Abstract
Nitrogen is an essential macronutrient for plant growth and survival. Here, the temporal and spatial sensing of nitrogen starvation is analyzed in Arabidopsis (Arabidopsis thaliana). The promoter for the high-affinity ammonium transporter, AtAmt1.1, is shown to be a valid indicator for nitrogen status in leaves and roots. An AtAmt1.1-Gal4 transgene using three 5× upstream activating sequence-driven reporters (luciferase, green fluorescent protein, and β-glucuronidase) facilitated in vivo profiling at the whole-plant and cellular levels. The effects of nitrogen supply, light duration, light intensity, and carbon on the expression of the AtAmt1.1 gene in the roots and aerial tissues are reported. Under nitrogen starvation, high expression is observed in the roots and, under nitrogen-sufficient conditions, high expression is observed in the leaves. This reciprocal regulation of AtAmt1.1 was confirmed by quantitative reverse transcription-polymerase chain reaction, which was also used to quantitate expression of the five other Amt genes in Arabidopsis. Although some of these show tissue specificity (roots or leaves), none exhibit reciprocal regulation like the AtAmt1.1-encoded high-affinity transporter. This robust reciprocal expression suggests that Arabidopsis undergoes rapid resource reallocation in plants grown under different nitrogen supply regimens. Ultimately, nitrogen starvation-mediated reallocation results in root architectural restructuring. We describe the precise timing and cellular aspects of this nitrogen limitation response.
Nitrogen is often the limiting macronutrient for plants. Because nitrogen incorporation into plant metabolites requires carbon skeletons, carbon and nitrogen metabolism are intricately linked. Carbon in the form of photosynthate and inorganic nitrogen from the soil are the chief forms assimilated by plants (Kang and Turano, 2003). Nitrate is the major and most readily available source of nitrogen for most higher plants (Marschner, 1995) and it is efficiently converted to ammonium by nitrate reduction before assimilation into organic compounds. Ammonium is also a direct source of nitrogen absorbed by plant roots (Ninnemann et al., 1994; Lauter et al., 1996), but, in general, the soil content of ammonium in unfertilized soils is 10 to 1,000 times lower than that of nitrate and is rarely higher than 50 μm (Marschner, 1995). Nevertheless, efficient uptake of ammonium is critical for optimal plant growth and development and this process is expected to be highly regulated in plants. Transporters of ammonium and nitrate exist in almost all organisms and it is likely that a combination of nitrate and ammonium ions are imported into plant roots (Bloom, 1994). For both nitrate and ammonium, low-affinity transport systems and high-affinity transport systems exist in plants (Forde and Clarkson, 1999). It is widely accepted that nitrate and ammonium uptake systems are controlled, in part, at the level of gene expression, but the nature or state of the key molecules that are sensed is debated (Rawat et al., 1999). Few nitrogen sensor proteins in plants have been identified (Hsieh et al., 1998; Kang and Turano, 2003; Yanagisawa et al., 2004; Chen et al., 2006), nor have in situ reporters for nitrogen starvation been demonstrated, other than chlorosis. In this article, we use nitrogen starvation reporters to better understand the physiology of complete nitrogen starvation in Arabidopsis (Arabidopsis thaliana).
Frommer and colleagues (Ninnemann et al., 1994) identified a high-affinity ammonium transporter (AtAmt1.1) in plants by complementing a yeast (Saccharomyces cerevisiae) strain (26972c) defective in the two NH4+ permeases (MEP1 and MEP2) with an Arabidopsis cDNA library (Minet et al., 1992). AtAmt1.1 functions as a saturable high-affinity transporter with a Km of 65 μm for methylamine, an ammonium analog. Using AtAmt1.1, two homologous cDNAs, AtAmt1.2 and AtAmt1.3, were identified (Gazzarrini et al., 1999), along with two other candidates, AtAmt1.4 and AtAmt1.5 (von Wiren et al., 2000a). Uptake studies in yeast reveal that AtAmt1.1 has the highest affinity for NH4+, which indicates that this gene is responsible for NH4+ uptake under severe nitrogen limitation (Gazzarrini et al., 1999).
During nitrogen limitation, there is strong correlation between high AtAmt1.1 mRNA levels in the roots and high ammonium uptake rates (Gazzarrini et al., 1999; Rawat et al., 1999; Gansel et al., 2001). Under nitrogen-limiting conditions, root AtAmt1.1 mRNA shows the highest increase among the AtAmt1.1, 1.2, and 1.3 genes, 7-fold higher when transferred from 1 to 0 mm NH4NO3 (Gazzarrini et al., 1999; Rawat et al., 1999). The tomato (Lycopersicon esculentum) and rice (Oryza sativa) Amt1.1 orthologs, LeAmt1.1 (von Wiren et al., 2000b) and OsAmt1.1 (Kumar et al., 2003), show similar expression in roots. When rice (Kumar et al., 2003) plants were transferred from 10 mm to 10 μm NH4+, root transcript levels of the OsAmt1.1 gene increased 5- to 7-fold. Conversely, upon resupplying ammonium to nitrogen-starved rice plants, Amt1.1 expression decreases 5- to 7-fold in the roots (Kumar et al., 2003). In a microarray study on Arabidopsis genes induced and repressed by nitrate, Amt1.1 was repressed 3.2-fold in 10 mm relative to 0.25 mm KNO3 (Wang et al., 2000). This is consistent with split-root studies (Gansel et al., 2001), which show that AtAmt1.1 expression is repressed 4-fold by 1 mm KNO3. The LeAmt1.1 gene was expressed preferentially in the root hair cells of tomato under all nitrogen conditions tested with a 3-fold decrease in the presence of 5 mm nitrate (Lauter et al., 1996). Whereas prior studies have focused on root expression of AtAmt1.1 mRNA, we decided to study the expression of AtAmt1.1 using real-time reporters to characterize the nitrogen starvation response throughout the plant. A Gal4 approach, whereby Gal4 is driven by the AtAmt1.1 upstream promoter control region, was developed. Three reporters, luciferase (LUC), green fluorescent protein (GFP), and β-glucuronidase (GUS) that are each activated by Gal4 via five repeats of the Gal4 upstream activating sequence (5×UAS) were used to visualize and quantify AtAmt1.1 expression temporally and spatially in response to a variety of environmental conditions. It is shown that such plant lines not only give an indication of physiological nitrogen status, but also they do so early in the process. We report reciprocal tissue expression patterns (leaves versus roots) under varying nitrogen conditions in addition to more comprehensive expression patterns for different plant tissues. Upon initiation of nitrogen stress, AtAmt1.1 expression in the roots first occurs in the root hairs and then spreads to the lateral and primary roots with the progression of nitrogen starvation. This expression is temporally associated with changes occurring in the root system architecture during the nitrogen starvation response, which is additionally correlated with AtAmt1.1 repression in leaves.
RESULTS
Construction of an AtAmt1.1-Gal4 Driver System for LUC, Red-Shifted GFP, and GUS
We constructed a reporter system for nitrogen stress that fulfilled multiple criteria. LUC was used for temporal studies because it has a short half-life (Thompson et al., 1991; Millar et al., 1992; Leeuwen et al., 2000) and can be imaged noninvasively in vivo. For detailed localization studies, we used a red-shifted (rs) GFP reporter fused in frame with the GUS open reading frame. Finally, we chose to drive these reporters with Gal4, which itself is expressed in response to nitrogen (i.e. with the AtAmt1.1 control region), so that all reporters could be assayed in a single plant. The three reporters are expressed when the GAL4 protein binds a 5×UAS, which precedes a minimal TATA upstream of each reporter's coding sequence. For the AtAmt1.1-driven Gal4 construct, 274 bp of the natural AtAmt1.1 promoter (upstream of the TATA) were used because the first ATG of the coding region for another gene (At4G13500.1) lies 274 bp upstream on the complementary strand (see Supplemental Fig. S1). The full 35S cauliflower mosaic virus enhancer instead of the AtAmt1.1 upstream region was used as a positive control (Engineer et al., 2005). Each T-DNA also has the 5×UAS LUC reporter and BASTA resistance. The Agrobacterium-mediated floral-dip protocol was used to transform Arabidopsis plants (ecotype Columbia) with AtAmt1.1, the full 35S Cauliflower mosaic virus, and the enhancerless Gal4 vectors.
