxins and activated by pollen coat proteins. Nature 410 220–223 [DOI] [PubMed] [Google Scholar]
- Cazalis R, Pulido P, Aussenac T, Pérez-Ruiz JM, Cejudo FJ (2006) Cloning and characterization of three thioredoxin h isoforms from wheat showing differential expression in seeds. J Exp Bot 57 2165–2172 [DOI] [PubMed] [Google Scholar]
- Chae HZ, Chung SJ, Rhee SG (1994) Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem 269 27670–27678 [PubMed] [Google Scholar]
- Cherest H, Surdin-Kerjan Y (1992) Genetic analysis of a new mutation conferring cysteine auxotrophy in Saccharomyces cerevisiae: updating of the sulfur metabolism pathway. Genetics 130 51–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of A. thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Cole GM, Stone DE, Reed SI (1990) Stoichiometry of G protein subunits affects the Saccharomyces cerevisiae mating pheromone signal transduction pathway. Mol Cell Biol 10 510–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coudevylle N, Thureau A, Hemmerlin C, Gelhaye E, Jacquot JP, Cung MT (2005) Solution structure of a natural CPPC active site variant, the reduced form of thioredoxin h1 from poplar. Biochemistry 44 2001–2008 [DOI] [PubMed] [Google Scholar]
- Devic M, Albert S, Delseny M, Roscoe TJ (1997) Efficient PCR walking on plant genomic DNA. Plant Physiol Biochem 35 331–339 [Google Scholar]
- Dietz KJ (2003) Plant peroxiredoxins. Annu Rev Plant Biol 54 93–107 [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Horling F, Konig J, Baier M (2002) The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J Exp Bot 53 1321–1329 [PubMed] [Google Scholar]
- Draculic T, Dawes IW, Grant CM (2000) A single glutaredoxin or thioredoxin gene is essential for viability in the yeast Saccharomyces cerevisiae. Mol Microbiol 36 1167–1174 [DOI] [PubMed] [Google Scholar]
- Florencio FJ, Yee BC, Johnson TC, Buchanan BB (1988) An NADP/thioredoxin system in leaves: purification and characterization of NADP-thioredoxin reductase and thioredoxin h from spinach. Arch Biochem Biophys 266 496–507 [DOI] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Gerard J, Jolivet Y, Gualberto J, Navrot N, Ohlsson PI, Wingsle G, Hirasawa M, Knaff DB, et al (2004. a) A specific form of thioredoxin h occurs in plant mitochondria and regulates the alternative oxidase. Proc Natl Acad Sci USA 101 14545–14550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelhaye E, Rouhier N, Jacquot JP (2004. b) The thioredoxin h system of higher plants. Plant Physiol Biochem 42 265–271 [DOI] [PubMed] [Google Scholar]
- González MC, Bevia O, Echevarria C, Vidal J, Cejudo FJ (1998) Expression and localization of phosphoenolpyruvate carboxylase in developing and germinating wheat grains. Plant Physiol 116 1249–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A (1979) Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J Biol Chem 254 9627–9632 [PubMed] [Google Scholar]
- Ishiwatari Y, Fujiwara T, McFarland KC, Nemoto K, Hayashi H, Chino M, Lucas WJ (1998) Rice phloem thioredoxin h has the capacity to mediate its own cell-to-cell transport through plasmodesmata. Planta 205 12–22 [DOI] [PubMed] [Google Scholar]
- Ishiwatari Y, Honda C, Kawashima I, Nakamura S, Hirano H, Mori S, Fujiwara T, Hayashi H, Chino M (1995) Thioredoxin h is one of the major proteins in rice phloem sap. Planta 195 456–463 [DOI] [PubMed] [Google Scholar]
