Abstract
Seventeen loci encode proteins of the preprotein and amino acid transporter family in Arabidopsis (Arabidopsis thaliana). Some of these genes have arisen from recent duplications and are not in annotated duplicated regions of the Arabidopsis genome. In comparison to a number of other eukaryotic organisms, this family of proteins has greatly expanded in plants, with 24 loci in rice (Oryza sativa). Most of the Arabidopsis and rice genes are orthologous, indicating expansion of this family before monocot and dicot divergence. In vitro protein uptake assays, in vivo green fluorescent protein tagging, and immunological analyses of selected proteins determined either mitochondrial or plastidic localization for 10 and six proteins, respectively. The protein encoded by At5g24650 is targeted to both mitochondria and chloroplasts and, to our knowledge, is the first membrane protein reported to be targeted to mitochondria and chloroplasts. Three genes encoded translocase of the inner mitochondrial membrane (TIM)17-like proteins, three TIM23-like proteins, and three outer envelope protein16-like proteins in Arabidopsis. The identity of Arabidopsis TIM22-like proteins is most likely a protein encoded by At3g10110/At1g18320, based on phylogenetic analysis, subcellular localization, and complementation of a yeast (Saccharomyces cerevisiae) mutant and coexpression analysis. The lack of a preprotein and amino acid transporter domain in some proteins, localization in mitochondria, plastids, or both, variation in gene structure, and the differences in expression profiles indicate that the function of this family has diverged in plants beyond roles in protein translocation.
Mitochondria and chloroplasts are estimated to contain 2,000 and 4,000 proteins, respectively (van Wijk, 2004; Millar et al., 2005). Despite the fact that both organelles contain a genome, the majority of proteins located in these organelles are encoded in the nucleus, synthesized in the cytosol, and imported into the respective organelle (Soll and Schleiff, 2004; Bedard and Jarvis, 2005; Lister et al., 2005). Although nonspecific import of some chloroplast proteins into mitochondria has been observed with in vitro protein uptake assays (Cleary et al., 2002; Rudhe et al., 2002; Chew et al., 2003a, 2003b), high fidelity of protein targeting appears to be maintained in vivo. The means by which this targeting specificity is maintained is unknown, but the presence of cytosolic targeting factors, targeting of mRNA, differences in predicted secondary structure of targeting signals, phosphorylation sites in targeting signals, and ability of organellar protein import systems to recognize precursor proteins all likely combine to achieve import specificity (Chew and Whelan, 2004).
The translocase of the outer mitochondrial membrane (TOM) complex transports mitochondrial proteins across the outer membrane to interact with either the translocase of the inner mitochondrial membrane (TIM)17, 23 complex or the TIM22 complex, depending on whether the protein is imported via the general or carrier import pathway, respectively (Neupert, 1997; Pfanner and Geissler, 2001). To date, the only characterized receptor present in plant mitochondria is TOM20 (Heins and Schmitz, 1996; Jansch et al., 1998) in contrast to several receptor subunits characterized in fungal and mammalian systems (Neupert, 1997; Pfanner and Geissler, 2001; Stojanovski et al., 2003). Translocase of the outer envelope of chloroplasts (TOC) and translocase of the inner envelope of chloroplasts (TIC) facilitate the import of chloroplast precursor proteins. TOC34, TOC64, and TOC159 have been characterized as the primary receptors for plastids (Soll and Schleiff, 2004; Bedard and Jarvis, 2005). Precursor proteins are translocated across the outer membranes of mitochondria and plastids via the β-barrel proteins TOM40 and TOC75, respectively, to interact with the TIM and TIC apparatus (Ahting et al., 2001; Hinnah et al., 2002). The pore-forming subunits of the TIMs are 23 and 22 (Bauer et al., 2000; Rehling et al., 2004), whereas the pore-forming subunit of TIC is thought to be TIC110, but other subunits are likely to be involved (Soll and Schleiff, 2004; Bedard and Jarvis, 2005). Although the primary recognition step is performed by outer membrane receptors, the inner membrane complexes also contain specific binding sites to achieve import. Thus, the vectorial movement of proteins from the cytosol to the matrix of mitochondria is proposed to follow the binding-chain hypothesis, where binding sites exist on both the TOM and TIM complexes (Rehling et al., 2001; Chacinska et al., 2005).
As TOM, TIM, TOC, and TIC play an essential role in protein translocation and maintaining import specificity, it is somewhat surprising that they may contain subunits derived from a common ancestor. The chloroplast receptor TOC64 and an outer mitochondrial membrane protein of 64 kD display 75% protein sequence similarity (Chew et al., 2004). Likewise, the outer envelope protein (OEP)16 in the outer envelope of chloroplasts belongs to the preprotein and amino acid transporter (PRAT) family of proteins (Rassow et al., 1999). The role of this protein is somewhat unclear; it has been shown that OEP16, localized to the outer envelope of pea (Pisum sativum) chloroplasts, forms a channel pore, selective for amino acids (Pohlmeyer et al., 1997). In contrast, it has been proposed that OEP16 from barley (Hordeum vulgare) acts as a precursor translocase for protochlorophyllide oxidoreductase A (POR; Reinbothe et al., 2004a, 2004b).
Examination of Arabidopsis (Arabidopsis thaliana) genome annotations reveals 17 genes that are annotated as either TIM17, 22, 23, or OEP16-like. Experimental data to define subcellular location of the encoded proteins exists for seven of these 17 proteins: AtTIM17-2 (At2g37410), AtTIM23-2 (At1g72750), At2g42210, At2g28900, At3g49560, At5g24650, and At5g55510, all identified in proteomic analyses (see Table I). We carried out in vitro and in vivo targeting studies to define the subcellular location of all PRAT protein in Arabidopsis. Complementation of the yeast (Saccharomyces cerevisiae) tim22 mutant was carried out to define this protein in plants. Immunological analysis was carried out to confirm dual targeting of a PRAT protein to mitochondria and chloroplasts. Analyses of the transcript abundance of this family of genes in comparison to genes encoding components of mitochondrial and chloroplastic protein import apparatus revealed organ and developmental regulation.
Table I.