Nitrogen-Regulated Reporter Expression in Amt-Gal4-LUC Plants
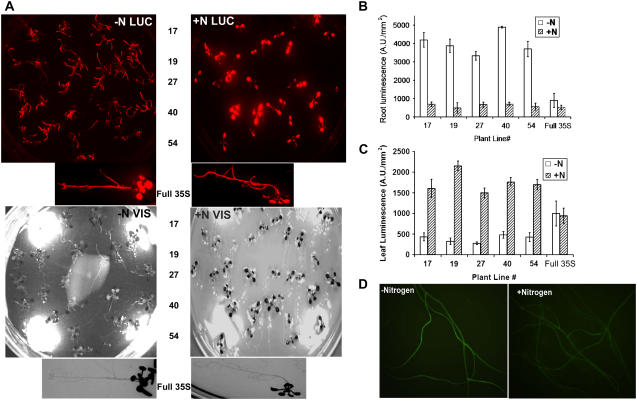
The plant lines in which Gal4 (and hence LUC) expression is driven by the AtAmt1.1 enhancer are designated Amt-Gal4-LUC lines. To establish whether the Amt-Gal4-LUC lines respond to nitrogen like the endogenous AtAmt1.1 gene, 87 transgenic lines were screened for nitrogen response. Eighty of these lines showed LUC increases under nitrogen starvation in the roots. The remaining seven lines showed very little expression. None of the 91 empty-vector lines (i.e. without the 274-bp AtAmt1.1 region from pRGK366; GenBank accession no. DQ666282) showed LUC expression. LUC expression in five representative Amt-Gal4-LUC lines and the full 35S control line (66-3E) under nitrogen replete (+N) and deficient (−N) conditions are shown in Figure 1A. Throughout this article, unless otherwise specified, +N or nitrogen-replete medium refers to 0.5× Murashige and Skoog medium, which contains 10.3 mm NH4NO3 and 9.4 mm KNO3 as the nitrogen source; −N or nitrogen-deficient medium refers to 0.5× Murashige and Skoog medium, which contains no nitrogen source.
Figure 1.
Nitrogen response of AtAmt1.1-driven Gal4-LUC and GFP biosensing lines. A (top), LUC (red) image of five independent Amt-Gal4-LUC lines and full 35S-driven Gal4 positive control line. Bottom shows visible images (VIS) of the plants. The image on the left shows plants undergoing nitrogen starvation for 7 d and the image on the right shows plants on +N medium for 7 d. Plants are grown under short-day conditions. B, Quantitation of the root LUC expression of plants shown in A. C, Quantitation of the leaf LUC expression shown in A. D, Root GFP images of two Amt-Gal4-LUC-GFP-GUS plants: images of −N-grown roots (left) and +N-grown roots (right). All images were acquired at the midpoint of the light period (i.e. 4 h into the light treatment).
Plants starved for nitrogen show higher root LUC expression than those grown on +N medium. When Amt-Gal4-LUC plants were starved for nitrogen for 7 d, the root LUC expression levels were 5- to 10-fold higher than those in the roots of +N plants (Fig. 1B). Surprisingly, LUC expression in leaves of +N plants was 5- to 8-fold higher than that of the −N plants (Fig. 1, A and C). Southern-blot and segregation analyses indicate that plants with single or multiple insertion loci show similar expression profiles (data not shown; see below). An average 7-fold increase in root expression under −N relative to an average 5-fold difference for leaves under +N represents a 35-fold reciprocal expression pattern.
To generate lines that also possessed the 5×UAS rsGFP-GUS reporter, we used a homozygous, single- locus line, RGK1 (hygromycin resistance), previously generated with this reporter (Engineer et al., 2005). RGK1 was crossed with the Amt-Gal4-LUC lines and BASTA and hygromycin-resistant lines were selected (designated as Amt-Gal4-LUC-GFP-GUS lines). Expression profiles similar to the Amt-Gal4-LUC lines were noted for the rsGFP reporter under +N and −N conditions. Roots expressed GFP under −N conditions (Fig. 1D), whereas leaves expressed GFP only under +N conditions (see below).
Validation of the Nitrogen Biosensor Reporter Plant Lines
Before using the Amt-Gal4-LUC plants to further analyze environmental responses and tissue-specific expression profiles, we wanted to confirm that Gal4-mediated LUC expression is correlated with endogenous AtAmt1.1 expression patterns. Quantitative real-time (qRT)-PCR was conducted on cDNA prepared from tissues of Amt-Gal4-LUC plants grown on +N and −N media. Tissues used to conduct these experiments were imaged for LUC expression prior to harvesting. All qRT-PCR reactions were carried out in the multiplexed format, with one set of primers for the housekeeping control gene (UBQ10) and the other set of primers for the endogenous AtAmt1.1gene. Samples included leaves and roots of −N- and +N-grown plants to test the reciprocal LUC expression pattern. Representative qRT-PCR results are shown in Supplemental Figures S2 to S4. Two Amt-Gal4-LUC lines were used for these analyses: line 17, with multiple insertions and line19-2, with a single insertion locus (see Table I, experiments 1–4 and 7). In each case tested, there is a positive correlation between AtAmt1.1 and LUC expression, indicating that the Amt-Gal4-LUC and Amt-Gal4-LUC-GFP-GUS plant lines accurately reflect endogenous AtAmt1.1 expression patterns and nitrogen nutritional states via reporter expression. These results suggest that the 274-bp upstream region used for reporter studies is sufficient to reproduce the endogenous expression patterns. Moreover, these results confirm the reciprocal regulation in leaves and roots in response to nitrogen.
Table I.
qRT-PCR assays of endogenous AtAmt1.1 expression in relation to AtAmt1.1-mediated LUC activity
Multiplexed qRT-PCR results for plants bearing single (line 19-2) and multiple (line 17) inserts of the AMT-GAL4-LUC T-DNA. Source tissue refers to tissues with distinct LUC expression profiles taken from plants grown on +N and/or −N media. LUC activation is quantitated and reported as a fold change/increase between the two source tissues being studied in any given replicate. qRT-PCR results are shown in the form of fold changes in endogenous AtAmt1.1 expression levels and are calculated from the ΔΔCt values (see “Materials and Methods”). A significant positive correlation (Pearson's R = 0.628095; P = 0.047689) is calculated for higher LUC expression and higher endogenous AtAmt1.1 gene expression in all cases tested. Numerical values represent the average value ± sd of four replicates.
| Experiment | Line No. | Source Tissue | LUC Activity | AtAmt1.1 Expression |
|---|---|---|---|---|
| 1 | 17 | Leaves of +N and −N plants | 10.3 ± 1.3× increase in +N leaves | 13.8 ± 1.4× increase in +N |
| 2 | 17 | Roots of +N and −N plants | 6.9 ± 2.4× increase in −N roots | 12.8 ± 3.2× increase in −N roots |
| 3 | 17 | Leaves and roots of −N plants | 6.1 ± 1.7× increase in roots | 13.4 ± 1.3× increase in roots |
| 4 | 17 | Leaves and roots of +N plants | 40.2 ± 14.7× increase in leaves | 16.8 ± 1.6× increase in leaves |
| 5 | 17 | Leaves of +N plants (low and high) | 9.1 ± 2.6× increase in high leaves | 6.5 ± 1.3× increase in high leaves |
| 6 | 19-2 | Leaves of +N plants (low and high) | 6.5 ± 3.2× increase in high leaves | 5.6 ± 1.4× increase in high leaves |
| 7 | 19-2 | Roots of +N and −N plants | 9.4 ± 2.2× increase in −N | 10.1 ± 1.5× increase in −N |
| 8 | 19-2 | Petioles and laminas of +N plants | 2.6 ± 0.3× increase in petioles | 2.3 ± 1.1× increase in stems |
To further test the robustness of the reporter system, we analyzed by qRT-PCR some tissues that expressed LUC under specialized conditions. For example, we observed that the petioles of +N-grown plants, which had not been exposed to light, had higher expression than the laminas (see Supplemental Fig. S5A). This differential expression disappears after supply of light for as little as 5 min. Analysis of the qRT-PCR results suggests that the endogenous AtAmt1.1 gene is also expressed higher in petioles than in laminas (Table I, experiment 8). We also noticed that, occasionally, a plant on +N media would show a leaf with low LUC expression (see Supplemental Fig. S5B). qRT-PCR studies conducted on the typical high-expression leaf and the low-expression outlier reveal that higher LUC expression is correlated with higher AtAmt1.1 expression (Table I, experiments 5 and 6). This result suggests that, occasionally, leaves will escape or avoid the +N response either because they are nitrogen limited or because the response itself is attenuated.
