Abstract
Analysis of the functions of Short Vegetative Phase (SVP)-like MADS-box genes in barley (Hordeum vulgare) indicated a role in determining meristem identity. Three SVP-like genes are expressed in vegetative tissues of barley: Barley MADS1 (BM1), BM10, and Vegetative to Reproductive Transition gene 2. These genes are induced by cold but are repressed during floral development. Ectopic expression of BM1 inhibited spike development and caused floral reversion in barley, with florets at the base of the spike replaced by tillers. Head emergence was delayed in plants that ectopically express BM1, primarily by delayed development after the floral transition, but expression levels of the barley VRN1 gene (HvVRN1) were not affected. Ectopic expression of BM10 inhibited spike development and caused partial floral reversion, where florets at the base of the spike were replaced by inflorescence-like structures, but did not affect heading date. Floral reversion occurred more frequently when BM1 and BM10 ectopic expression lines were grown in short-day conditions. BM1 and BM10 also inhibited floral development and caused floral reversion when expressed in Arabidopsis (Arabidopsis thaliana). We conclude that SVP-like genes function to suppress floral meristem identity in winter cereals.
During the life cycle of a plant, the shoot apical meristem progresses through three phases of development: vegetative, inflorescence, and floral (Poethig, 1990). In each phase, the apical meristem produces a different set of organs. The vegetative meristem produces leaves, the inflorescence meristem produces leaves and floral meristems to form the inflorescence, and the floral meristem produces the organs that form the flower. The different phases of meristem development are controlled by genes that establish and maintain meristem identity.
The shift from vegetative to inflorescence meristem identity, the floral transition, marks the beginning of the reproductive growth phase and is an important determinant of flowering time. In Arabidopsis (Arabidopsis thaliana), the Short Vegetative Phase (SVP) gene encodes a MADS-box transcription factor that delays the floral transition (Hartmann et al., 2000). Mutations that disrupt SVP cause early flowering (Hartmann et al., 2000), whereas ectopic expression of SVP results in late flowering. Ectopic expression of SVP also inhibits floral meristem identity, causing floral abnormalities such as the conversion of sepals and petals to leaf-like structures (Brill and Watson, 2004; Masiero et al., 2004) and causing inflorescence-like structures to develop within flowers (Brill and Watson, 2004). The development of inflorescences within flowers indicates that meristematic cells within the flower have lost floral identity and have formed an inflorescence instead of floral organs, a phenomenon known as floral reversion (Tooke et al., 2005). Presumably, ectopic expression of SVP causes floral reversion by interfering with a mechanism that maintains floral meristem identity.
The Arabidopsis gene, AGAMOUS-LIKE 24 (AGL24), is closely related to SVP (Yu et al., 2002; Michaels et al., 2003). Unlike SVP, AGL24 promotes the floral transition. Mutations that disrupt AGL24 cause late flowering, whereas overexpression of AGL24 accelerates flowering (Yu et al., 2002; Michaels et al., 2003). AGL24 is expressed during vegetative development and is induced by treatments that accelerate floral transition, such as vernalization (prolonged exposure to low temperatures), long days, or the application of gibberellins (Yu et al., 2002; Michaels et al., 2003). These data suggest that AGL24 acts to promote floral transition in response to vernalization and long-day conditions (Yu et al., 2002; Michaels et al., 2003). Although AGL24 has the opposite effect on flowering time compared to SVP, plants that ectopically express AGL24 exhibit floral abnormalities similar to those caused by ectopic expression of SVP, and ectopic expression of AGL24 also causes floral reversion. Thus, AGL24 promotes the floral transition but inhibits floral meristem identity. It has been suggested that AGL24 promotes inflorescence meristem identity (Yu et al., 2004).
Cereals and related grass species follow a pattern of development similar to that of other plants. When floral transition occurs, the shoot apex elongates and floral primordia form, resulting in the appearance of distinctive double ridges. Then, as the shoot apex differentiates into a compact inflorescence, the floral primordia give rise to spikelets that produce the floral meristems that develop into florets (flowers). In winter cereals, such as wheat (Triticum aestivum) and barley (Hordeum vulgare), the floral transition can be accelerated by long days or by prolonged cold treatment (vernalization) and is also subject to genetic variation that affects the number of leaves that develop prior to the floral transition (Boyd et al., 2003).
The induction of the floral transition by vernalization is mediated by the VRN1 gene in winter cereals and related grass species such as Lolium perenne (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003; Andersen et al., 2006). VRN1 encodes an APETALA1 (AP1)-like MADS-box gene that is induced by exposure to long periods of cold and promotes the floral transition (Danyluk et al., 2003; Trevaskis et al., 2003; Yan et al., 2003). It is not known how cold causes activation of VRN1 expression during vernalization. One suggestion is that an SVP-like gene, Vegetative to Reproductive Transition gene 2 (VRT2, or HvVRT2 for the barley homolog), mediates induction of VRN1 in vernalized plants (Kane et al., 2005). In wheat, VRT2 is expressed in vegetative plants. During cold treatment, VRT2 expression decreases gradually in contrast to the gradual increase in expression of VRN1 (Kane et al., 2005). The expression patterns of VRT2 and VRN1 during cold treatment of wheat are consistent with the hypothesis that VRT2 represses VRN1 and that down-regulation of VRT2 by cold allows expression of VRN1. There is, however, no direct evidence that VRT2 regulates VRN1. In the vernalization-responsive grass L. perenne, overexpression of the VRT2 homolog, L. perenne MADS-Box gene 10 (LpMADS10), does not affect the timing of flowering (Petersen et al., 2006). Thus, it remains unclear whether SVP-like genes regulate flowering time in grasses.
We have investigated the potential roles of SVP-like genes in the vernalization response of barley, an important crop and a useful diploid model for winter cereal crops. We examined the phenotypes of transgenic plants with altered expression levels for two of the barley SVP-like genes and examined whether SVP-like genes are likely to regulate expression of the barley VRN1 gene (HvVRN1). We show that in barley, SVP-like genes regulate meristem identity but are unlikely to mediate derepression of HvVRN1 during vernalization.