Summary of data on expression and subcellular localization of the PRAT gene family and encoded proteins
The locus and gene model are taken from TAIR 6. The number of amino acids is taken from sequenced cDNA in our laboratory. The presence of a PRAT domain is indicated. *, Lacks one amino acid of this domain or spacing differs from that defined in Rassow et al. (1999); P, Molecular mass predicted; A, molecular mass apparent as determined by SDS-PAGE. Location defined by in vitro and in vivo protein uptake assays, by antibody decoration (AB), and proteome analyses: M, Mitochondria; C, chloroplast; T, tonoplast; PM, plasma membrane; NT, no targeting observed. Cluster in which each gene was found in leaf development: C1/C, defined chloroplast components; C2/-M and C3/M, defined mitochondrial components. Aramemnon, Consensus number of transmembrane regions from nine predictors and the predicted location using the Aramemnon database (http://aramemnon.botanik.uni-koeln.de); with the predicted location, either the consensus was given or the only predictor is listed: M, Mitochondria; C, chloroplast; S, secreted; ?, no prediction; NM, not a membrane protein (Schwacke et al., 2003). References indicate detection of components in previous proteomic analyses. At1g18320 and At3g10110 are identical and are listed together. At4g16160 was not expressed during leaf development. ND, Not determined.
| Locus | Name | Gene Model | No. Amino Acid | Molecular Mass P versus A | PRAT | In Vitro | In Vivo | AB | Leaf | Proteome | Aramemnon | References |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| At1g17530 | AtTIM23-1 | 1 | 187 | 19/18 | +* | M | M | C3/M | 3 ? | |||
| At1g18320 | AtTIM22 | 1 + 1 | 173 | 18/18 | + | M | M | ND | 3 C or S | |||
| At3g10110 | ||||||||||||
| At1g20350 | AtTIM17-1 | 1 | 218 | 23/27 | + | M | M | C3/M | NM | |||
| At1g72750 | AtTIM23-2 | 2 | 188 | 20/20 | +* | M | M | C3/M | M | 3 ? | Murcha et al. (2003); Heazlewood et al. (2004); Lister et al. (2004) | |
| At2g28900 | OEP16 like | 1 | 148 | 16/16 | + | C | C | C1/C | C | 2 C | Pohlmeyer et al. (1997); Ferro et al. (2003); Froehlich et al. (2003) | |
| At2g37410 | AtTIM17-2 | 2 | 243 | 25/31 | + | M | M | C3/M | M | 3 C | Murcha et al. (2003, 2005); Brugiere et al. (2004); Lister et al. (2004) | |
| At2g42210 | 4 | 159 | 17/18 | + | M | M | C2/M | M | 4 ? | Heazlewood et al. (2004) | ||
| At3g04800 | AtTIM23-3 | 1 | 188 | 20/20 | + | M | M | C3/M | 4 M | |||
| At3g25120 | 1 | 189 | 20/20 | − | M | M | C2/M | 4 S | ||||
| At3g49560 | 1 | 261 | 28/31 | − | M + C | C | C | C1/C | C | NM | Ferro et al. (2003); Froehlich et al. (2003) | |
| At3g62880 | OEP16 like | 2 | 136 | 14/16 | +* | NT | C | C3/M | 3 C | |||
| At4g16160 | OEP16 like | 2 | 176 | 18/20 | +* | C | C | ND | 3 C | |||
| At4g26670 | 1 | 210 | 22/24 | − | NT | NT | C | C1/C | 4 C | |||
| At5g11690 | AtTIM17-3 | 1 | 133 | 14/20 | + | M | M | C2/M | 3 M | |||
| At5g24650 | 1 | 259 | 28/31 | − | M + C | M | M + C | C2M | C, T, PM | 4 M | Ferro et al. (2003); Froehlich et al. (2003); Marmagne et al. (2004); Szponarski et al. (2004) | |
| At5g55510 | 1 | 214 | 20/22 | − | NT | C | C2/M | C | NM | Ferro et al. (2003) |
RESULTS
The PRAT Gene Family
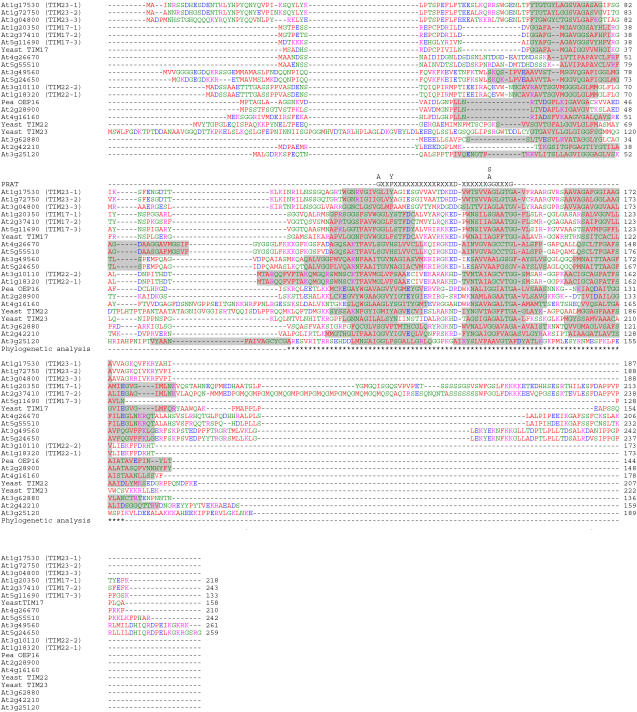
A query of the most recent genome annotation for Arabidopsis searched with the text term TIM17 yields 17 distinct loci (The Arabidopsis Information Resource [TAIR] 6; http://www.arabidopsis.org). This represents a large increase in family size in comparison to yeast, where single genes encode the TIM17, 22, and 23 proteins that define this family. The predicted proteins range in size from 133 in TIM17-3 (At5g11690) to 261 amino acids in the protein encoded by At3g49560 (Table I; Fig. 1). Transmembrane-spanning regions were analyzed using the Dense Alignment Surface transmembrane predictor (Cserzo et al., 1997) and compared to previously defined transmembrane regions in yeast TIM17, 22, and 23 (Rassow et al., 1999) and the proposed transmembrane structure of pea OEP16 (Linke et al., 2004). Although this family of proteins is defined as having four transmembrane regions, in the Aramemnon database four transmembrane regions were only predicted for five of the 17 proteins using the consensus prediction of nine predictors. Some of these proteins were not even defined as membrane proteins (Schwacke et al., 2003). However, because many predictors also fail to predict four transmembrane regions in the corresponding yeast proteins and, even for OEP16, where detailed investigation strongly supports four transmembrane regions, only two or three are predicted; thus, it is likely that more than five of the proteins have four transmembrane regions. A PRAT domain, previously used to define this family, was present in seven of the 17 Arabidopsis predicted proteins, whereas four contained a degenerate PRAT domain, with one additional amino acid spacing or with one of the consensus amino acids missing (Fig. 1; Table I; Rassow et al., 1999). An examination of the rice (Oryza sativa) genome yields a similar picture, with 24 genes encoding proteins in this family (Supplemental Fig. S1).