Reciprocal Leaf/Root Expression Patterns of the AtAmt1.1 Gene in Response to Nitrogen throughout Day/Night Cycles
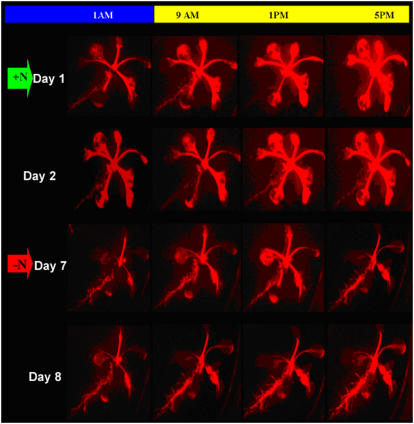
To determine whether reciprocal AtAmt1.1 expression occurs throughout the day, LUC imaging of a single plant was carried out four times daily. One Amt-Gal4-LUC 17 seedling was followed during this assay, which consisted of imaging the plant at the beginning of the light period, midpoint of the light period, end of the light period, and midpoint of the dark period over the course of an 8-d experiment (Fig. 2). A 2-week-old seedling grown on +N medium was shifted to fresh +N medium and imaged for 2 d before a shift to −N medium. The seedling was imaged after 4 d on −N (Fig. 2, red arrow). As expected, LUC expression in roots is repressed under +N conditions, whereas the leaves show strong LUC expression, which increases during the day. When shifted to −N medium, leaf expression is repressed and root expression is activated, which also increases during the day. These increases in organ-specific, light-dependent expression of LUC are easily observed in each of the horizontal images of Figure 2, from left to right. The same results were obtained with line 19 (data not shown). This diurnal effect has been noted previously for AtAmt1.1 root expression (Gazzarrini et al., 1999) and is consistent with a light source control in nitrogen sensing (see below).
Figure 2.
Grid of LUC images (red) showing the progression of a single AtAmt1.1-driven Gal4-LUC plant line 17 through growth on +N and −N media over the course of 8 d. The bar at the top of the image indicates time during the diurnal cycle (yellow and blue indicate light and dark periods, respectively) at which the images were taken (columns). The top two rows show images of the plant grown on +N medium and the bottom two rows show images of the plant after nitrogen starvation. The plant was initially grown on +N medium for 2 weeks and then shifted to fresh +N medium and imaged for LUC (green arrow, days 1 and 2). The plant was shifted to −N medium after day 2 and allowed to undergo nitrogen starvation for 4 d after which LUC imaging was continued (red arrow, days 7 and 8).
Light Intensity and Duration Impacts Expression Patterns
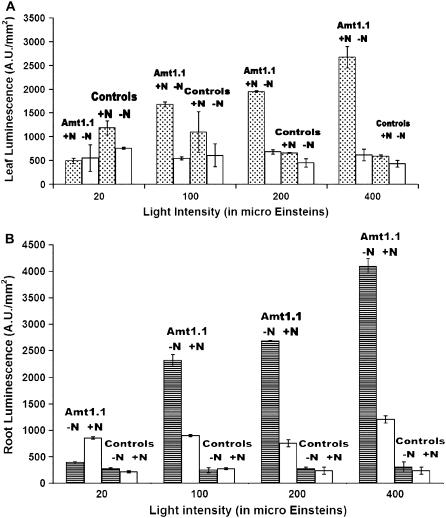
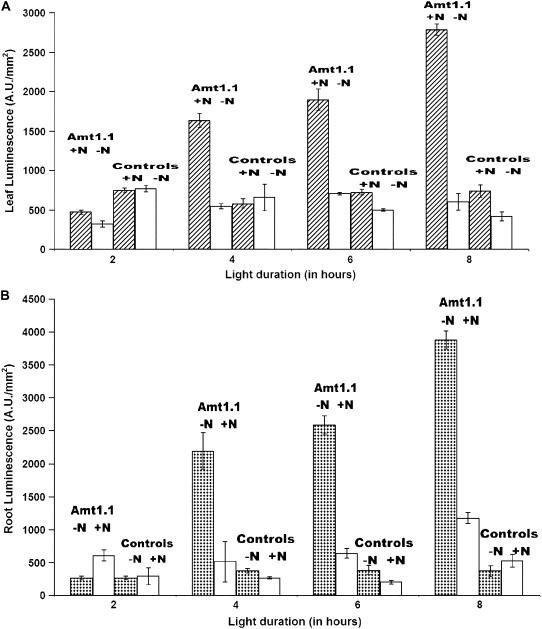
Although we have just reported the expression patterns of plants under 8-h days, our early experiments with the Amt-Gal4-LUC plants used 16-h, long-day conditions. For 16-h days, we observed a nitrogen response (LUC expression), but only a couple-fold difference largely because root expression in the +N roots was not as repressed. When 8-h, short-day conditions (100 μE) were used, as reported above, the nitrogen effect in roots is increased. Our hypothesis is that increased exposure to light (as in the long-day conditions) increases photosynthetic activity in the plants, which creates a greater demand for nitrogen uptake and assimilation. To further test this, the effects of light intensity and duration on AtAmt1.1 expression in Amt-Gal4-LUC plants was studied. Sets of 10 plants from each of three independent Amt-Gal4-LUC lines and one full 35S line were subjected to a range of light intensities (20, 100, 200, or 400 μE) and also to a range of light durations (2, 4, or 8 h of light/24-h diurnal cycle). Identical sets of plants were subjected to media with and without nitrogen for these conditions. For the light intensity experiments, short-day (8 h) conditions were used and plants were imaged for LUC expression at the midpoint of the light cycle. As shown in Figure 3, there is a positive, statistically significant correlation between higher light intensities and higher LUC expression in +N leaves (Fig. 3A) and −N roots (Fig. 3B). For the light duration experiments, plants were imaged at the end of the specific light period. Again, for these daylength experiments, the more light supplied, the higher the LUC expression (Fig. 4, A [for leaves] and B [for roots]). The full 35S plants do not show significant differences (Figs. 3 and 4, controls). These results indicate that there is a delicate balance between light and the nitrogen starvation response of ammonium transport. This is consistent with the up-regulation observed for other ion transporters in response to photosynthesis and sugars (Lejay et al., 2003).
Figure 3.
Quantification of LUC expression in leaves (A) and roots (B) of AtAmt1.1-driven Gal4-LUC line 19-2 (denoted by Amt1.1) grown on +N and −N media in short-day conditions, as indicated, over a range of light intensities. Error bars represent sd (n = 10). Expression profiles of full 35S-driven Gal4 control lines is also shown (denoted by controls). LUC imaging was conducted at the midpoint of the light period.
Figure 4.
Quantification of LUC expression in leaves (A) and roots (B) of AtAmt1.1-driven Gal4-LUC line 19-2 (denoted by Amt1.1) grown on +N and −N media, as indicated, over a range of light durations (at 100 μE of light). Error bars represent sd (n = 10). Data are representative of other Amt-Gal4-LUC lines. Expression profiles of full 35S-driven Gal4 control lines is also shown (denoted by controls).
Temporal and Spatial Expression in Leaves of Amt-Gal4-LUC Plants Using rsGFP and GUS Reporters
AtAmt1.1-mediated LUC expression in leaves is up-regulated during nitrogen sufficiency and light supply (duration and intensity) and down-regulated in the absence of nitrogen under all growth conditions. We next determined cell-specific AtAmt1.1-mediated reporter expression in aerial tissues through the life cycle using the rsGFP and GUS reporters of Amt-Gal4-LUC-GFP-GUS plants.
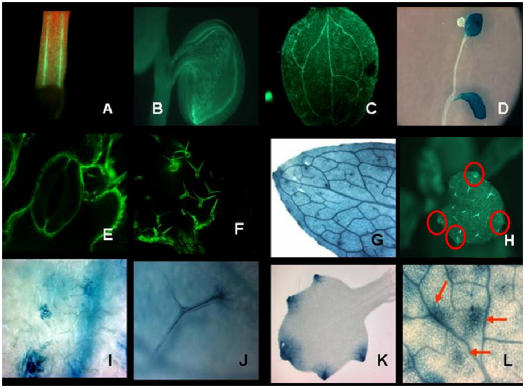
Two-day-old germinating seedlings constantly show GFP expression in the hypocotyl and cotyledons on +N medium, with the strongest expression in the vasculature (Fig. 5, A and B). This is also the case when seeds are germinated on −N medium (data not shown). This suggests that initial expression reflects nitrogen reserves being distributed in the germinating seedling. Because very low GUS or rsGFP is observed in −N plant leaves, as expected, all studies on aerial tissues are from +N plants. In mature plants, expression is seen in the leaf vasculature (Fig. 5, C and G), trichomes (Fig. 5, F and J), mesophyll cells (Fig. 5, D, G, and L), guard cells (Fig. 5, E and I), and, notably, in hydathodes (Fig. 5, G, H, and K). Leaf expression on +N medium is observed in source and sink leaves. Leaf vasculature shows higher expression than the mesophyll cells (Fig. 5, C and G). Trichomes and patches of cells surrounding the trichomes show higher expression than the mesophyll cells (Fig. 5L, arrows). In very young emerging leaves, expression was primarily seen in the hydathodes and in the vasculature immediately surrounding the hydathodes, but not in the remaining parts of the leaf (Fig. 5K). On inflorescence stalks, the stems themselves do not show expression, but the leaves do (Fig. 5D). In all cases, the LUC, rsGFP, and GUS reporters correlated in their expression patterns and intensities.