RESULTS
Three SVP-Like Genes Are Expressed in Vegetative Tissues of Barley But Are Repressed during Spike Development
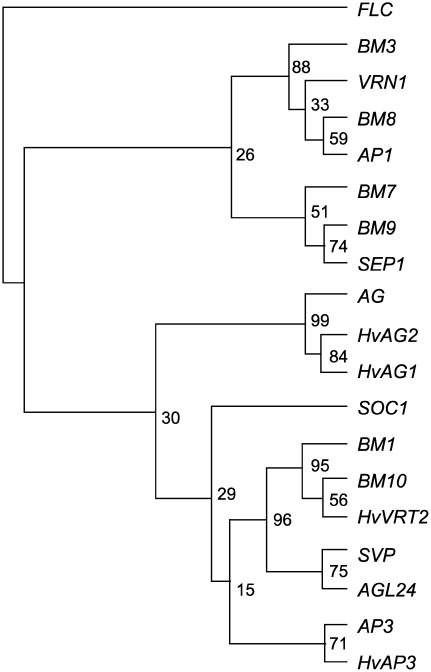
Three SVP-like genes were isolated from a barley cDNA library: Barley MADS1 (BM1; Schmitz et al., 2000), HvVRT2 (Kane et al., 2005), and a third gene, designated BM10 (GenBank EF043040; Supplemental Fig. S1). Phylogenetic analysis shows that SVP-like genes of barley are more closely related to each other than to either SVP or AGL24, suggesting that SVP-like genes have diverged separately in monocots and dicots (Fig. 1). Among the barley SVP-like genes, BM10 is more closely related to HvVRT2 (77% amino acid sequence identity) than BM1 (51% amino acid sequence identity). All three genes are expressed in a range of vegetative organs (Supplemental Fig. S2).
Figure 1.
Phylogeny of barley MADS-box genes. A consensus phylogenetic tree created using the protein parsimony method of the PHYLIP package (http://evolution.genetics.washington.edu/phylip.html). Bootstrap support values from 1,000 bootstrap replicates are shown for each node as percentages. Analysis was performed on an alignment of the amino acid sequences of the MADS-box domains (the region equivalent to amino acids 1–62 from alignment in Supplemental Fig. S1) of: AGAMOUS (AG, NP 567569), AGL24 (NM118587), SVP (NM127820), BM10 (EF043040), HvVRT2 (DQ201168), BM1 (AJ249142), SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1, NM130128), SEPALLATA1 (SEP1, NM121585), FLOWERING LOCUS C (FLC, NM121052), AP3 (NM115294), AP1 (NM105581), HvVRN1 (VRN1, AJ249144), BM3 (AJ249143), Hordeum vulgare AGAMOUS-like 1 (HvAG1, AF486648), Hordeum vulgare AGAMOUS-like 2 (HvAG2, AF486649), BM7 (AJ249145), BM8 (AJ249146), BM9 (AJ249147), and Hordeum vulgare APETALA3 (HvAP3, AY541065).
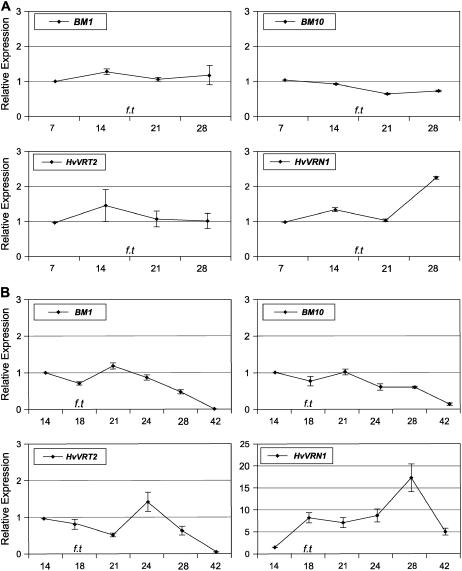
The relationship between the floral transition and expression of the SVP-like genes was examined in an early flowering spring barley variety (cv Golden Promise). Expression levels of BM1, BM10, and HvVRT2 were monitored at weekly intervals in whole plants during the first 4 weeks of development. The floral transition, as indicated by the double ridge stage of apex development, occurred after approximately 18 d. HvVRT2 and BM1 were expressed at similar levels throughout the developmental time course (Fig. 2A). BM10 expression decreased slightly after 21 d of growth (to 0.6-fold of starting level). In comparison, expression of HvVRN1 increased after 21 d (Fig. 2A).
Figure 2.
Developmental regulation of BM1, BM10, HvVRT2, and HvVRN1. A, Relative expression levels of BM1, BM10, HvVRT2, and HvVRN1 assayed by qRT-PCR and normalized to ACTIN (see “Materials and Methods”) in RNA from plants of different ages (cv Golden Promise, minus roots). Expression level is shown relative to the initial time point (7-d-old plants). B, Relative expression levels of BM1, BM10, HvVRT2, and HvVRN1 in isolated shoot apices harvested from plants of different ages. Expression is shown relative to the initial time point (RNA from apices of 14-d-old plants). f.t denotes the time point of the floral transition, as indicated by the appearance of double ridges on the shoot apex. Error bars show se.
Expression levels of BM1, BM10, and HvVRT2 were then monitored in the developing shoot apex. Expression levels of all three genes were similar in vegetative apices (14 d, vegetative meristem) and in apices at the double ridge stage (18 d, inflorescence meristems) but were reduced at later stages of apical development (42 d, floral meristems; Fig. 2B). At this stage, floral organ differentiation was visible (Supplemental Fig. S3). Expression of HvVRN1 increased around the time of floral transition (10-fold) and remained high throughout subsequent stages of apex development (Fig. 2B).
BM1, BM10, and HvVRT2 Are Not Regulated by Day Length
Day length influences the timing of floral transition in cereals. Long days have been reported to repress HvVRT2 expression during cold treatment (Kane et al., 2005). We examined the influence of day length on expression of BM1, BM10, and HvVRT2 during long-day induction of the floral transition in a day-length responsive spring barley variety (cv Icheon Naked). Plants were grown in short days then shifted to inductive long-day conditions. Plants shifted to long days underwent floral transition, as indicated by the presence of double ridges at the shoot apex, within 2 weeks, whereas plants maintained in short days remained vegetative. Plants in the long-day treatment flowered 60 d earlier than plants maintained in short days (head emergence after 50 ± 1 d versus 111 ± 1 d). After 7 d, expression levels of BM1, BM10, and HvVRT2 were similar in both treatments (Fig. 3A). In comparison, expression of HvFT, a key gene in the photoperiod response pathway (Turner et al., 2005), was strongly induced by the long-day treatment (Fig. 3B). Expression of HvVRN1 was also induced by long days (Fig. 3A), as has been reported previously for this barley variety (Trevaskis et al., 2006). These data suggest that, in barley, transcriptional repression of SVP-like genes is not required to promote floral transition in response to long-day conditions.
Figure 3.