Figure 1.
Multiple sequence alignment of the predicted proteins of the PRAT family. Proteins were aligned using ClustalX with gaps introduced to maximize alignment and the TIM17, 22, and 23 proteins from yeast and OEP16 from pea included as a comparison. Predicted transmembrane regions are highlighted in gray; the PRAT domain is indicated above the sequence where X indicates any amino acid. The positions used for phylogenetic analysis are marked with asterisks below the sequences. Residues are colored according to properties: red, hydrophobic (AVFPMILW); blue, acidic (DE); magenta, basic (RHK); green, hydrophilic (STYCNGQ).
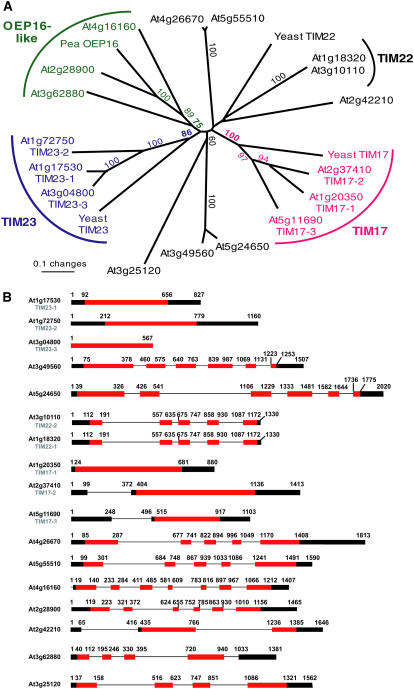
Phylogenetic analysis of the predicted Arabidopsis proteins with yeast TIM17, 22, 23, and OEP16 from pea is shown in Figure 2A. Three genes previously designated as encoding TIM17 (AtTIM17-1 [At1g20350], AtTIM17-2 [At2g37410], and TIM17-3 [At5g11690]) form a distinct group, which is close to yeast TIM17, and range in size from 133 amino acids in TIM17-3 (At5g11690) to 243 amino acids in TIM17-2 (At2g37410; Murcha et al., 2003). A clearly distinct grouping, previously designated TIM23, can also be observed with yeast TIM23 (AtTIM23-1 [At1g17530], AtTIM23-2 [At1g72750], and TIM23-3 [At3g04800]; Murcha et al., 2003). Three predicted proteins (At2g28900, At3g62880, and At4g16160) branch with pea OEP16, with sequence identity varying from 21% to 52% (Fig. 2A; Supplemental Figs. S2 and S3). These three groupings are maintained if the rice sequences are included (Supplemental Fig. S2). The situation with TIM22 is not as clear; if only proteins of Arabidopsis are analyzed, TIM22 of yeast branches with three proteins, namely, At1g18320, At3g10110, and At2g42210, but this grouping is not supported by bootstrapping (Fig. 2A). If the 24-member family of proteins from rice is included in the analysis, yeast TIM22 clusters with the two proteins encoded by At1g18320 and At3g10110 and a rice protein (Os03g18500), with some bootstrap support (Supplemental Fig. S2). Therefore, in Arabidopsis, TIM22 is probably represented by At1g18320 and At3g10110. It becomes apparent by the poor bootstrap support of the basal nodes of both trees that the relationship of the three defined groups mentioned above and the residual proteins cannot be resolved.
Figure 2.
Phylogenetic analysis and gene structure of genes encoding PRAT proteins. A, Neighbor-joining tree of the Arabidopsis PRAT protein family with yeast TIM17, 22, 23, and pea OEP16 included for comparison. The tree was constructed using 82 amino acid positions in the region of transmembrane helices 2 to 4 as marked in Figure 1. Only bootstrap values above 50% are shown. B, Gene structure of the various genes belonging to the TIM17, 22, and 23 family of proteins in TAIR 6. [See online article for color version of this figure.]
Some genes display very high protein sequence identity, indicating they have likely arisen from recent duplications. At1g18320 and At3g10110 encode predicted proteins with 100% identity, but are not located on segments of chromosomes previously annotated to be duplicated (Arabidopsis Genome Initiative, 2000). The coding, 5′-, and 3′-untranslated regions are identical and are approximately 500 bp upstream of the annotated transcriptional start site. This high level of nucleic acid sequence identity is maintained for approximately 2 kb, until At3g10113 (downstream of At3g10110) and At1g18330 (downstream of At1g18320; data not shown). At4g26670 and At5g55510 encode predicted proteins that display 79% sequence identity and At3g49560 and At5g24650 encode predicted proteins that display 83% sequence identity. The former pair, as well as At1g17350 (TIM23-1) and At1g72750 (TIM23-2), which display 83% protein identity, are located in previously annotated duplicated regions of the Arabidopsis genome (Arabidopsis Genome Initiative, 2000). Analysis of the gene structures reveals that the genes defined as TIM17 or TIM23 contain no introns in the coding regions, but, for the other members, the number of introns varies from one in the coding region (At2g42210) to six (At4g16160), leading to quite variable gene structures (Fig. 2B).