Figure 5.
GFP and GUS localization studies of leaves from plants grown on +N medium. A, B, C, E, F, and H, GFP images; D, G, I, J, K, and L, GUS images. A, Hypocotyl vasculature of 2-d-old seedling (yellow depicts GFP expression). B, Cotyledon vasculature of seedling shown in A. C and G, Leaf venation of mature leaves. E and I, Guard cells of stomata. F and J, Trichomes. H, Hydathodes. K, Hydathodes on a newly emerging leaf. L, Trichomes with the cell patches (red arrows) that express AtAmt1.1 surrounding the trichomes. Tissues used for GFP or GUS imaging were harvested or imaged at the midpoint of the light period.
Temporal and Spatial Expression Profiles of AtAmt1.1 in Roots and Root Architectural Changes of −N-Grown Plants
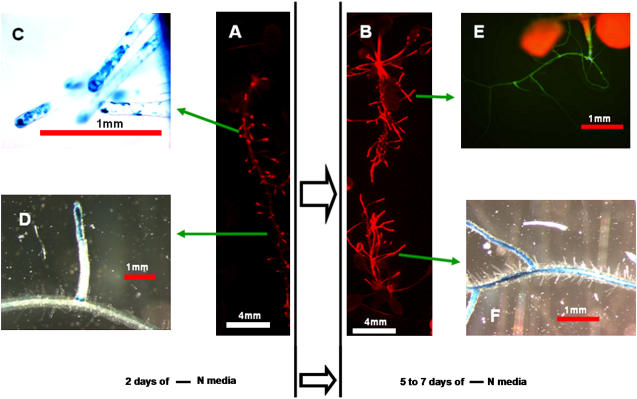
We noticed that, upon shifting to −N conditions, increases in root LUC expression paralleled architectural changes in the root system. To characterize the onset of root expression in more detail, taking into account the simultaneous architectural changes, we used the LUC, rsGFP, and GUS reporters. When +N-grown plants are shifted to −N medium, within 48 h they exhibit a punctate LUC expression pattern only in the distal parts of the root system (Fig. 6A). Microscopic observations with rsGFP and GUS reveal that this expression is in new and existing root hairs in these regions (Fig. 6C, for GUS). The punctate pattern also results from expression in the tips of newly emerging lateral roots and the junctions of primary/lateral roots from which they arise (Fig. 6D). As nitrogen starvation progresses further (5–7 d), LUC, rsGFP, and GUS expression intensify through the entire root system, including the root hairs and the steles of the primary and lateral roots (see Fig. 6, B, E, and F).
Figure 6.
Temporal and spatial root AtAmt1.1 expression as nitrogen starvation progresses from 2 d poststarvation (A, C, D) to 5 to 7 d poststarvation (B, E, F). A and B, LUC expression is depicted in red. GUS images of root hairs (C) and newly emerging lateral roots from the primary roots (D). E and F, GFP and GUS images, respectively, showing expression in the entire root system, including the steles of primary and lateral roots. Lengths are indicated with bars (in millimeters). All images were acquired at the midpoint of the light period.
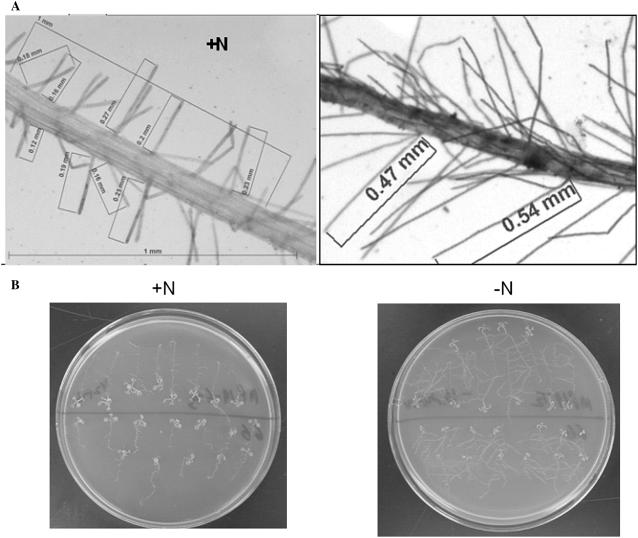
Beginning within 48 h of nitrogen starvation, we observed that root system architectural changes accompany AtAmt1.1-mediated reporter expression (see Fig. 7, A and B). To quantify these changes, we analyzed the roots of individual plants undergoing nitrogen starvation (Table II). Although there is a very slight increase in the numbers of lateral roots and root hairs after 2 d, both show an increase in length (approximately 2-fold for root hairs and 1.3-fold for lateral roots) as compared to plants grown simultaneously on +N medium (Fig. 7A; Table II). After 5 to 7 d of nitrogen starvation, the lengths and numbers of lateral roots and root hairs increase significantly (Fig. 7B; Table II). The timing of these is correlated with the aforementioned reciprocal regulation of AtAmt1.1. Furthermore, in hydroponic experiments conducted in our laboratory, we find similar increases in root system architecture for mature plants. This reciprocal AtAmt1.1 expression was also evident when plants were grown on a series of nitrogen content media (0, 5, 50, 500, and 5,000 μm ammonium nitrate), with a gradual shift from leaf to root expression (Supplemental Fig. S6). This result shows that 0.5 mm ammonium nitrate is sufficient to repress expression in the roots and induce expression in the leaves.
Figure 7.
Root architectural transformations in response to nitrogen limitation. A, Arabidopsis seedlings were germinated on +N medium for 10 d and then shifted to +N and −N plates simultaneously. Images taken 2 d into the treatment show roots grown on +N (left) and −N (right). B, Root system structural changes with nitrogen starvation 7 d after initiation of starvation. Two-week-old seedlings were transferred to +N and −N media for 7 d. Images for B were acquired at the midpoint of the light cycle.
Table II.
Root architectural transformations in response to nitrogen limitation
Root system architectural changes with nitrogen starvation (2 and 5 d after initiation of starvation). The length and number of lateral roots (Lat. Rt.) and root hairs (Rt. Hairs; per millimeter of primary root) were determined. Results show mean values (±sd) from measurements on 10 plants for each treatment.
| Nitrogen | Lat. Rt. No. | Lat. Rt. Length | Rt. Hair No. | Rt. Hair Length |
|---|---|---|---|---|
| 2 d after Treatment | ||||
| Minus | 6.5 ± 2.0 | 0.78 ± 0.12 cm | 33.7 ± 2.6 | 0.52 ± 0.06 mm |
| Plus | 4.6 ± 1.6 | 0.45 ± 0.06 cm | 30.6 ± 2.2 | 0.19 ± 0.04 mm |
| 5 d after Treatment | ||||
| Minus | 12.9 ± 2.1 | 1.37 ± 0.20 cm | 52.1 ± 3.3 | 0.74 ± 0.08 mm |
| Plus | 7.5 ± 1.4 | 0.48 ± 0.05 cm | 41.2 ± 3.0 | 0.23 ± 0.04 mm |
Effects of Different Nitrogen Sources and Supply on Whole-Plant Amt1.1 Expression Using Split-Root Studies
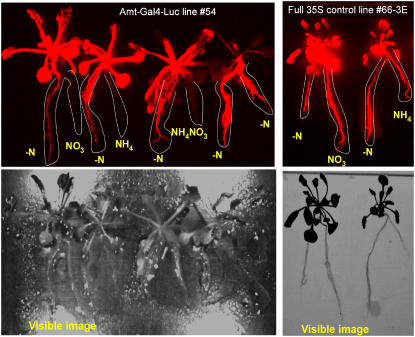
Amt-Gal4-LUC seedlings were used in split-root studies to interrogate responses to various sources and concentrations of nitrogen and the local versus systemic responses. Gansel et al. (2001) have used the split-root system to analyze root expression of AtAmt1.1 mRNA. Roots of each Amt-Gal4-LUC plant were divided such that one-half was subjected to −N medium and the other half was supplied with −N medium containing 1 mm of potassium nitrate, ammonium nitrate, or ammonium chloride for different plants (Fig. 8). Controls included −N medium for both halves of an Amt-Gal4-LUC plant and the full 35S line (66-3E), which was subjected to various nitrogen media as well. The pH in all treatments was adjusted to 5.7. LUC expression was quantified 48 h after the initial treatment. The full 35S line showed equal LUC activity in both halves of the roots and in the leaves regardless of the presence or source of nitrogen. In contrast, Amt-Gal4-LUC plants showed regulated LUC expression with higher expression in the −N halves and lower expression in the nitrogen-supplied halves. Differences ranged from 5- to 10-fold between the two halves of the roots of the same plant. We conclude that ammonium or nitrate at 1 mm levels can repress local root expression of AtAmt1.1. The leaves of the Amt-Gal4-LUC plants whose root system had been supplied with a source of nitrogen also expressed LUC strongly. The plant that was not supplied with nitrogen to either half of its root system did not exhibit leaf expression (see Fig. 8, plant on far right of the left image). This suggests that the effect on leaves is induced systemically via the supply from the roots. We also conclude that 1 mm or greater of nitrogen is sufficient for leaf induction of AtAmt1.1 and that this supply does not require the entire root system for induction.