The effect of short or long day lengths on expression levels of BM1, BM10, and HvVRT2. A, Relative expression levels of BM1, BM10, HvVRT2, and HvVRN1, assayed by qRT-PCR, in RNA from plants (cv Icheon Naked, minus roots) that were maintained in short days (8 h dark/16 h light) or shifted to long days (16 h light/ 8 h dark) for 7 d. Expression is shown relative to the initial time point (14-d-old plants in short days). B, Expression levels of HvFT, normalized to ACTIN, in the same RNA samples. Error bars show se. Asterisks indicate P values of Student's t tests: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
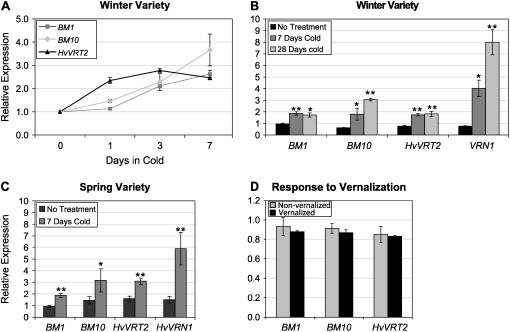
BM1, BM10, and HvVRT2 Are Induced by Cold
It has been reported that VRT2 is repressed during cold treatment in wheat (Kane et al., 2005). We found that BM1, BM10, and HvVRT2 were induced by cold in a vernalization-responsive winter barley (cv Sonja: Hvvrn1, HvVRN2). Cold induction of BM1 and HvVRT2 occurred within 3 d (Fig. 4A; Supplemental Fig. S4A), but longer cold treatments did not cause further increases in expression (Fig. 4B). Expression levels of BM10 also increased within 3 d of cold treatment and increased further with longer cold treatments (Fig. 4, A and B). Cold treatment also induced HvVRN1 expression, as has been reported previously for winter barley varieties (Trevaskis et al., 2003). In a spring barley, (cv Golden Promise: HvVRN1, Hvvrn2), cold treatment also induced BM1, BM10, and HvVRT2. This correlated with induction of HvVRN1 (Fig. 4C; Supplemental Fig. S4B). These data are not consistent with HvVRT2 being a repressor of HvVRN1.
Figure 4.
BM1, BM10, and HvVRT2 are induced by cold. A, Relative expression levels of BM1, BM10, and HvVRT2, assayed by qRT-PCR, in a winter variety (cv Sonja, minus roots) after 0, 1, 3, or 7 d cold treatment (4°C). Expression is shown relative to the initial time point (14-d-old plants grown in standard glasshouse conditions). B, Relative expression levels of BM1, BM10, HvVRT2, and HvVRN1, assayed by qRT-PCR, in a winter barley variety (cv Sonja, minus roots) maintained in normal glasshouse conditions for 7 d (no treatment) or after 7 or 28 d of cold treatment. Expression is shown relative to the initial time point (14-d-old plants in standard glasshouse conditions). C, Expression levels of BM1, BM10, HvVRT2, and HvVRN1, assayed by qRT-PCR, in plants of a spring barley cultivar (cv Golden Promise, minus roots) that were maintained in normal glasshouse conditions (no treatment) or shifted to cold conditions (4°C) for 7 d. Expression is shown relative to the initial time point (14-d-old plants in normal glasshouse temperatures). Error bars show se. D, Expression levels of BM1, BM10, and HvVRT2 assayed by qRT-PCR in plants (cv Sonja, minus roots) that were imbibed and vernalized for 9 weeks then reacclimatized to standard glasshouse conditions for a week. Expression is shown relative to control nonvernalized plants at the same developmental stage (two leaves, vegetative shoot apex). Differences were nonsignificant by Student's t test. Error bars show se. Asterisks indicate P values of Student's t tests: *, P < 0.05; **, P < 0.01.
The effect of vernalization on BM1, BM10, and HvVRT2 expression levels was examined in a vernalization-responsive barley winter variety (cv Sonja). BM1, BM10, or HvVRT2 expression levels did not differ between vernalized and nonvernalized plants (Fig. 4D). Expression of HvVRN1 was strongly induced by the same treatment (250 ± 40.3) and heading date was accelerated (Trevaskis et al., 2003). Thus, it seems unlikely that repression of HvVRT2 is required for induction of HvVRN1 or the acceleration of floral transition by vernalization.
VRN1 Does Not Regulate SVP-Like Genes in Winter Cereals
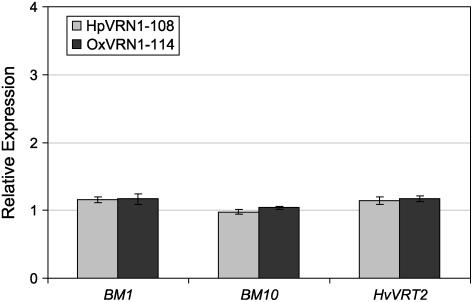
Cold treatment of plants induced expression of both HvVRN1 and SVP-like genes. We examined whether HvVRN1 might regulate expression of SVP-like genes. Expression levels of BM1, BM10, and HvVRT2 were compared between transgenic plants (cv Golden Promise) that overexpress VRN1 or have reduced levels of HvVRN1 transcript. Reverse transcription (RT)-PCR quantification showed that these lines had HvVRN1 expression levels that were 21- ± 0.25-fold or 0.30- ± 0.01-fold that of wild-type plants, respectively, a difference that is clearly visible by RNA gel-blot analysis (Supplemental Fig. S5A). These changes did not affect expression of BM1, BM10, or HvVRT2 (Fig. 5). Similarly, in wheat, VRT2 expression did not vary between spring and winter near-isogenic lines of the Triple Dirk series (Pugsley, 1971), which express VRN1 to different levels (Trevaskis et al., 2003; Supplemental Fig. S5B). It is unlikely that VRN1 directly regulates expression of VRT2.
Figure 5.
HvVRN1 does not regulate transcription of BM1, BM10, orHvVRT2. Expression levels of BM1, BM10, and HvVRT2, assayed by qRT-PCR, in RNA from transgenic plants that carry either a RNAi construct against HvVRN1 (line HpVRN1-108) or a VRN1 overexpression construct (line OxVRN1-114). Expression is shown relative to a wild-type plant (14-d-old plants minus roots). Similar results were obtained in a second independently transformed line for each construct. Error bars show se. Differences were nonsignificant by Student's t test.
BM1 or BM10 Does Not Regulate Expression of HvVRN1
To investigate the functions of SVP-like genes in barley, we altered the expression levels of BM1 and BM10 (HvVRT2-like) in transgenic barley. These genes were chosen because they are phylogenetically distinct (Fig. 1) and have different expression profiles during cold treatment. Initially, the effects of reducing the levels of BM1 or BM10 expression levels were investigated using gene-specific RNA interference (RNAi) constructs. More than 50 independent transgenic lines were produced for each construct. Some lines showed reduced expression levels for the targeted genes (Supplemental Fig. S6), but none of the lines generated for either construct showed any phenotypic abnormalities or any change in heading date (Supplemental Fig. S7).