To determine whether the expansion of this family of proteins is restricted to Arabidopsis, plants, or has taken place in other eukaryotic lineages, we examined the number of genes encoding mitochondrial protein import components in a variety of organisms. A bioinformatic approach was undertaken to identify homologs to the yeast protein import machinery in Arabidopsis, rice, Neurospora crassa, Homo sapiens, Mus musculus, Rattus norvegicus, Drosophila melanogaster, and Caenorhabditis elegans. The results were entered into a relational database that can be accessed at http://www.plantenergy.uwa.edu.au/MPRIC. Homologs to the majority of yeast import components were identified in all organisms, with the exception of TOM5 (found in yeast, Arabidopsis, and rice), TOM6 (found in yeast, Arabidopsis, and N. crassa), MDM10/TOB38/TIM12/HOT13p/SOM1/MBA1 (found only in yeast), TOM34 (found only in animals and N. crassa), TOM70/TOM72 (not found in plants), TIM18 and TIM54 (found only in yeast and N. crassa), PAM17 (found only in yeast, N. crassa, and C. elegans), and IMP1/IMP2 (not found in N. crassa). Whereas the genomes of yeast and N. crassa encode only a single isoform of each component (except for ERV1 and OXA1 in N. crassa, and mtHSP70 in yeast and N. crassa), plant and animal genomes often encode the import components in small multiple-gene families. This is particularly evident for TOM7/TOM20/TOM40/PAM18 (plants and animals), TIM8/TIM9 (animals), TIM17 (plants and animals), and TIM22/TIM23 (plants). Of particular interest is the large size of the TIM17/TIM22/TIM23 family in both Arabidopsis and rice and the relatively large number of genes encoding members of this family that cannot be assigned or whose function is unknown, indicating that these gene families have undergone extensive expansion in plants. Examination of the PRAT family of proteins in rice indicates that there are orthologous genes for all those found in Arabidopsis, indicating that this family of proteins diverged before the split of the monocot/dicot lineages (http://www.plantenergy.uwa.edu.au/MPRIC; Supplemental Figs. S1 and S2). The number of genes in each grouping differs, indicating duplication after lineage divergence, but strongly suggesting that the large size of this family of proteins is a general feature of angiosperms. Rice genes encoded by Os03g30220, Os03g30230, and Os04g30740 do not appear to have orthologs in Arabidopsis. Examination of the protein alignment (Supplemental Fig. S1) indicates that they are not highly conserved in the region containing the PRAT domain and thus they may represent pseudogenes or incorrect annotations.
Subcellular Localization of PRAT Proteins
Because proteins encoded by different members of this family have been reported in chloroplasts and mitochondria (Table I), we determined the subcellular localization of all the encoded proteins in vitro using protein uptake assays into isolated mitochondria and chloroplasts, and in vivo using green fluorescent protein (GFP) tagging. As controls, we used the small subunit of Rubisco (rbcS) for in vitro chloroplast import (Anderson and Smith, 1986) and the maize (Zea mays) phosphate translocator (ZmPic) and adenine nucleotide translocator (ZmANT) for in vitro mitochondrial import (Bathgate et al., 1989; Winning et al., 1992) because this family of proteins is imported via the carrier import pathway (Neupert, 1997; Pfanner and Geissler, 2001; Murcha et al., 2005). For in vivo GFP localization, we used the red fluorescent protein (RFP) fused to the targeting signal for mitochondrial alternative oxidase (Aox; Aox-RFP) or fused to the targeting signal of rbcS (rbcS-RFP). Arabidopsis suspension cells were transiently transformed with a PRAT protein fused to GFP and Aox-RFP or rbcS-RFP. Fluorescence was collected at the appropriate wavelength and images were merged to determine the location of the PRAT-GFP fusion protein.
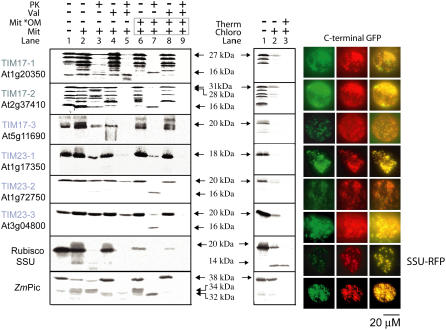
Mitochondrial localization was defined for the three proteins belonging to the TIM17 and TIM23 families using both in vitro uptake assays and in vivo GFP localization of proteins (Fig. 3). In vitro incubation of radiolabeled proteins with purified mitochondria resulted in a protease-protected band (Fig. 3, lane 3). This band was either abolished or diminished in intensity if the ionophore valinomycin was added to the import reaction prior to commencement (Fig. 3, lane 5). In yeast, TIM17, 22, and 23 are imported via the carrier import pathway, where stage II or IIIa intermediates are bound to the import receptor or translocated through the TOM40 pore, respectively. Such intermediates are resistant to protease digestion even in the absence of a membrane potential because they have not yet come into interaction with the inner membrane. Thus, to verify that the protease-protected protein is inserted into the inner membrane, we ruptured the outer membrane to allow access of added protease to the inner membrane. Inner membrane proteins are still protected from added protease, or produce inner membrane-protected fragments, whereas import intermediates are digested (Pfanner and Geissler, 2001; Truscott et al., 2002, 2003). Treatment of outer membrane-ruptured mitochondria with protease resulted in a protease-protected band that was absent if valinomycin was present in the import assay (Fig. 3, lanes 7 versus 9). The exception was TIM17-3 (At5g11690), where a protease-protected band was only observed with intact mitochondria (Fig. 3, lane 3 versus lane 7). Because this protein contains only 133 amino acids, it is possible that the lack of protection in outer membrane-ruptured mitochondria is due to the fact that it does not have the four transmembrane-spanning regions typical of this family; the protein ends in the middle of the region where the fourth transmembrane region is predicted for yeast TIM17 (Fig. 1). Mitochondrial localization was confirmed for all TIM17 and TIM23 proteins because linking GFP to the C terminus yielded a mitochondrial pattern as determined by colocalization of the GFP signal with Aox-RFP. Some differences were observed with the GFP signal; whereas TIM17-3 (At5g11690) yielded a typical mitochondrial pattern of numerous small particles, the GFP pattern in other constructs, although concentrated in small particles, was more intense and appeared to encompass a greater surface area of the cell. This may be due to the fact that the GFP moiety may, in some cases, still remain outside the mitochondrion; as for TIM17-2 (At2g37410), the C terminus portion is in the outer membrane accessible by externally added protease (Murcha et al., 2005). Furthermore, the ability of GFP to dimerize may contribute to this altered mitochondrial pattern. However, in all cases, the GFP pattern overlapped with the Aox-RFP pattern. Furthermore, there was no evidence of import into purified chloroplasts, there were no protease-protected products detected, and binding appeared to be very low (Fig. 3).