Figure 8.
Split-root experiments with AtAmt1.1-driven Gal4-LUC plants (line 54, left) and full 35S-driven Gal4 positive control plants (line 66-3E, right). Top shows LUC images (red) and bottom shows visible images of those plants. Images were taken 48 h after initiation of the treatment. Root systems of each plant were split into two halves and provided no nitrogen (−N) or 1 mm of potassium nitrate (NO3), ammonium chloride (NH4), or ammonium nitrate (NH4NO3). The two halves of each root system are highlighted with a white line. Images acquired at the midpoint of the light period.
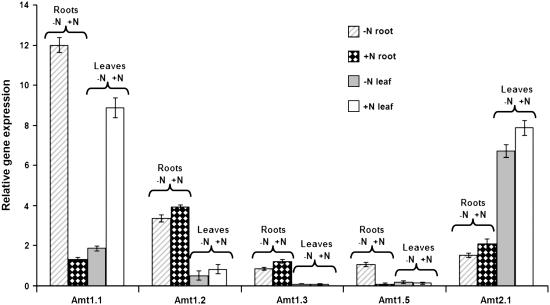
Expression of Other Amt Genes (Amt 1.2, 1.3, 1.4, 1.5, and 2.1)
To investigate whether other high-affinity ammonium transporters in Arabidopsis are reciprocally controlled, we conducted simultaneous qRT-PCR analyses on leaf and root tissues of plants grown with and without nitrogen (Fig. 9). Our qRT-PCR results confirmed previously published data of northern blots, showing that Amt1.2 is expressed in the roots and leaves and that the expression in the roots is independent of nitrogen supply to the plants (Gazzarrini et al., 1999; Loque and von Wiren, 2004); Amt2.1 is expressed in the roots and leaves, with leaf expression higher than root, and independent of nitrogen supply (Sohlenkamp et al., 2000, 2002; Kaiser et al., 2002). New results shown here include the following: None of the other five Amt genes show reciprocal regulation in response to nitrogen starvation. The Amt1.2 gene is expressed at a 5-fold higher level in roots than in leaves. Although expressed at lower overall levels than Amt1.2, the Amt1.3 gene is also higher (10-fold) in roots than in leaves. The Amt1.5 gene was expressed at low levels in all tissues except for the −N-grown roots, where it was expressed 8-fold higher. Thus, Amt1.5 appears to be a second member of the family that responds to nitrogen starvation in the roots. Because the Amt2.1 gene showed 4-fold higher expression in leaves than roots for both nitrogen treatments, Amt2.1 may have a primary function in leaves (see “Discussion”). Our results did not show a nitrogen response for root Amt2.1 and Amt1.3. We failed to detect expression for the Amt1.4 gene, which might suggest this is a pseudogene or that our growth conditions did not encompass the nutritional and/or developmental stimulus required for its expression.
Figure 9.
Expression (qRT-PCR) profiles for Amt genes in leaves and roots of plants grown with and without nitrogen. Results are relative to the housekeeping control gene UBQ10 and represent the average of three independent biological replicates. Error bars indicate se.
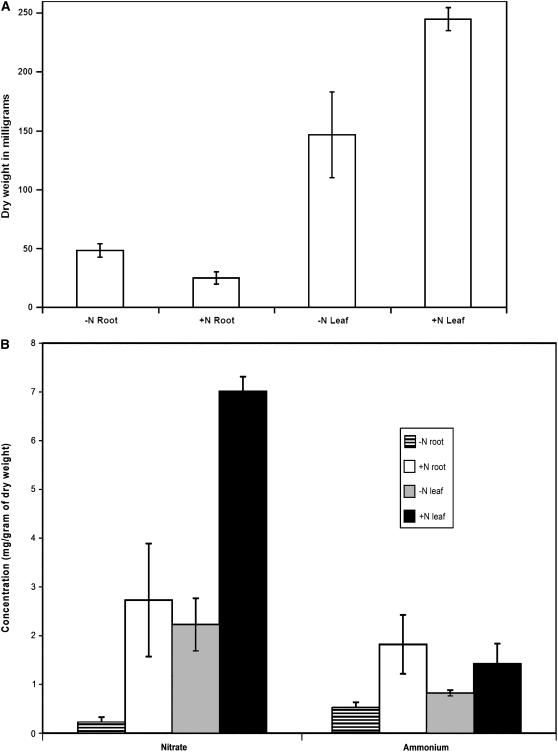
Nitrate and Ammonium Levels in Leaf and Root Tissues
Previous results have indicated a reduction in nitrate and ammonium levels in roots and leaves after 4 d of nitrogen limitation under hydroponic growth conditions (Kaiser et al., 2002). We analyzed nitrate and ammonium levels in leaves and roots of plants grown with and without nitrogen (based on dry weight) after 5 d of nitrogen starvation under our conditions. There was significantly more nitrate in the leaves and roots of plants grown with nitrogen as opposed to those grown without nitrogen (Fig. 10). The roots of plants grown on nitrogen had 12-fold and leaves 3-fold more nitrate than those grown without nitrogen. The roots of +N-grown plants also had 3.5-fold more ammonium than those of −N plants. The leaves of +N plants only showed a modest (1.7-fold) increase in ammonium levels as compared to those of −N plants. Our results corroborate previously published results of Kaiser et al. (2002). The modest increase of ammonium and the dramatic increase of nitrate in the leaves of +N- over −N-grown plants suggests that the internal signal for leaf activation of AtAmt1.1 under +N growth conditions could be nitrate (and not ammonium) or a metabolite such as Gln.
Figure 10.
Total dry weights (A) and free nitrate and ammonium pools (B) in leaves and roots of plants grown with and without nitrogen. Two-week-old plants grown on nitrogen were transferred to growth media with (+N) and without (−N) nitrogen for 5 d. A, Dry weight of tissues used to quantitate nitrate and ammonium levels in B (complete tissues from 20 seedlings were used for A and B). B, Levels of free nitrate and ammonium as indicated in leaves and roots of +N and −N plants. Results are an average of three biological replicates. Error bars indicate se.
DISCUSSION
This study describes a reporter system that we could use to analyze the nitrogen starvation response in Arabidopsis. We have intentionally focused on complete nitrogen starvation, using this biosensor line as a guide to address cell type responses and root anatomical changes. Key responses occurred at low nitrogen (<50 μm) concentrations in the medium, indicating that these are likely not general stress responses. Clearly, the expression characteristics of the high-affinity ammonium transporter, AtAmt1.1, are more complicated than previously envisioned, but understanding the uptake and distribution of nitrogen in the plant will require such in situ analyses and more. Using a variety of reporters for biosensing could lead to a more practical understanding of the nitrogen starvation response and how best to avoid or cope with this limitation.
AtAmt1.1 as a Nitrogen Starvation Biosensor
Plant systems exhibit varied morphological and biochemical responses to nutrient stress. High-affinity nutrient transporters are an integral part of responses to specific nutrient starvation. Enhanced expression and activity of these transporters during nutrient starvation enables the plant to scavenge for and utilize the nutrients that are critical for growth and survival. In agricultural systems, nitrogen supply is second only to water supply in terms of major factors that affect yield and productivity (Marschner, 1995). After a period of nitrogen starvation, Arabidopsis preferentially imports ammonium, the more reduced form of nitrogen over the inorganic nitrate (Gazzarrini et al., 1999). Our results on AtAmt1.1 suggest that expression of this ammonium transporter can be used at the whole-plant level to rapidly report nitrogen stress. The AtAmt1.1-Gal4-based system reproduces expression patterns of the endogenous AtAmt1.1 reported in studies that analyzed AtAmt1.1 mRNA in the roots.