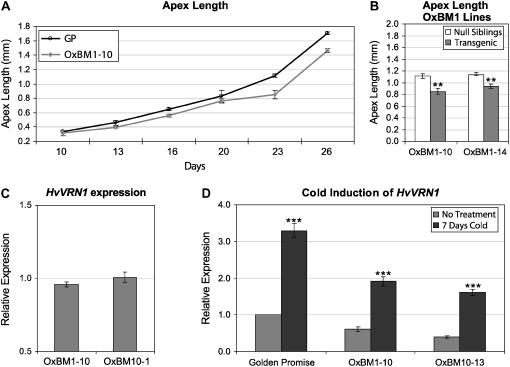
As reduced expression of BM1 or BM10 did not elicit any abnormal phenotypes, BM1 or BM10 were expressed under the control of the constitutive maize (Zea mays) UBIQUITIN promoter (Supplemental Fig. S8). The BM1 overexpression construct (OxBM1) delayed head emergence by approximately 10 d, whereas the BM10 overexpression construct (OxBM10) did not influence heading date (Supplemental Fig. S7). Comparison of apex lengths, a measure of floral development, between plants that overexpress BM1 and control siblings showed that overexpression of BM1 delayed the floral transition by approximately 3 d (Fig. 6, A and B) and that the delay in head emergence in BM1 overexpression lines (approximately 10 d) must be due primarily to delayed floral development after the floral transition. HvVRN1 expression levels were similar in plants that ectopically express BM1 or BM10 and sibling null control lines (Fig. 6C), and HvVRN1 expression could be induced by cold treatment in plants that ectopically express BM1 or BM10 (Fig. 6D). Thus, the delay of heading date caused by OxBM1 is not due to transcriptional repression of HvVRN1.
Figure 6.
Ectopic expression of BM1 delays the floral transition, but HvVRN1 levels are not altered. A, Apex length (average length in millimeters of apices from five plants) of wild-type plants, cv Golden Promise (GP), or transgenic plants homozygous for OxBM1 (line OxBM1-10) harvested at different time points. B, Comparison of the average length of the shoot apex after 23 d for homozygous transgenic and sibling null control plants from two independent BM1 overexpression lines. C, HvVRN1 expression levels in BM1 (line 10) or BM10 (line 1) overexpression lines (14-d-old plants minus roots). Expression levels were assayed by qRT-PCR and are shown relative to the level of expression in control sibling null plants. D, HvVRN1 expression, as assayed by qRT-PCR, in plants (whole plants minus roots) homozygous for the BM1 (OxBM1-10) or BM10 (line 13) overexpression construct that were grown for 14 d then either moved to cold conditions for 7 d or maintained in standard glasshouse conditions (no treatment). Expression is shown relative to the level of expression in Golden Promise plants from the control treatment. Error bars show se. Asterisks indicate P values of Student's t tests: **, P < 0.01; ***, P < 0.001. Similar results were obtained in a second independently transformed line for each construct.
Ectopic Expression of BM1 or BM10 Inhibits Floral Development and Causes Floral Reversion
Ectopic expression of BM1 caused a number of phenotypes in the spike where BM1 is not normally expressed. The rachis internodes were elongated giving the spike a concertina phenotype (Fig. 7, A and B) and the vestigial florets normally found on two-row cultivars were reduced in size (Fig. 7B). In the fertile florets, the palea and lemma were elongated and more leaf like (Fig. 7C), whereas the lodicules were smaller than in wild-type plants (Fig. 7D). These phenotypes are all indicative of a loss of floral characteristics and a shift toward stem characteristics; unlike the spike, the internodes of cereal stems are elongated and the nodes bear leaves but not flowers.
Figure 7.
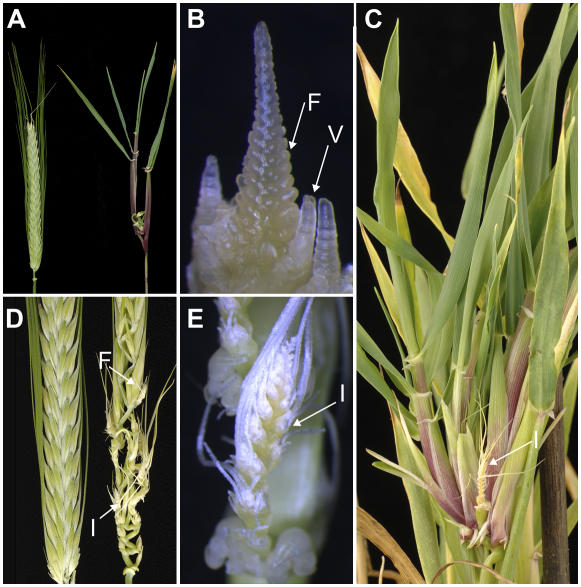
Abnormal spike morphology in plants transformed with OxBM1 or OxBM10 constructs. Spike phenotypes caused by ectopic expression of BM1 or BM10. A, A spike from a wild-type plant (cv Golden Promise, left) compared to spikes from a transgenic plant homozygous for OxBM1 (line OxBM1-10, middle) or OxBM10 (line OxBM10-13, right). Spikes were taken at head emergence. B, Comparison of dissected inflorescence stems from a wild-type plant (left) and a plant overexpressing BM1 (OxBM1-10, right) showing the elongation of rachis internodes (ri) and the reduced size of the vestigial florets (vf). C, A comparison of florets from a wild-type plant (left) and a plant overexpressing BM1 (OxBM1-10, right) showing the elongation of the lemma (l). D, As above, but the lemma has been removed to reveal the elongation of the palea (p) and the reduction in size of the lodicules (ld).
In a small number of T0 lines (2/50 for BM1 lines), tillers developed at the base of spikes at positions where florets would normally be located (Fig. 8, A–C). This suggests that ectopic expression of BM1 converts a floral meristem to a vegetative meristem to give rise to tillers from within the spike. This effect is analogous to floral reversion in Arabidopsis. Floral reversion occurred at a higher frequency when plants were grown in short days. For example, when 10 plants overexpressing BM1 (line 10) were grown in short days, four failed to produce heads and six produced heads that showed profuse ectopic tillering, with multiple tillers arising from within the primary spike (Fig. 8C). No ectopic tillering was observed when the same line was grown in long days or in a control sibling null grown in short days. Spike development was severely disrupted when ectopic tillering occurred. The apex progressed past floral transition, but often florets did not develop fully (Fig. 8, B and C) and plants were infertile when grown in short days, even when florets did develop.