Figure 3.
In vitro and in vivo subcellular localizations of Arabidopsis TIM17 and TIM23 proteins. The ability to import proteins in vitro was assessed by incubating radiolabeled precursor proteins with mitochondria or chloroplasts under conditions that support protein uptake into each organelle. The uptake of rbcS was used as a positive control for chloroplast import and the uptake of the phosphate translocator from maize (ZmPic) was used as a positive control for mitochondrial import. Attaching GFP in frame to the C-terminal end of the protein and transformation of suspension cells by biolistic bombardment and visualization by fluoresence microscopy assessed in vivo targeting ability. The targeting signals of Aox and rbcS attached to the RFP (Aox-RFP and rbcS-RFP) were used as controls for mitochondria and plastids, respectively. For mitochondrial import, in vitro import into mitochondria was followed by rupture of the outer membrane to verify insertion into the inner membrane. Addition of compounds to the import assay are indicated; the presence of Mit and Mit*OM (lanes 6–9; boxed) indicates that mitochondria were ruptured after the import assay, but prior to protease treatment. For chloroplast import, precursors were incubated with purified chloroplast followed by thermolysin treatment as indicated. Unless otherwise indicated, RFP (middle) is the pattern obtained with Aox-RFP. Mit, Mitochondria; Mit*OM, outer membrane-ruptured mitochondria; Val, valinomycin; PK, proteinase K; Chloro, chloroplasts; Therm, thermolysin. Sizes are indicated as apparent molecular mass in kilodaltons.
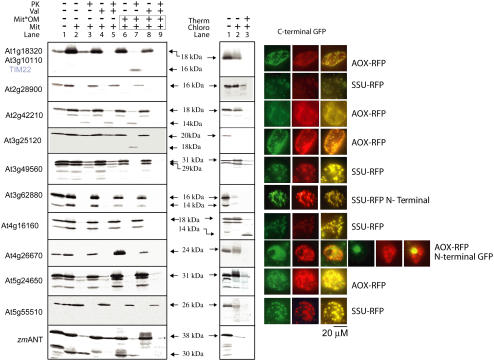
Similar analysis was carried out with the proteins encoded by the 11 remaining genes (Fig. 4). Because At1g18320 and At3g10110 are 100% identical, only one cDNA was used in the analysis. Mitochondrial localization was defined for the proteins encoded by At1g18320/At3g10110, At2g42210, and At3g25120. All three proteins yielded a typical mitochondrial pattern when tagged with GFP and there was no protease protection upon incubation with chloroplasts. For At1g18320/At3g10110 and At3g25120, it was concluded that the proteins were located in the mitochondrial inner membrane, based on the fact that a distinct inner membrane protease-protected band was observed with outer membrane-ruptured mitochondria (Fig. 4, lane 7). At2g42210 displayed some unusual features upon import into mitochondria. Incubation of the 18-kD protein with mitochondria and protease treatment produced an additional product with an apparent molecular mass of 14 kD (Fig. 4, lane 3). An identical pattern was observed when valinomycin was present in the import assay and when the outer membrane was removed before protease treatment (Fig. 4, lanes 4–7). Notably, the addition of valinomycin and rupture of the outer membrane prior to protease treatment resulted in complete digestion (Fig. 4, lanes 8 and 9). Thus, the similarity in the protected fragments in the presence and absence of valinomycin may be due to the fact that, in the presence of valinomycin, insertion into the outer membrane results in the same region being protected as insertion into the inner membrane in the absence of valinomycin.
Figure 4.
In vitro and in vivo subcellular localizations of unknown Arabidopsis PRAT proteins. N terminus indicates that GFP was fused in frame to the N-terminal end of the protein. The adenine nucleotide translocator from maize was used as a control for mitochondrial import.
The OEP16-like proteins, encoded by At2g28900, At3g62880, and At4g16160, were designated as targeted to chloroplasts. This was based on the fact that the pattern of GFP observed was identical to rbcS-RFP (Fig. 4). In the case of At3g62880, plastidic targeting only took place when GFP was fused to the N-terminal region of the encoded protein. In comparison to the mitochondrial pattern, in these cells, the oval-shaped GFP fluorescence spots were larger (>2 μm) and fewer in number. Additionally, none of these proteins were imported into mitochondria, based on the observation that no protease-protected products were observed under any conditions. Import into isolated chloroplasts yielded protected products for At2g28900 and At4g16160; notably, the latter had an apparent molecular mass of 14 kD compared to the precursor protein of 22 kD (Fig. 4). The protein encoded by At3g62880 did not yield protease-protected products upon incubation with chloroplasts (Fig. 4). In fact, the protein did not even bind to chloroplasts.