Leaf Expression of AtAmt1.1
Expression studies on the AtAmt1.1 gene have been limited to mRNA profiling in roots. Although AtAmt1.1 expression in the aerial parts of the plant has been suggested (Ninnemann et al., 1994; Gazzarrini et al., 1999; Shelden et al., 2001; Kaiser et al., 2002), no studies have reported the expression in the leaves in response to nitrogen. We observe a dramatic reciprocal shift in AtAmt1.1 expression from leaves to roots, as plants are shifted from +N to −N media (Figs. 1 and 2). We can speculate on the physiological sense of this reciprocal regulation. During plant growth on +N medium, actively photosynthesizing leaf mesophyll cells produce carbon skeletons that require assimilation of nitrogen. The demand for plant nitrogen uptake is satisfied by the low-affinity uptake system in the roots, which transports the nitrogen from the medium to the shoots via the xylem. Under such conditions, we see high expression of the AtAmt1.1 gene in the leaf vasculature and, albeit lower, in mesophyll cells (see Figs. 1, 2, and 5). It is possible that cells within the vasculature (e.g. xylem parenchyma) express the AtAmt1.1 gene to satisfy their need for ammonium. The stronger expression pattern in the vasculature may also be due to the density of cells in the vasculature as compared to the spongy mesophyll. Another possible role for AtAmt1.1 is the shuttling of NH4+ lost from cells during photorespiration back into the mesophyll cells. When nitrogen is assimilated into the carbon skeletons in the form of amino acids, it can be sent via the phloem to the roots to sustain growth and metabolic activity in the roots.
Root System Restructuring during Nitrogen Starvation and the Concomitant Reciprocal Shift in AtAmt1.1 Expression from Leaves to Roots
Previous studies have shown the inhibitory effects of high Suc-to-nitrogen ratios on lateral root initiation (Malamy and Ryan, 2001) and the stimulatory effects of localized nitrogen-rich patches on plant root system architecture (Drew, 1975; Zhang and Forde, 2000). We studied the nitrogen starvation response in root system restructuring concurrently with expression changes of AtAmt1.1. We quantitated the lengths and numbers of new lateral roots and root hairs in the early stages (48 h after the onset of nitrogen stress; 0 μm nitrogen). Lengths of root hairs and lateral roots increased, but their numbers remained relatively constant compared to +N-grown plants. Similar results with root hair elongation were observed when Arabidopsis plants are given low phosphorus (Bates and Lynch, 1996; Sanchez-Calderon et al., 2006). Possibly, this elongation reflects an active, immediate increase in ammonium uptake potential as the first response. As starvation progressed 5 to 7 d after the application of nitrogen stress, when leaf AtAmt1.1 expression is maximally repressed and root expression is optimally activated, the numbers and lengths of both root hairs and lateral roots increased significantly. When we used very low amounts of nitrogen in the medium (5 μm ammonium nitrate) as opposed to no nitrogen, we still observed similar root proliferation with a gradual inverse relationship to nitrogen levels supplied (data not shown). We suggest that, at the onset of nitrogen starvation, the existing root organs (lateral roots and root hairs) elongate for optimal import and to probe the rhizosphere in the vicinity of the existing root system to locate nitrogen-rich soil patches. A role for root hairs in the sensing response to nitrogen deficiency has been suggested (Shin et al., 2005). Failing to encounter any nitrogen (as in our system), Arabidopsis subsequently undertakes the more costly process of increasing the numbers of lateral roots and root hairs. The latter process requires resource reallocation in the plant for growth and cell division.
Roles for the Amt Family of Genes in Ammonium Acquisition and Transport
The Arabidopsis family of high-affinity ammonium transporters has six members encoded by the genes Amt1.1, 1.2, 1.3, 1.4, 1.5, and 2.1 (Gazzarrini et al., 1999; Sohlenkamp et al., 2000). The Amt1.1 to 1.5 genes show high sequence similarity to each other and the Amt2.1 gene is distantly related to the Amt1 family (Loque and von Wiren, 2004). Previous nitrogen supply related Amt mRNA studies have only focused on roots to study some members of these genes (Gazzarrini et al., 1999; Kaiser et al., 2002). With the exception of Amt2.1 (Sohlenkamp et al., 2000, 2002), no studies report how expression of Amt genes is affected by nitrogen supply simultaneously in roots and leaves. To gain a fuller understanding of how these genes are regulated by nitrogen supply, we characterized the gene expression profiles for all six of the Amt genes in Arabidopsis in leaves and roots simultaneously when plants were grown with and without nitrogen (Fig. 9). The qRT-PCR data for each Amt gene allows comparisons to each other, where AtAmt1.1 is shown to be expressed the highest under −N (roots) and +N (leaves). Amt1.1 is the only Amt gene that shows a dramatic reciprocal increase in expression in Arabidopsis with nitrogen supply (high in roots of −N- and leaves of +N-grown plants).
Similar to Amt1.1, Amt1.5 also shows an increase (8-fold) in expression in −N roots (although at much lower overall levels), suggesting a role in ammonium acquisition in the roots under nitrogen starvation conditions. The higher levels of Amt1.2 and 1.3 in the roots (as opposed to leaves and independent of the nitrogen treatment) suggest housekeeping roles for these genes primarily in the roots and may point to a mechanism whereby the plant is always ready to transport ammonium in the roots. This feature, combined with nitrogen starvation inducibility for Amt1.1 and 1.5, may be sufficient to mask any deficiencies in ammonium transport in the roots caused by the loss of one of the Amt genes, such as in the amt1.1 mutant (Kaiser et al., 2002). Such compensation may not occur in the leaves as only Amt1.1 and 2.1 are expressed at high levels in the leaves. In fact, the amt1.1 knockout shows a thickening of the mesophyll tissue and a decrease in the intercellular airspace between mesophyll cells (Kaiser et al., 2002). Our results help explain these changes because Amt1.1 appears to be one of two major ammonium transporters in leaves.
MATERIALS AND METHODS
Plasmid Construction
The vector pRGK335 (Engineer et al., 2005) was used to create a cloning system (pRGK366) with dark-blue/white screening capabilities and a multiple cloning site (MCS). The lacZ and MCS used in creating the pRGK366 cloning vector was obtained from the plasmid pNEB193 via PCR: lacZ-MCS forward primer, TGGATATCGGTGTCGGGGCTGGCTGGCTTA; lacZ-MCS reverse primer, CCGATATCTTT/GACACTTTATGCTTCCGGC.
The lacZ promoter was modified to produce dark-blue colonies instead of light-blue colonies by changing the T to a G in the −35 region (see reverse primer for underlined base pair). As β-galactoside-mediated activation is not required in our system, such a change was beneficial for getting dark-blue/white screening capabilities. In addition to this change, the cleaved-amplified polymorphic binding regions in the lacZ′ promoter were also excluded to reduce the distance from the start site of Gal4 transcription and the TATA box of the minimal 35S promoter (164 bp). Restriction sites for EcoR-V were engineered into the primer (boxed regions). The PCR product was digested with EcoR-V and the pRGK335 plasmid was digested with PmeI. Blunt-end ligations of the two products resulted in pRGK366.
The AtAmt1.1 promoter region was isolated by PCR from genomic DNA from the Columbia ecotype of Arabidopsis (Arabidopsis thaliana): AtAmt1.1 forward primer, TCGGCGCGCCTTCCAACAACTATATGGATGTGATA; AtAmt1.1 reverse primer, TTGGATCCAAAGAAGGAAGCTAAAGGCTAGGGTT.
The PCR primers had restriction sites for AcsI and BamHI engineered into them (boxed regions). The PCR product and the vector pRGK366 were digested with AcsI and BamHI and ligated. The resulting plasmid pRGK 367 (GenBank accession no. DQ666283) had the AtAmt1.1 promoter region in the right orientation and was confirmed by sequencing and restriction digests.
Construction of the positive control vector, the rsGFP, and the vector containing rsGFP-GUS are as described previously (Engineer et al., 2005). The Amt-Gal4-LUC-GFP-GUS lines were created by pollinating Amt-Gal4-LUC lines with lines bearing the pGFP-GUS construct. Having two independent T-DNA responder elements to the AtAmt1.1 promoter in the Amt-Gal4-LUC-GFP-GUS lines ensures that reporter expression represents endogenous gene expression patterns (when all three reporters correlate in expression profiles) and reduces the likelihood of isolating plant lines that exhibit random reporter expression due to positional effects of the T-DNA integration site in the genome. All constructs were verified by sequence analysis and restriction digests with multiple enzymes.