Figure 8.
Floral reversion in plants transformed with OxBM1 or OxBM10 constructs. A, A spike from a short-day-grown wild-type plant (left) compared to a spike from a plant overexpressing BM1 (OxBM1-10, right) grown in the same conditions. B, The shoot apex (250× magnification) from a short-day-grown plant overexpressing BM1 (OxBM1-10) showing the conversion of floral primordia (F) to vegetative meristems (V). C, The severe floral reversion phenotype caused by ectopic expression of BM1 in short days (OxBM1-10). Multiple tillers can be seen radiating from the base of the inflorescence (I) that has not developed fully. D, Comparison of a wild-type spike (left) and a spike from a BM10 overexpression line (OxBM10-13, right) showing conversion of florets (F) to inflorescences (I). E, A magnified view (90× magnification) of inflorescences (I) that have developed in the place of florets in the same line.
Overexpression of BM10 caused very similar spike and floret phenotypes in long-day-grown plants (Fig. 7A). Moreover, the florets at the base of the spike frequently failed to develop in BM10 overexpression lines and reverted to inflorescence-like structures (Fig. 8, D and E), suggesting that expression of BM10 causes the floral meristem to revert to an inflorescence, a partial floral reversion. As was the case for BM1 overexpression lines, overexpression of BM10 also caused complete floral reversion in short days, giving rise to ectopic tillers from within the spike. The same phenotypes did not occur in null sibling controls.
Expression of Barley SVP-Like Genes in Arabidopsis Causes Floral Reversion Phenotypes
When BM1 and BM10 were expressed in Arabidopsis (C24 ecotype) under the control of the cauliflower mosaic virus 35S promoter (35S), floral abnormalities similar to those caused by ectopic expression of SVP resulted. These included conversion of petals and sepals to leaf-like structures, enlargement of the gynoecium, and production of stipitate ovaries, where the base of the ovary elongates (Fig. 9, A–C). The presence of leaves and elongation of internodes are features of the inflorescence and are indicative of a shift toward inflorescence identity in the flower. The flowers of Arabidopsis plants expressing BM1 or BM10 also underwent frequent floral reversion events, where an inflorescence formed within the flower (Fig. 9, D–F). This occurred in two different locations within flowers: at the end of the gynoecium or more commonly within the axil of the sepal. Sometimes this occurred more than once from a single flower (Fig. 9, D and E).
Figure 9.
Floral reversion and abnormal floral phenotypes in Arabidopsis plants that express BM1 or BM10. A, Flower from an Arabidopsis plant transformed with the 35S∷BM1 construct showing the conversion of sepals and petals to leaf-like organs and floral reversion, where an inflorescence has arisen from within the flower. B, The flower from an Arabidopsis plant transformed with the 35S∷BM10 construct showing the conversion of sepals and petals to leaf-like organs. C, Stipitate ovary and enlargement of the gynoecium in a flower from an Arabidopsis plant expressing BM10. D, Multiple floral reversion events in a plant expressing BM1 plant. E and F, Floral reversion events in plants expressing BM10.
DISCUSSION
We have investigated the relationship between the timing of the floral transition and expression levels of three SVP-like genes in barley. We found no evidence that transcriptional repression of these SVP-like genes is required for floral transition to occur. All three genes are expressed at similar levels in vegetative and inflorescence meristems. Moreover, we found no evidence that transcriptional repression of BM1, BM10, or HvVRT2 is required to accelerate the floral transition in response to vernalization or long days. Expression of all three genes did decrease at later stages of apex development, suggesting that reduced expression levels of BM1, BM10, and HvVRT2 may be required for floral development.
Ectopic expression of BM1 or BM10 caused a shift toward vegetative characteristics in the spikes of barley, consistent with the hypothesis that down-regulation of SVP-like genes is important during floral development. Ectopic expression of Oryza sativa MADS gene 22 and LpMADS10 cause similar inhibition of floral characteristics when ectopically expressed in rice (Oryza sativa) or L. perenne, respectively, but neither gene affects heading date (Sentoku, personal communication; Petersen et al., 2006). It has been suggested that these genes must regulate meristem identity (Sentoku et al., 2005; Petersen et al., 2006).
In long days, ectopic expression of BM1 caused floral primordia to revert to vegetative meristems in some lines, resulting in the appearance of ectopic tillers within the spike. Similarly, ectopic expression of BM10 caused floral meristems to revert to inflorescence-like structures. Thus, both genes disrupt floral meristem identity and cause floral reversion when expressed ectopically in barley. For both genes, the effect of ectopic expression was more extreme in short days and often completely blocked normal floral development after floral transition. These floral reversion phenotypes strongly support the hypothesis that SVP-like genes regulate meristem identity in cereals and related grass species. The slight delay of the floral transition caused by ectopic expression of BM1 shows that this gene may also inhibit inflorescence meristem identity in long days and could influence the timing of the floral transition in some conditions or in some genetic backgrounds. The absence of abnormal phenotypes in barley plants with reduced expression of BM1 or BM10 suggests there is functional redundancy between SVP-like genes in barley, although it is also possible that none of the lines generated (over 50 for each construct) had sufficiently reduced expression of the target gene to reveal any phenotypes.
Expression of BM1 or BM10 caused floral reversion when expressed in Arabidopsis (a dicot) and barley (a monocot). Expression of other SVP-like genes (e.g. SVP and AGL24) is known to cause floral reversion in Arabidopsis. The ability of SVP-like genes to trigger floral reversion is not an artifact of ectopic expression; in tomato (Lycopersicon esculentum), indeterminant sympodial growth is achieved through successive floral reversions, where an inflorescence develops below the flower on an otherwise determinant floral branch (Szymkowiak and Irish, 2006). A SVP-like gene, JOINTLESS, is required to generate an inflorescence meristem (the sympodial meristem) below the flower and is essential for sympodial growth (Szymkowiak and Irish, 2006). Floral reversion is also caused by mutations that inhibit floral meristem identity, such as ap1 (for review, see Tooke et al., 2005). In the ap1 mutant, floral reversion occurs more frequently in short-day conditions (Okamuro et al., 1997). In barley, we found that short-day conditions increased the frequency and severity of floral reversion in plants that ectopically express BM1 or BM10. This suggests that day length conditions influence floral meristem identity in Arabidopsis and barley.