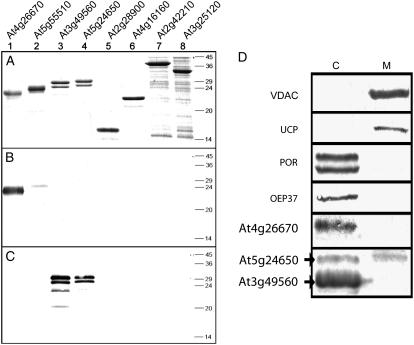
In vitro targeting assays did not yield definitive subcellular localization with the proteins encoded by At3g49560 and At5g24650. With isolated mitochondria, protease-protected products were consistently detected even in the presence of valinomycin (Fig. 4). Notably, both proteins are digested to completion upon protease treatment with outer membrane-ruptured mitochondria. This may indicate that the proteins were inserted into the outer membrane. On the other hand, incubation with isolated chloroplasts also resulted in protease protection, indicating successful import (either in the inner or outer membrane of the chloroplasts). However, for both organelles, it cannot be excluded that the observed protease protection results from unspecific binding of the proteins on the surface of the organelles. With GFP targeting, two patterns were evident; for At3g49560, clear plastidic localization was evident, whereas for At5g24650, mitochondrial localization was concluded based on colocalization with Aox-RFP. To verify the subcellular localization of the proteins encoded by At3g49560 and At5g24650, antibodies were raised against the overexpressed protein encoded by At3g49560. Because this protein and the protein encoded by At5g24650 display 83% identity, the antibody cross-reacts with both proteins, but not with a variety of other overexpressed PRAT proteins (Fig. 5, A and C). The protein encoded by At5g24650 has a small, but detectable, higher apparent molecular mass than that encoded by At3g49560 (Fig. 5A). Western-blot analyses with antibodies raised against the protein encoded by At3g49560 detect a prominent band in chloroplasts and a weaker band with a slightly higher molecular mass (Fig. 5D). Probing mitochondria with this antibody only detects the higher band (i.e. the protein encoded by At5g24650). Probing the mitochondrial fraction with antibodies to other chloroplast proteins such as POR of chloroplasts and OEP37 reveals no cross-reaction, indicating no significant contamination of purified mitochondria with chloroplasts. Thus, given that only the upper band is detected and that there is no significant contamination, it was concluded that the protein encoded by At5g24650 was located in mitochondria and chloroplasts and the protein encoded by At3g49560 was located in chloroplasts.
Figure 5.
Immunological analysis of mitochondria and chloroplasts with antibodies raised to proteins encoded by At3g49560 and At4g26670. Immunological analysis of PRAT proteins encoded by At4g26670, At5g55510, At3g49560, At5g24650, At2g28900, At4g16160, At2g42210, and At3g25120 (lanes 1–8). A, Coomassie-stained gel (1 μg protein/lane). B, Western blot with antibodies raised against the protein encoded by At4g26670 (0.1 μg protein/lane). C, Western blot with antibodies raised against the protein encoded by At3g49560 (0.1 μg protein/lane). Lanes 1 to 6, Full-length proteins fused with a C-terminal His tag expressed in Escherichia coli and purified using a nickel-nitrilotriacetic acid column. Lanes 7 to 8, Crude E. coli lysates containing partial proteins fused with an N-terminal glutathione S-transferase tag. D, Mitochondrial and chloroplastidic proteins were separated by SDS-PAGE and probed with various antibodies. VDAC, Mitochondrial voltage-dependent anion channel; UCP, mitochondrial inner membrane uncoupling protein.
Subcellular localization of the protein encoded by At4g26670 could not be defined by in vivo and in vitro targeting studies. C-terminal GFP tagging did not yield a pattern that corresponded to either mitochondrial or plastidic localization; N-terminal GFP tagging indicated nuclear localization, a pattern frequently observed with GFP if it is not targeted to any localization in the cell. Utilization of an RFP peroxisomal-targeted protein indicated that the protein was not targeted to peroxisomes (data not shown; Pracharoenwattana et al., 2005). Western-blot analysis of isolated chloroplasts and mitochondria using an antibody raised against the At4g26670 protein yielded a strong band in the chloroplast fraction, strongly suggesting a chloroplastidic location for this protein (Fig. 5D). The antibody raised against At4g26670 did not recognize the protein encoded by At5g55510 (Fig. 5B), which encodes for a 79% identical protein. This protein, however, showed a clear plastidic pattern when tagged with C-terminal GFP (Fig. 4).
Functional Identification of TIM22 from Arabidopsis
Because TIM22 is an essential protein in yeast and the identity of the ortholog was unclear in Arabidopsis, the ability of various PRAT proteins imported into isolated mitochondria to complement a yeast mutant for lacking a functional TIM22 protein was tested (Sirrenberg et al., 1996). The endogenous yeast TIM22 gene was placed under the Gal promoter so that its expression was dependent on the presence of Gal in the growth medium. The ability of three Arabidopsis genes to complement this strain was tested, At1g18320/At3g10110, At2g42210, and At3g25120. Culturing of yeast on Gal-free medium resulted in only At1g18320/At3g10110 supporting growth (Fig. 6). Thus, only At1g18320/At3g10110 could support a TIM22 function under the conditions tested. Previously, we have shown that the Arabidopsis orthologs for TIM17 and TIM23 can also complement yeast mutants (Murcha et al., 2003). Thus, it appears that the function of these translocases is well conserved across wide phylogenetic gaps and suggests that At2g42210 and At3g25120 may not play roles in protein transport.
Figure 6.
Complementation of yeast lacking a functional TIM22 protein. The ability of mitochondrially targeted PRAT proteins from Arabidopsis to complement yeast lacking TIM22 was tested using a replacement strategy. Expression of the yeast TIM22 was dependent on the presence of Gal in the medium. The ability of the Arabidopsis genes to complement was tested by expressing them under a constitutive promoter in yeast and growing in a medium lacking Gal. The ability to complement was evidenced by growth. The yeast gene (ScTIM22) acts as a positive control and the empty vector acts as a negative control.