- Issakidis-Bourguet E, Mouaheb N, Meyer Y, Miginiac-Maslow M (2001) Heterologous complementation of yeast reveals a new putative function for chloroplast m-type thioredoxin. Plant J 25 127–135 [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A (1983) Transformation of intact yeast cells treated with alkali cations. J Bacteriol 153 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo JL, Chueca A, Jacquot JP, Hermoso R, Lazaro JJ, Sahrawy M, Gorge JL (1997) High-yield expression of pea thioredoxin m and assessment of its efficiency in chloroplast fructose-1,6-bisphosphatase activation. Plant Physiol 114 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kwon SJ, Kang SW, Rhee SG, Kim K (1999) Purification and characterization of a second type thioredoxin peroxidase (type II TPx) from Saccharomyces cerevisiae. Biochemistry 38 776–783 [DOI] [PubMed] [Google Scholar]
- Kobrehel K, Wong JH, Balogh A, Kiss F, Yee BC, Buchanan BB (1992) Specific reduction of wheat storage proteins by thioredoxin h. Plant Physiol 99 919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon SJ, Park JW, Choi WK, Kim IH, Kim K (1994) Inhibition of metal-catalyzed oxidation systems by a yeast protector protein in the presence of thioredoxin. Biochem Biophys Res Commun 201 8–15 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Laloi C, Mestres-Ortega D, Marco Y, Meyer Y, Reichheld JP (2004) The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol 134 1006–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2ΔΔCT method. Methods 25 402–408 [DOI] [PubMed] [Google Scholar]
- Maeda K, Finnie C, Svensson B (2004) Cy5 maleimide labelling for sensitive detection of free thiols in native protein extracts: identification of seed proteins targeted by barley thioredoxin h isoforms. Biochem J 378 497–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx C, Wong JH, Buchanan BB (2003) Thioredoxin and germinating barley: targets and protein redox changes. Planta 216 454–460 [DOI] [PubMed] [Google Scholar]
- McInnis SM, Emery DC, Porter R, Desikan R, Hancock JT, Hiscock SJ (2006) The role of stigma peroxidases in flowering plants: insights from further characterization of a stigma-specific peroxidase (SSP) from Senecio squalidus (Asteraceae). J Exp Bot 57 1835–1846 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Reichheld J, Vignols F (2005) Thioredoxins in Arabidopsis and other plants. Photosynth Res 86 419–433 [DOI] [PubMed] [Google Scholar]
- Meyer Y, Vignols F, Reichheld JP (2002) Classification of plant thioredoxins by sequence similarity and intron position. Methods Enzymol 347 394–402 [DOI] [PubMed] [Google Scholar]
- Minet M, Dufour ME, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2 417–422 [DOI] [PubMed] [Google Scholar]
- Montrichard F, Renard M, Alkhalfioui F, Duval FD, Macherel D (2003) Identification and differential expression of two thioredoxin h isoforms in germinating seeds from pea. Plant Physiol 132 1707–1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Berlett BS, Poston JM, Stadtman ER (1997) The yeast peptide methionine sulfoxide reductase functions as an antioxidant in vivo. Proc Natl Acad Sci USA 94 9585–9589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaheb N, Thomas D, Verdoucq L, Monfort P, Meyer Y (1998) In vivo functional discrimination between plant thioredoxins by heterologous expression in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA 95 3312–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller EGD (1991) Thioredoxin deficiency in yeast prolongs-S phase and shortens the G1 interval of the cell-cycle. J Biol Chem 266 9194–9202 [PubMed] [Google Scholar]
- Munhoz DC, Netto LES (2004) Cytosolic thioredoxin peroxidase I and II are important defenses of yeast against organic hydroperoxide insult: catalases and peroxiredoxins cooperate in the decomposition of H2O2 by yeast. J Biol Chem 279 35219–35227 [DOI] [PubMed] [Google Scholar]
- Pagano EA, Chueca A, Lopez-Gorge J (2000) Expression of thioredoxins f and m, and of their targets fructose-1,6-bisphosphatase and NADP-malate dehydrogenase, in pea plants grown under normal and light/temperature stress conditions. J Exp Bot 51 1299–1307 [PubMed] [Google Scholar]