SVP-like genes may be components of a conserved regulatory mechanism that controls meristem identity in both dicots and monocots. This may involve interactions between SVP-like and AP1-like MADS-box proteins. In Arabidopsis, SVP and AGL24 interact with AP1 and members of a LEUNIG-SEUSS corepressor complex to repress AGAMOUS during floral development (Gregis et al., 2006). Similar interactions between SVP-like and AP1-like proteins may be important for floral development in cereals and other grasses. In yeast (Saccharomyces cerevisiae) two-hybrid assays, LpMADS10 interacts with LpVRN1 (LpMADS1; Ciannamea et al., 2006; Petersen et al., 2006), and TaVRT2 interacts with VRN1 (Kane et al., 2005).
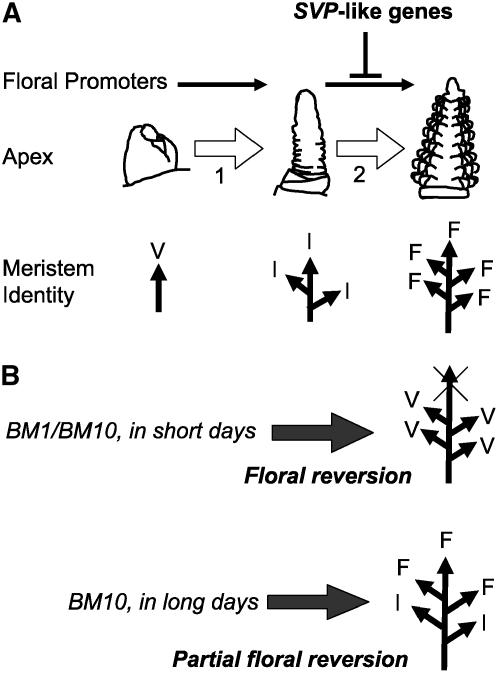
If SVP-like genes inhibit floral meristem identity in cereals, what role does this play during normal plant development? BM1 and BM10 may be expressed in vegetative and inflorescence tissues to prevent premature transition from inflorescence to floral meristem identity (Fig. 10). It seems likely that these SVP-like genes would delay the formation of floral meristems more during winter when temperatures drop and days are short, inducing the expression of BM1 and BM10 and enhancing the inhibition of floral meristem identity by these genes. These genes may therefore play an important role to repress floral development during winter, possibly to counteract induction of flowering by vernalization. According to this hypothesis, BM1 and BM10 act antagonistically to genes that promote floral meristem identity and developmental down-regulation of BM1 and BM10 some time after floral transition is required to allow floral development to proceed. HvVRT2, which is similar in sequence to BM10 and has a similar expression pattern to BM1 and BM10, may have a similar function.
Figure 10.
Schematic representation of the regulation of meristem identity in winter cereals by SVP-like genes. A, The shoot apex normally proceeds through two phase transitions: from vegetative to inflorescence, and from inflorescence to floral. Long days and vernalization accelerate these phase transitions through the activity of floral promoters such as FT, VRN1, or gibberellins. SVP-like genes are expressed in the vegetative and inflorescence apex to inhibit floral meristem identity and prevent precocious development of floral organs, particularly under short days and in cold conditions. B, Ectopic expression of BM1 or BM10 inhibits floral meristem identity and retards floral development. In short days, where floral promoters are less active, ectopic expression of BM1 or BM10 has a stronger effect and causes the floral meristems to revert to vegetative meristems. In long days, where floral promoters promote floral development this is sufficient to weaken floral characteristics and cause occasional floral reversion (BM1) or partial floral reversion (BM10).
Kane et al. (2005) reported that in short-day conditions, HvVRT2 expression decreased after 77 d of cold treatment. This may have been caused by prolonged cold treatment (vernalization) or by the floral transition, which occurred at 70 d. We found that cold induced HvVRT2 expression in vegetative barley plants and that this correlated with induction of HvVRN1. This is not consistent with the suggestion that derepression of HvVRT2 is required for cold induction of HvVRN1. Another mechanism proposed to account for induction of HvVRN1 by cold involves the HvVRN2 floral repressor locus (Yan et al., 2004; Fu et al., 2005). We found that HvVRN1 is induced by cold in the spring barley Golden Promise, which lacks HvVRN2, suggesting that cold induction of HvVRN1 can occur independently of HvVRN2. Golden Promise also lacks the regulatory region in the first intron of HvVRN1, which is required for repression in nonvernalized plants (Fu et al., 2005), suggesting that this region is not critical for cold induction of HvVRN1. Identification of the regulatory components that mediate cold induction of VRN1 will be an important step in understanding the vernalization response in winter cereals.
In summary, the phenotypes of barley and Arabidopsis plants that ectopically express BM1 or BM10 are consistent in showing inhibition of floral organ development and reversion of the floral meristems. Thus, the primary function of SVP-like genes in cereals seems to be in determining meristem identity. This is likely to be a conserved function of SVP-like genes per se.
MATERIALS AND METHODS
Plant Growth
Plants were grown in pots of soil in sunlit glasshouses under long days (16 h light/8 h dark, with supplementary lighting used when natural light levels dropped below 200 μE) or short days (8 h light/16 h dark). Glasshouses had an average temperature of 19°C. For vernalization treatment, plants were imbibed in soil and placed at 4°C (12 h light/12 h dark) for 9 weeks, then reacclimatized to normal glasshouse temperatures for 1 week. Nonvernalized control plants were grown for 2 weeks at normal glasshouse temperatures to reach a developmental stage equivalent to the vernalized plants at the time of harvest. Plants were harvested at the middle of the light period for each treatment. For cold treatment, plants were harvested directly from cold growth chamber (4°C, 12 h light/12 h dark).
Library Screening
cDNA was synthesized from RNA isolated from vegetative tissues of a winter barley (Hordeum vulgare cv Igri). cDNA was packaged into the Stratagene λ-ZAP phagemid vector (Sydney) and screened at high or low hybridization stringency with probes from the MADS-box domain of the FLOWERING LOCUS C (FLC) gene of Arabidopsis (Arabidopsis thaliana; GenBank NM121052) according to established methods (Ausubel et al., 1994). Sequence alignments were generated using the ClustalW program (Chenna et al., 2003).
RNA Extraction and Gel-Blot Analysis
Total RNA was extracted using the method of Chang et al. (1993). Twenty micrograms of RNA was separated in formaldehyde gels and blotted to Hybond-N membrane (Amersham Biosciences) as described in Ausubel et al. (1994). RNA blots were hybridized with riboprobes and washed using the protocol of Dolferus et al. (1994). Riboprobes were synthesized using the Promega T7 transcription kit. Blots were visualized in a phosphoimager for varying lengths of time, dependant on signal intensity. The following primers were used to amplify gene-specific riboprobe fragments: BM1, 5′-ACTGCAGGCGTCCAAGATG-3′ and 5′-CCACACTATGCTATATATTCCC-3′; BM10, 5′-GGAAGTAACAGTAGTTGTC-3′ and 5′-ATACCAACAAGGTACATA-3′; HvVRT2, 5′-TCATCTGACTCTGTGATGACGG-3′ and 5′-AAGACAGTTACTGAATACCCAGGC-3′; HvVRN1, 5′-GGTGAGCCACATCAACGGCTGA-3′ and 5′-TTACATGGTAGATTACTCGTACAGCC-3′. In each case, the T7 polymerase promoter was incorporated into the reverse primer sequence.