Transcript Abundance of PRAT Genes Relative to Other Components of Mitochondrial and Chloroplast Protein Import Apparatus
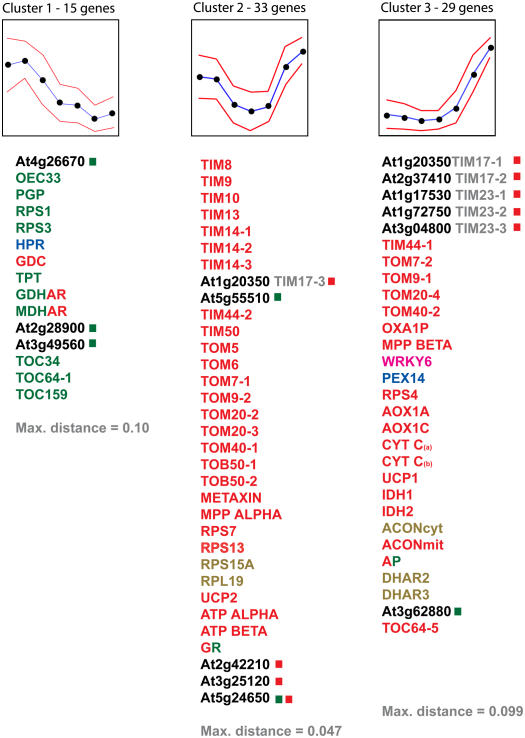
Because this family of genes was initially defined in yeast as being essential for the process of protein import into mitochondria (Pfanner and Geissler, 2001), and OEP16 is proposed to be involved in the import of POR A into chloroplasts (Reinbothe et al., 2004b, 2005), we examined the transcript abundance of all members of this family in comparison to transcripts encoding components of the mitochondrial, chloroplast, and peroxisome protein import apparatus. We also included several other metabolic components whose subcellular localization has been defined (Supplemental Table S1). We used quantitative reverse transcription (QRT)-PCR analysis to examine transcript abundance over development in leaves from 0 weeks (just emerged) to 6 weeks (senescence) for 77 genes (Lister et al., 2004; Clifton et al., 2005). A distinct pattern emerged when analyzed using self-organizing map analysis (Fig. 7; Golub et al., 1999; Tamayo et al., 1999). For a number of genes encoding components defined as being located in chloroplasts, transcript abundance was initially high, after which it declined steadily (Fig. 7; Supplemental Table S1). This pattern was also observed with genes encoding the P-subunit of Gly decarboxylase and hydroxypyruvate reductase, located in mitochondria and peroxisomes, respectively, and playing a well-characterized role in the photorespiratory cycle (Siedow and Day, 2002). Three genes of the PRAT family, At2g28900, At3g49560, and At4g26670, exhibited a similar pattern. For At2g28900 and At4g26670, this is in agreement with chloroplastidic location as defined above (Figs. 4 and 5). Also, for the protein encoded by At3g49560, a chloroplastidic expression pattern is consistent with immunological analysis (Fig. 5). Cluster groups 2 and 3 were dominated by genes encoding mitochondrial components and yet contained the gene At3g62880 of the PRAT family encoding a protein defined as having a chloroplast location. Notably, the undefined PRAT proteins encoded by At2g42210 and At3g25120, defined as mitochondrial by in vitro and in vivo targeting studies, displayed a pattern of expression consistent with mitochondrial location. So does the protein encoded by At5g24650; this further supports dual localization for the protein. According to available public microarray data (see below), At4g16160 is not expressed in rosette leaves; analysis of transcript abundance by QRT-PCR is in agreement with this, where expression was extremely low or not detectable (data not shown). Due to the 100% sequence identity between At1g18320 and At3g10110, it was not possible to determine the expression of either unambiguously.
Figure 7.
Self-organizing map analysis of the expression patterns of genes encoding mitochondria (red), chloroplast (green), dual-targeted (red and green), cytosolic (brown), peroxisomal (blue), and nuclear (magenta) proteins. QRT-PCR analysis was carried out on RNA isolated from second rosette leaves at seven stages of development: just emerged 0, 1, 2, 3, 4, 5, and 6 weeks old. The amount of mRNA for each transcript was normalized where the maximal value was set to 100 and other values were expressed relative to it. The blue line represents the pattern of transcript abundance with variation indicated by red lines. Members of the PRAT family are indicated in black with a colored square used to indicate the subcellular location as defined in Figures 3 to 5. Because At1g18320 and At3g10110 display 100% nucleic acid sequence homology, it was not possible to develop a QRT-PCR assay that was gene specific and thus they were omitted from this analysis. Transcripts of At4g16160 could not be detected because this gene is not expressed in rosette leaves. Gene abbreviations are listed in Supplemental Table S1.
DISCUSSION
Table I summarizes our current knowledge of proteins encoded by the PRAT family of genes in Arabidopsis. Three proteins encoded by genes of this family can be described as TIM17, TIM23, and OEP16, respectively, based on sequence similarity and subcellular localization. However, even with these relatively clear cases, it cannot be assumed that their function is orthologous to other species; AtTIM23-1 (At1g17350), AtTIM23-2 (At1g72750), and At4g16160 and At3g62880 (both OEP16-like) lack a defined PRAT domain that may indicate divergence of function. Furthermore, expression profiles of the latter two genes differ from the third OEP16 gene (At2g28900) in that they do not display a chloroplastidic pattern of expression during leaf development.
The identity of TIM22 in Arabidopsis cannot be unambiguously defined by sequence analysis alone. Based on mitochondrial localization of the predicted proteins, it could be encoded by At1g18320/At3g10110, At2g42210, or At3g25120. All other possible candidates are located in chloroplasts in both organelles or are of unclear localization. Phylogenetic comparison favors the protein encoded by At1g18320/At3g10110 and, because this protein can complement a yeast mutant for TIM22, it strongly suggests that these loci encode TIM22 in Arabidopsis.
In this study, we have used immunodecoration, transcript pattern, and GFP tagging to define chloroplastidic location for the protein encoded by At3g49560 in agreement with two independent proteomic approaches (Ferro et al., 2003; Froehlich et al., 2003). At5g24650 displays 83% sequence identity to the protein encoded by At3g49560 and has also been shown to occur in chloroplasts by proteomic approaches (Ferro et al., 2003; Froehlich et al., 2003). In this study, four different independent approaches indicate that it also is located in mitochondria: (1) in vitro protein uptake assays; (2) in vivo GFP localization; (3) immunological localization; and (4) pattern of transcript abundance consistent with mitochondrial localization. Thus, we concluded on the basis of this study and previous proteomic analysis that the protein encoded by At5g24650 is dual targeted to mitochondria and chloroplasts. Different experimental approaches can reveal or detect proteins in different locations. Proteome and immunological analyses reveal the protein encoded by At5g24650 to be located in chloroplasts, whereas immunological, in vivo targeting and expression analysis reveal mitochondrial localization. Examination of the subcellular localization of proteins defined by mass spectrometry-based proteomics and comparison to localization defined by GFP indicate that there are many proteins where localization defined by proteome analysis and GFP tagging differs (Heazlewood et al., 2005). For the 547 proteins defined to be mitochondrial by mass spectrometry, GFP-targeting data exist to confirm 18, but in vivo GFP data indicate that 25 are located in other organelles. Likewise, in chloroplasts for the 1,017 proteins defined to be located in chloroplasts, in vivo GFP targeting confirms 26, but differs for 60. Although some of these discrepancies may be due to contamination of organelles used for proteome analysis, it is likely that many are due to dual targeting of proteins to mitochondria and chloroplasts. Proteome analysis of subcellular organelles can only detect proteins in the organelle analyzed and thus is not suitable as an experimental approach to define dual-targeted proteins. Although GFP tagging has the potential to reveal dual targeting where GFP can be detected in both organelles, some dual-targeted proteins are only targeted to one organelle when attached to GFP, such as a σ factor in maize (Beardslee et al., 2002), and dual-targeted ascorbate peroxidase and monodehydroascorbate reductase (Chew et al., 2003b). This may be due to a variety of factors, such as GFP (passenger) protein affecting targeting ability, the ability of organelles in transformed tissue to import dual-targeted proteins, and the sorting/partitioning of dual-targeted protein between the two organelles. Immunological analyses overcome the technical limitations of the above approaches.