Plant Growth Conditions
For all experiments mentioned in this article, seeds were always sterilized under constant agitation with ethanol:bleach (70:30) for 5 min followed by 5 min in 100% ethanol and drying on filter paper. Seeds were sparsely sprinkled on 0.6% agar plates (bacteriological agar; Acumedia, catalog no. 7178A) containing 0.5× Murashige and Skoog salts and 1× vitamins, vernalized overnight at 4°C, and then grown under an 8-h light/16-h dark cycle for 2 weeks in a Percival growth chamber. Light intensity in the chamber was maintained at 100 μE and light/dark temperatures were set at 24°C/22°C, respectively. The pH was always maintained at 5.7 in all agar (0.6%) and liquid media. After 2 weeks of growth, sets of 10 plants were transferred aseptically to different media (freshly prepared) and treatments mentioned in the various experiments in this article. For this transfer to different media, care was taken to maintain uniformity in the size and health of all selected plants. Seedlings with stunted or vitreous shoots or with stunted or highly branched roots were not selected. Healthy green seedlings with the first set of true leaves having emerged and expanded were chosen. Crowding effects were minimized by evenly spacing out the transferred seedlings on the agar plates or liquid medium. As a rule, a 150-mm × 30-mm agar plate always contained 100 mL of media (+N or −N), typically with no more than 10 plants that are 2 weeks old per plate. If a set of plants is left for more than 7 d on an agar plate containing +N medium, the nitrogen in the medium is depleted and AtAmt1.1 expression is initiated in the −N zones of the root system. For media containing varying amounts of nitrate, ammonium, and ammonium nitrate, modified Murashige and Skoog medium containing no nitrogen was used and the required amount of ammonium nitrate was added to the medium. For light intensity experiments, varying intensities were achieved by raising or lowering the agar plates of plants relative to the light source or by using black mesh to cover the plate to admit less light. For light duration experiments, after exposing the plants to light for the specific number of hours, the plates were wrapped in a black felt cloth and then by aluminum foil and placed in the chamber to maintain constant temperature conditions. For split-root experiments, root systems of 3-week-old mature plants were gently separated into two equal halves and placed on the edges of two adjoining containers bearing liquid Murashige and Skoog medium, one with nitrogen and one without.
Imaging of Plant Reporters
For imaging LUC expression in plant tissues, the luciferin substrate (d-luciferin firefly potassium salt; Biosynth AG; catalog no. l-8220) was sprayed on plants at a concentration of 1 mm and the plants were subsequently imaged under a CCD darkbox camera system by Fuji (LAS1000). One milliliter of luciferin was sprayed evenly for each 150-mm × 30-mm agar plate. Sterile luciferin solutions were always prepared and used fresh (within 24 h of preparation). Whole-plant images were captured with the provided software (Image-Pro) from Fuji Labsciences and quantitated using the Fuji Image Gauge software (version 3.41). LUC expression in tissues was calculated as relative luminescence per area of the image by digitally selecting the regions of interest on the captured LUC image and then measuring the area and luminescence in that region with the help of image gauge software. Background and dark-frame subtraction was carried out for each image. All figures with LUC expression are shown in false-color red. All LUC imaging was carried out under identical instrument settings, such as exposure time, distance from lens, aperture size, focus setting, and range scope setting. For GUS staining, plants were incubated in 5-bromo-4-chloro-3-indolyl-β-glucuronic acid for 4 h and then fixed in ethanol-glacial acetic acid for 4 h. GUS images were captured using a standard light microscope. GFP imaging was conducted directly in the plants without fixing the tissues and a Zeiss compound fluorescence scope was used. For confocal studies, a Nikon instrument was used.
Multiplexed qRT-PCR Experiments
Plant samples were imaged for LUC activity, washed, blotted dry, weighed, frozen in liquid nitrogen, and ground to a fine powder. Total RNA was extracted using TRIzol reagent (Sigma-Aldrich) according to the manufacturer's specifications. RNA was treated with Invitrogen Amp grade DNase-I according to the manufacturer's instructions (catalog no. 18068-015). First-strand cDNA synthesis was carried out on 2 μg of this RNA for each sample using Invitrogen SuperScript III reverse transcriptase (catalog no. 18080). cDNA was diluted 4-fold and 2 μL were used per 25-μL reaction in subsequent qRT-PCR experiments. Light upon extension primers for the qRT-PCR experiments were designed by Invitrogen's online light upon extension designer software and the sequences (5′→3′) and fluorophore designations were as follows: AtAmt1.1 forward, GGATGAGATGGCCGGTATGG; AtAmt1.1 reverse, CACGATACCAGAAGGAGAAGGAGATCG[FAM]G; AtAmt1.2 forward, CGGCAAAAGCTGAAGTCGGAGC[ALX546]G; AtAmt1.2 reverse, GCCACCATAGCTTCTTCGCTTT; AtAmt1.3 forward, CGGGCAAACCCACCGAGTAACC[ALX546]G; AtAmt1.3 reverse, TGCACAGCAGCTCTAACCACAC; AtAmt1.4 forward, CGGAGTACATATAAGCCAACCCTC[ALX546]G; AtAmt1.4 reverse, TACCGGCGGAGGATGAGATAG; AtAmt1.5 forward, CGGAGGTCTCTCACTGGTTCTGGTCTC[ALX546]G; AtAmt1.5 reverse, CAGTGCCAAACAAACGGTCTTC; AtAmt2.1 forward, CGGTTAGTAAGCGAAAC[ALX546]G; AtAmt2.1 reverse, ATTCCGGCGGTTATGTTATTCA; UBQ10 forward, GTACGTCGCAGCTTGAGGATGGACGTA[JOE]C; UBQ10 reverse, ACGAAGACGCAGGACCAAGT. FAM, 6-Carboxy-fluorescein; JOE, 6-carboxy-4′,5′-dichloro-2′,7′-dimethyoxy-fluoresein; ALX546, Alexa fluor 546.
Reactions were carried out with Sigma Jumpstart Taq ReadyMix for qRT-PCR (catalog no. D7440) according to the manufacturer's instructions with the exception of using a final concentration of 4.5 mm MgCl2 for the Gal4 gene and 6.5 mm MgCl2 for the LUC gene. Cycling conditions included 94°C for 120 s followed by 40 cycles of 94°C for 15 s, 65°C for 30 s, and 72°C for 30 s on the Cepheid Smart Cycler system. Every multiplexed PCR reaction contained the primer pair for the UBQ10 gene as an internal control (final concentration of 100 nm) in addition to the primer pair for one of the Amt genes (final concentration of 200 nm). For each sample of cDNAs, reactions were carried out in triplicate. A melt curve analysis for each gene product followed the PCR reaction. PCR products were run on a 5% agarose gel with a reference 10-bp DNA ladder (Invitrogen) to confirm the specificity and size of the PCR products (data not shown). For each PCR tube, the Δ-threshold cycle (ΔCt) for the sample Amt1.1, 1.2, 1.3, 1.4, 1.5, or 2.1 gene was calculated relative to the UBQ10 gene Ct and then the ΔCts were averaged for the triplicates. Fold differences in gene expression between samples were calculated by first determining the ΔΔCt values between samples and then using the formula: fold change = 2(ΔΔCt). At least three biological replicates were carried out for each set of experiments. To calculate PCR efficiencies for each gene, 10-fold serial dilutions of the template were used in reactions and R2 values were computed.
Statistical Analysis of qRT-PCR and LUC Expression Results
To correlate the expression patterns of LUC and the endogenous gene expression of AtAmt1.1, Pearson's correlation test was used. The analysis indicated a positive correlation of 0.628095 with a P value of 0.047698.
Quantification of Nitrate and Ammonium Levels
Plants were germinated and grown on +N medium for 2 weeks. Sets of 20 plants were then transferred to +N and −N media. After 5 d, leaf and root tissues were separately collected and frozen in liquid nitrogen. Leaf and root samples were lyophilized for 24 h at −50°C. Nitrate and ammonium analysis was conducted by the University of Missouri, Columbia's agricultural extension plant and soil testing lab using UV spectrophotometry and the QuickChem automated flow injection ion analyzer.