RT-PCR and Real-Time PCR Analysis
Quantitative RT-PCR (qRT-PCR) was performed on a Rotor-Gene 2000 Real-Time Cycler (Corbett Research). Products were sequenced to ensure that products were gene specific. The ACTIN gene was used as a housekeeping control to correct for uneven amounts of sample and control cDNA templates. Relative expression levels of genes of interest were compared between cDNA samples using the Comparative Quantification analysis method (Rotogene-5 software, Corbett Research). This method uses information about the start of the exponential phase of amplification (take-off point) and the average amplification efficiency of each primer set to enable direct concentration comparison between different samples generating a relative concentration. Quantification for each primer set and cDNA template combination was performed in triplicate and included a no-template control to ensure results were not influenced by primer-dimer formation or DNA contamination. In all cases, primers specifically amplify cDNA and do not amplify genomic DNA products. Three technical repeats were performed on each cDNA sample. Two biological repeats were assayed, giving similar trends. Data from one biological repeat is presented. The following primers were used: BM1, 5′-AGAGGAGAACGCAAGGCTAAAGG-3′ and 5′-AGTTGAAGAGTGATAATCCGAGCCTGAG-3′; BM10, 5′-GCTCATCGTCTTCTCCTCCAC-3′ and 5′-CTCCTCGCCTCTCATCTGTC-3′; HvVRT2, 5′-AAGCTCTCCCAGTTCGCCAGCTCC-3′ and 5′-TTAGTCCGTCAAGTTCCTCACC-3′; HvVRN1, 5′-GCATAAGTTGGTTCTTCCTGGCTCTG-3′ and 5′-GCCTCATCATCTTCTCCACCAA-3′; HvFT, 5′-GCGACCCCAACCTTAGAGAG-3′ and 5′-CTCGGCAAAGTCCCTGGTG-3′. ACTIN primers have been described previously (Trevaskis et al., 2006).
Plant Transformation
For Arabidopsis transformation, overexpression constructs were made by fusing cDNA clones of BM1 and BM10 to the cauliflower mosaic virus 35S promoter, and the resulting overexpression cassettes were ligated into the pART27 binary vector (Gleave, 1992). Arabidopsis plants were transformed with the floral dip method (Clough and Bent, 1998). T1 and T2 generations were screened by PCR using gene-specific primers (same primer sets as for qRT-PCR). For barley transformation, overexpression constructs were made by fusing cDNA clones of BM1, BM10, or VRN1 (GenBank BE431095) to the maize (Zea mays) ubiquitin promoter (Christensen et al., 1992), and the resulting overexpression cassettes were ligated into the pWBVEC8 binary vector (Wang et al., 1998). RNAi constructs were made by amplifying fragments designed to specifically silence BM1 (5′-AGGGGAGGAGAGGATGGC-3′ and 5′-GCTTCGGGTAATTTCCAGCC-3′), BM10 (5′-GGCAAGCTAGTGGTTGTTGATACCG-3′and 5′-ACGGGTCCTAGCAAATGCCAAACAG-3′), or HvVRN1 (5′-TGGATCCGGTACCGCAACAAGATCAGACTCAGCC-3′ and 5′-TCTAGATATCAAATTCACATAAACAACATCC-3′), which were inserted into the pSTARLING vector (http://www.pi.csiro.au/rnai/vectors.htm). The resulting hairpin cassettes were then ligated into the pWBVEC8 binary backbone. Barley plants were transformed using Agrobacterium transformation of excised embryos (Tingay et al., 1997; Mathews et al., 2001). An average of 50 independent transformants was produced for each construct. T1, T2, and T3 plants were screened for segregation of transgenes using primers that amplify from the hygromycin selectable marker gene (5′-AAAAGCCTGAACTCACCGC-3′ and 5′-TCGTCCATCACAGTTTGCC-3′). Expression analysis was carried out on plants homozygous for the transgene and sibling null control lines, which did not inherit the transgene. For each construct, expression analysis was carried out on at least two independently transformed lines that had phenotypes typical of the majority of T1 transformants.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number EF043040.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. An alignment of amino acid sequences encoded by SVP-like genes from barley and Arabidopsis.
Supplemental Figure S2. BM1, BM10, HvVRT2, and HvVRN1 transcript levels in different organs of barley.
Supplemental Figure S3. Apex development in the spring variety Golden Promise.
Supplemental Figure S4. Cold induction of HvVRT2 and HvVRN1.
Supplemental Figure S5. HvVRN1 expression levels do not influence expression levels of BM1, BM10, or HvVRT2.
Supplemental Figure S6. Effective silencing of BM1 and BM10 by RNAi constructs.
Supplemental Figure S7. Days to head emergence in transgenic barley plants.
Supplemental Figure S8. Ectopic expression of BM1 or BM10.
Supplementary Material
Acknowledgments
We thank Sandra Stops for her excellent technical assistance, Rod King for his help training B. Trevaskis to dissect apices from barley, and Rod King, Jake Jacobsen, John Watson, Masumi Robertson, and Jean Finnegan for reading draft versions of the manuscript and providing constructive criticism.