- Reichheld JP, Mestres-Ortega D, Laloi C, Meyer Y (2002) The multigenic family of thioredoxin h in Arabidopsis thaliana: specific expression and stress response. Plant Physiol Biochem 40 685–690 [Google Scholar]
- Rouhier N, Gelhaye E, Sautiere PE, Brun A, Laurent P, Tagu D, Gerard J, de Fay E, Meyer Y, Jacquot JP (2001) Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiol 127 1299–1309 [PMC free article] [PubMed] [Google Scholar]
- Russel M, Model P, Holmgren A (1990) Thioredoxin or glutaredoxin in Escherichia coli is essential for sulfate reduction but not for deoxyribonucleotide synthesis. J Bacteriol 172 1923–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahrawy M, Hecht V, Lopez-Jaramillo J, Chueca A, Chartier Y, Meyer Y (1996) Intron position as an evolutionary marker of thioredoxins and thioredoxin domains. J Mol Evol 42 422–431 [DOI] [PubMed] [Google Scholar]
- Schagger H, Vonjagow G (1987) Tricine sodium dodecyl-sulfate polyacrylamide-gel electrophoresis for the separation of proteins in the range from 1 kDa to 100 kDa. Anal Biochem 166 368–379 [DOI] [PubMed] [Google Scholar]
- Schobert C, Baker L, Szederkenyi J, Grossmann P, Komor E, Hayashi H, Chino M, Lucas WJ (1998) Identification of immunologically related proteins in sieve-tube exudate collected from monocotyledonous and dicotyledonous plants. Planta 206 245–252 [Google Scholar]
- Serrato AJ, Cejudo FJ (2003) Type-h thioredoxins accumulate in the nucleus of developing wheat seed tissues suffering oxidative stress. Planta 217 392–399 [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Crespo JL, Florencio FJ, Cejudo FJ (2001) Characterization of two thioredoxins h with predominant localization in the nucleus of aleurone and scutellum cells of germinating wheat seeds. Plant Mol Biol 46 361–371 [DOI] [PubMed] [Google Scholar]
- Szederkenyi J, Komor E, Shobert C (1997) Cloning of the cDNA for glutaredoxin, and abundant sieve-tube exudate protein from Ricinus communis L and characterization of the glutathione-dependent thiol-reduction system in sieve tubes. Planta 202 913–916 [DOI] [PubMed] [Google Scholar]
- Verdoucq L, Vignols F, Jacquot JP, Chartier Y, Meyer Y (1999) In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J Biol Chem 274 19714–19722 [DOI] [PubMed] [Google Scholar]
- Vignols F, Bréhélin C, Surdin-Kerjan Y, Thomas D, Meyer Y (2005) A yeast two-hybrid knockout strain to explore thioredoxin-interacting proteins in vivo. Proc Natl Acad Sci USA 102 16729–16734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignols F, Mouaheb N, Thomas D, Meyer Y (2003) Redox control of Hsp70-Co-chaperone interaction revealed by expression of a thioredoxin-like Arabidopsis protein. J Biol Chem 278 4516–4523 [DOI] [PubMed] [Google Scholar]
- Walz C, Juenger M, Schad M, Kehr J (2002) Evidence for the presence and activity of a complete antioxidant defence system in mature sieve tubes. Plant J 31 189–197 [DOI] [PubMed] [Google Scholar]
- Wangensteen OS, Chueca A, Hirasawa M, Sahrawy M, Knaff DB, Gorge JL (2001) Binding features of chloroplast fructose-1,6-bisphosphatase-thioredoxin interaction. Biochim Biophys Acta 1547 156–166 [DOI] [PubMed] [Google Scholar]
- Wong JH, Kim YB, Ren PH, Cai N, Cho MJ, Hedden P, Lemaux PG, Buchanan BB (2002) Transgenic barley grain overexpressing thioredoxin shows evidence that the starchy endosperm communicates with the embryo and the aleurone. Proc Natl Acad Sci USA 99 16325–16330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Motohashi K, Kasama T, Hara Y, Hisabori T (2004) Target proteins of the cytosolic thioredoxins in Arabidopsis thaliana. Plant Cell Physiol 45 18–22 [DOI] [PubMed] [Google Scholar]