Root Measurements
For root architectural studies, 2-week-old seedlings grown on +N media were used. Sets of 10 plants were transferred to +N and −N media. Root measurements were carried out 2 and 5 d after this transfer. For lateral root lengths, the total length of all lateral roots from each plant was divided by the total number of lateral roots for that plant. For root hair number measurements, the root hairs in a 1-mm section of the root were counted. This 1-mm section was typically 3 mm from the root tip. Root hair length measurements were also conducted on this 1-mm section.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ666282 (pRGK366) and DQ666283 (pRGK367).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Amt-Gal4-LUC vector and AtAmt1.1 promoter control region.
Supplemental Figure S2. Graph showing qRT-PCR results from a sample run (four replicates) on leaves of +N- and −N-grown plants.
Supplemental Figure S3. Graph showing qRT-PCR results from a sample run (four replicates) on roots of +N- and −N-grown plants.
Supplemental Figure S4. PCR efficiencies for the AtAmt1.1 and UBQ10 genes.
Supplemental Figure S5. LUC expression profiles of +N leaf tissues used in qRT-PCR experiments (see Table I).
Supplemental Figure S6. Expression profiles for plants grown on media with varying amounts of ammonium nitrate.
Supplementary Material
Acknowledgments
We wish to thank Dr. Daniel Schachtmann for helpful comments on the manuscript and Dr. Howard Berg for help with microscopy. We would also like to thank Karen Fitzsimmons and Melissa Curran for their technical support.
This work was supported by the Monsanto/Washington University Collaborative Agreement (to R.G.K.) and, in part, by the Schneiderman graduate student fellowship (to C.B.E.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert G. Kranz (kranz@biology.wustl.edu).
The online version of this article contains Web-only data.
References
- Bates TR, Lynch JP (1996) Stimulation of root hair elongation in Arabidopsis thaliana by low phosphorus availability. Plant Cell Environ 19 529–538 [Google Scholar]
- Bloom AJ (1994). In KJ Boote, JM Bennett, TR Sinclair, GM Paulsen, eds, Physiology and Determination of Crop Yield. ASA/CSA, Madison, WI, pp 303–310
- Chen YM, Ferrar TS, Lohmeir-Vogel E, Morrice N, Mizuno Y, Berenger B, Ng KKS, Muench DG, Moorhead GBG (2006) The PII signal transduction protein of Arabidopsis thaliana forms an arginine-regulated complex with plastid N-acetyl glutamate kinase. J Biol Chem 281 5726–5733 [DOI] [PubMed] [Google Scholar]
- Drew MC (1975) Comparison of the effects of a localized supply of phosphate, nitrate, ammonium and potassium on the growth of the seminal root system, and in the shoot, in barley. New Phytol 75 479–490 [Google Scholar]
- Engineer C, Fitzsimmons K, Schmuke J, Dotson S, Kranz R (2005) Development and evaluation of a Gal4-mediated LUC/GFP/GUS enhancer trap system in Arabidopsis. BMC Plant Biol 5 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde BG, Clarkson DT (1999) Nitrate and ammonium nutrition of plants: physiological and molecular perspectives. Adv Bot Res 30 1–90 [Google Scholar]
- Gansel X, Munos S, Tillard P, Gojon A (2001) Differential regulation of the NO3− and NH4+ transporter genes AtNrt2.1 and AtAmt1.1 in Arabidopsis: relation with long-distance and local controls by N status of the plant. Plant J 26 143–155 [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Lejay L, Gojon A, Ninnemann O, Frommer WB, Wiren NV (1999) Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 11 937–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh M-H, Lam H-M, van de Loo FJ, Coruzzi G (1998) A PII-like protein in Arabidopsis: putative role in nitrogen sensing. Proc Natl Acad Sci USA 95 13965–13970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser BN, Rawat SR, Siddiqi MY, Masle J, Glass ADM (2002) Functional analysis of an Arabidopsis T-DNA knockout of the high-affinity NH4+ transporter AtAMT1;1. Plant Physiol 130 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Turano FJ (2003) The putative glutamate receptor 1.1 (AtGLR1.1) functions as a regulator of carbon and nitrogen metabolism in Arabidopsis thaliana. Proc Natl Acad Sci USA 100 6872–6877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Silim SN, Okamoto M, Siddiqi MY, Glass ADM (2003) Differential expression of three members of the AMT1 gene family encoding putative high-affinity NH4+ transporters in roots of Oryza sativa subspecies indica. Plant Cell Environ 26 907–914 [DOI] [PubMed] [Google Scholar]
- Lauter F-R, Ninnemann O, Bucher M, Riesmeier JW, Frommer WB (1996) Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc Natl Acad Sci USA 93 8139–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuwen WV, Hagendoorn MJM, Ruttnik T, Poecke RV, Plas LHWVD, Krol ARVD (2000) The use of the luciferase reporter system for in planta gene expression studies. Plant Mol Biol Rep 18 143a–143t [Google Scholar]
- Lejay L, Gansel X, Cerezo M, Tillard P, Muller C, Krapp A, von Wiren N, Daniel-Vedele F, Gojon A (2003) Regulation of root ion transporters by photosynthesis: functional importance and relation with hexokinase. Plant Cell 15 2218–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loque D, von Wiren N (2004) Regulatory levels for the transport of ammonium in plant roots. J Exp Bot 55 1293–1305 [DOI] [PubMed] [Google Scholar]
- Malamy JE, Ryan KS (2001) Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol 127 899–909 [PMC free article] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants. Academic Press, San Diego
- Millar AJ, Short SR, Chua NH, Kay SA (1992) A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet M, Dufour M, Lacroute F (1992) Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J 2 417–422 [DOI] [PubMed] [Google Scholar]
- Ninnemann O, Jauniaux J-C, Frommer WB (1994) Identification of a high affinity NH4+ transporter from plants. EMBO J 13 3464–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat SR, Salim SN, Kronzuker HJ, Siddiqi MY, Glass ADM (1999) AtAMT1 gene expression and NH4+ uptake in roots of Arabidopsis thaliana: evidence for regulation by root glutamine levels. Plant J 19 143–152 [DOI] [PubMed] [Google Scholar]
- Sanchez-Calderon L, Lopez-Bucio J, Chacon-Lopez A, Gutierrez-Ortega A, Hernandez-Abreu E, Herrera-Estrella L (2006) Characterization of low phosphorus insensitive mutants reveals a crosstalk between low phosphorus-induced determinate root development and the activation of genes involved in the adaptation of Arabidopsis to phosphorus deficiency. Plant Physiol 140 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelden MC, Dong B, Bruxelles GLD, Trevaskis B, Whelan J, Ryan PR, Howitt SM, Udvardi MK (2001) Arabidopsis ammonium transporters, AtAMT1;1 and AtAMT1;2, have different biochemical properties and functional roles. Plant Soil 231 151–160 [Google Scholar]
- Shin R, Berg RH, Schachtman DP (2005) Reactive oxygen species and root hairs in Arabidopsis root response to nitrogen, phosphorus and potassium deficiency. Plant Cell Physiol 46 1350–1357 [DOI] [PubMed] [Google Scholar]
- Sohlenkamp C, Shelden M, Howitt S, Udvardi M (2000) Characterization of Arabidopsis AtAMT2, a novel ammonium transporter in plants. FEBS Lett 467 273–278 [DOI] [PubMed] [Google Scholar]
- Sohlenkamp C, Wood CC, Roeb GW, Udvardi MK (2002) Characterization of Arabidopsis AtAMT2, a high-affinity ammonium transporter of the plasma membrane. Plant Physiol 130 1788–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JF, Hayes LS, Lloyd DB (1991) Modulation of firefly luciferase stability and impact on studies of gene-regulation. Gene 103 171–177 [DOI] [PubMed] [Google Scholar]
- von Wiren N, Gazzarrini S, Gojon A, Frommer WB (2000. a) The molecular physiology of ammonium uptake and retrieval. Curr Opin Plant Biol 3 254–261 [PubMed] [Google Scholar]
- von Wiren N, Lauter F-R, Ninnemann O, Gillissen B, Walch-Liu P, Engels C, Jost W, Frommer WB (2000. b) Differential regulation of three functional ammonium transporter genes by nitrogen in root hairs and by light in leaves of tomato. Plant J 21 167–175 [DOI] [PubMed] [Google Scholar]
- Wang R, Guegler K, LaBrie ST, Crawford NM (2000) Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12 1491–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Akiyama A, Kisaka H, Uchimiya H, Miwa T (2004) Metabolic engineering with Dof1 transcription factor in plants: improved nitrogen assimilation and growth under low-nitrogen conditions. Proc Natl Acad Sci USA 101 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG (2000) Regulation of Arabidopsis root development by nitrate availability. J Exp Bot 51 51–59 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.