This work was supported by the Commonwealth Scientific and Industrial Research Organization (CSIRO; postdoctoral fellowship to B.T.), and by Graingene, a research alliance between CSIRO, Grains Research and Development Corporation, Australian Wheat Board Limited, and Syngenta Seeds (to M.T.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Liz Dennis (liz.dennis@csiro.au).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Andersen JR, Jensen LB, Asp T, Lubberstedt T (2006) Vernalization response in perennial ryegrass (Lolium perenne L.) involves orthologues of diploid wheat (Triticum monococcum) VRN1 and rice (Oryza sativa) HD1. Plant Mol Biol 60 481–494 [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brenton R, Kingston RE, Moore DD, Siedman JG, Smyth JA, Struhl K (1994) Current Protocols in Molecular Biology. Wiley and Sons, New York
- Boyd WJR, Li CD, Grime CR, Cakir M, Potipibool S, Kaveeta L, Men S, Jalal Kamali MR, Barr AR, Moody DB, et al (2003) Conventional and molecular genetic analysis of factors contributing to variation in the timing of heading among spring barley (Hordeum vulgare L.) genotypes grown over a mild winter growing season. Aust J Agric Res 54 1277–1301 [Google Scholar]
- Brill EM, Watson JM (2004) Ectopic expression of a Eucalyptus grandis SVP orthologue alters the flowering time of Arabidopsis thaliana. Funct Plant Biol 31 217–224 [DOI] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney K (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Report 11 113–116 [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18 675–689 [DOI] [PubMed] [Google Scholar]
- Ciannamea S, Kaufmann K, Frau M, Nougalli Tonaco IA, Petersen K, Nielsen KK, Angenent GC, Immink RGH (2006) Protein interactions of MADS box transcription factors involved in flowering in Lolium perenne. J Exp Bot 57 3419–3431 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735–743 [DOI] [PubMed] [Google Scholar]
- Danyluk J, Kane NA, Breton G, Limin AE, Fowler DB, Sarhan F (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol 132 1849–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolferus R, Jacobs M, Peacock WJ, Dennis ES (1994) Differential interactions of promoter elements in the stress response of the Arabidopsis alcohol dehydrogenase gene. Plant Physiol 105 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D, Szücs P, Yan L, Helguera M, Skinner JS, Von Zitzewitz J, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273 54–65 [DOI] [PubMed] [Google Scholar]
- Gleave AP (1992) A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20 1203–1207 [DOI] [PubMed] [Google Scholar]
- Gregis V, Sessa A, Colombo L, Kater MM (2006) AGL24, SHORT VEGETATIVE PHASE, and APETALA1 redundantly control AGAMOUS during early stages of flower development in Arabidopsis. Plant Cell 18 1372–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann U, Hohmann S, Nettesheim K, Wisman E, Saedler H, Huijser P (2000) Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J 21 351–360 [DOI] [PubMed] [Google Scholar]
- Kane NA, Danyluk J, Tardif G, Ouellet F, Laliberte JF, Limin AE, Fowler DB, Sarhan F (2005) TaVRT-2, a member of the StMADS-11 clade of flowering repressors, is regulated by vernalization and photoperiod in wheat. Plant Physiol 138 2354–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiero S, Li MA, Will I, Hartmann U, Saedler H, Huijser P, Schwarz-Sommer Z, Sommer H (2004) INCOMPOSITA: a MADS-box gene controlling prophyll development and floral meristem identity in Antirrhinum. Development 131 5981–5990 [DOI] [PubMed] [Google Scholar]
- Mathews PR, Wang MB, Waterhouse PM, Thornton S, Fieg S, Gubler F, Jacobsen JV (2001) Marker gene elimination from transgenic barley, using co-transformation with adjacent “twin T-DNAs” on a standard Agrobacterium transformation vector. Mol Breed 7 195–202 [Google Scholar]
- Michaels SD, Ditta G, Gustafson-Brown C, Pelaz S, Yanofsky M, Amasino RM (2003) AGL24 acts as a promoter of flowering in Arabidopsis and is positively regulated by vernalization. Plant J 33 867–874 [DOI] [PubMed] [Google Scholar]
- Okamuro JK, Szeto W, Lotys-Prass C, Jofuku KD (1997) Photo and hormonal control of meristem identity in the Arabidopsis flower mutants apetala2 and apetala1. Plant Cell 9 37–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen K, Kolmos E, Folling M, Salchert K, Storgaard M, Jensen CS, Didion T, Nielsen KK (2006) Two MADS-box genes from perennial ryegrass are regulated by vernalization and involved in the floral transition. Physiol Plant 126 268–278 [Google Scholar]
- Poethig RS (1990) Phase change and the regulation of shoot morphogenesis in plants. Science 250 923–930 [DOI] [PubMed] [Google Scholar]
- Pugsley AT (1971) A genetic analysis of the spring-winter habit of growth in wheat. Aust J Agric Res 22 21–31 [Google Scholar]
- Schmitz J, Franzen R, Ngyuen TH, Garcia-Maroto F, Pozzi C, Salamini F, Rohde W (2000) Cloning, mapping and expression analysis of barley MADS-box genes. Plant Mol Biol 42 899–913 [DOI] [PubMed] [Google Scholar]
- Sentoku N, Kato H, Kitano H, Imai R (2005) OsMADS22, an STMADS11-like MADS-box gene of rice, is expressed in non-vegetative tissues and its ectopic expression induces spikelet meristem indeterminacy. Mol Genet Genomics 273 1–9 [DOI] [PubMed] [Google Scholar]
- Szymkowiak EJ, Irish EE (2006) JOINTLESS suppresses sympodial identity in inflorescence meristems of tomato. Planta 223 646–658 [DOI] [PubMed] [Google Scholar]
- Tingay S, McElroy E, Kalla R, Fieg S, Wang M, Thornton S, Brettell R (1997) Agrobacterium mediated barley transformation. Plant J 11 1369–1376 [Google Scholar]
- Tooke F, Ordidge M, Chiurugwi T, Battey N (2005) Mechanisms and function of flower and inflorescence reversion. J Exp Bot 56 2587–2599 [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Bagnall DJ, Ellis MH, Peacock WJ, Dennis ES (2003) MADS box genes control vernalization-induced flowering in cereals. Proc Natl Acad Sci USA 100 13099–13104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevaskis B, Hemming MN, Peacock WJ, Dennis ES (2006) HvVRN2 responds to daylength, whereas HvVRN1 is regulated by vernalization and developmental status. Plant Physiol 140 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A, Beales J, Faure S, Dunford RP, Laurie DA (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310 1031–1034 [DOI] [PubMed] [Google Scholar]
- Wang MB, Li Z, Mathews PR, Upadyaya NM, Waterhouse PM (1998) Improved vectors for Agrobacterium tumefaciens-mediated transformation of monocot plants. Acta Horta 461 401–407 [Google Scholar]
- Yan L, Loukoianov A, Blechl A, Tranquilli G, Ramakrishna W, SanMiguel P, Bennetzen JL, Echenique V, Dubcovsky J (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303 1640–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L, Loukoianov A, Tranquilli G, Helguera M, Fahima T, Dubcovsky J (2003) Positional cloning of the wheat vernalization gene VRN1. Proc Natl Acad Sci USA 100 6263–6268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Ito T, Wellmer F, Meyerowitz EM (2004) Repression of AGAMOUS-LIKE 24 is a crucial step in promoting flower development. Nat Genet 36 157–161 [DOI] [PubMed] [Google Scholar]
- Yu H, Xu Y, Tan EL, Kumar PP (2002) AGAMOUS-LIKE 24, a dosage-dependent mediator of the flowering signals. Proc Natl Acad Sci USA 99 16336–16341 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.