Because the proteins encoded by At3g49560 and At5g24650 are 83% identical and are targeted to chloroplasts or chloroplasts and mitochondria, respectively, it indicates that small changes in amino acid sequence can alter subcellular localization. Examination of the upstream sequence of At5g24650 for a possible noncanonical start codon, previously reported to be responsible for dual targeting of DNA polymerase to mitochondria and chloroplasts (Christensen et al., 2005), revealed an in-frame stop codon just 20 bp from the start ATG, and six stop codons in the 300 bp upstream of the start codon. Thus, current evidence does not reveal any additional N-terminal region that may explain the dual targeting of the protein encoded by At5g24650. A comparison of the proteins encoded by the two genes does reveal that the protein encoded by At5g24650 is predicted to form a helix-coil-helix in the first 35 amino acids and that the first helix contains several positive-charged residues. In contrast, in the protein encoded by At3g49560, the predicted helix-coil-helix occurs between amino acids 15 and 50 and, noticeably, the first helix predicted lacks any positive residues. Finally, the protein encoded by At3g49560 also contains four Gly residues at positions 4 to 7; previously, it has been shown that insertion of Gly residues into a mitochondrial targeting signal abolished mitochondrial targeting ability (Tanudji et al., 1999). These differences may account for the different targeting ability of these proteins. Changing a single amino acid in the targeting signal of peroxisomal 3-ketoacyl-CoA thiolase resulted in dual targeting to mitochondria and peroxisomes (Tsukamoto et al., 1994). Proteomic analyses indicate that the protein encoded by At5g24650 is present in the tonoplast and plasma membranes (Marmagne et al., 2004; Szponarski et al., 2004). Given the absence of major mitochondrial and plastidic contaminants in these studies, any functional characterization of the protein encoded by At5g24650 needs to take into account that these organelles may also be affected in addition to mitochondria and plastids. However, the location of the protein encoded by At5g24650 in these membranes needs to be confirmed by immunological means.
Despite TIM17, TIM23, and the proposed role for OEP16 in amino acid or POR A transport, the function of all other members of the entire PRAT family of proteins is unknown. However, examination of transcript abundance profiles during leaf development suggests that at least some members of this family may play an important role in exchange of metabolites or other molecules between mitochondria and plastids. Three genes, At5g55510, At5g24650, and At3g62880, display transcript abundance profiles that were distinct from those that define a typical chloroplast pattern. The latter pattern is dominated by photosynthetic function and this may indicate that the proteins encoded by these genes are involved in nonphotosynthetic plastidic functions. In agreement with this proposal is a recent report that details the expression of At4g16160 in the maturation phase of seeds and pollen grains, both noted to be desiccation-tolerant tissues (Drea et al., 2006). Previously, a protein related to the PRAT family of proteins has been located in complex I of the N. crassa and bovine electron transport chain (Nehls et al., 1991; Carroll et al., 2002), further indicating that PRAT proteins play a role in a variety of processes. The diverse functions and subcellular localizations of PRAT proteins characterized to date suggest roles in a wide variety of transport processes. Because some mitochondrially targeted PRAT proteins failed to complement a deletion in tim22, this suggests roles other than in protein translocation. Thus, expansion of members of this family of proteins, even members that are targeted to the same organelle, seems to have been accompanied by expansion of function.
CONCLUSION
It is well documented that proteins encoded by genes derived from one organelle can end up in another organelle (Martin and Herrmann, 1998). This has been observed with proteins involved with all types of function in mitochondria and chloroplasts and, in some cases, such proteins are even dual targeted (Peeters and Small, 2001; Duchene et al., 2005). However, it is now apparent that even the machinery involved in dictating the organelle proteomes have mixed ancestry, being located in mitochondria and plastids, and dual targeted. This has also been observed with other components involved with protein import, such as peptidases involved in degrading targeting signals (Bhushan et al., 2003; Stahl et al., 2005). This cautions against presumptions on subcellular localizations, even for components intimately involved in the process of protein import into organelles. It indicates that the process of recognition of proteins on the organellar surface, in some instances, or for subsets of proteins may take place by very similar components.
MATERIALS AND METHODS
BLAST and PSI-BLAST algorithms were used to search protein databases for proteins displaying significant homology to import components characterized in yeast (Saccharomyces cerevisiae; Altschul et al., 1997). Hits from this search with expectation values ≤10−5 were then used to query yeast protein databases. If the hit with the greatest confidence in this search was the yeast import component sequence used in the initial search, the protein was termed a putative protein import component. Proteins were aligned using ClustalX (Thompson et al., 1994, 1997). Phylogenetic trees were constructed with the program PAUP (Swofford, 2002). Only the conserved region of the proteins around the PRAT domain was used in the phylogenic analyses. Data were bootstrap resampled 100 times. Gene structures were obtained from TAIR 6 and three individual cDNA clones were sequenced.
In vitro protein import assays into isolated mitochondria from Arabidopsis (Arabidopsis thaliana) were carried out as previously described (Lister et al., 2004). Outer membrane-ruptured mitochondria were prepared by osmotic shock (Murcha et al., 2005). In vitro protein import assays into isolated chloroplasts from pea (Pisum sativum) were carried out as previously outlined (Rudhe et al., 2002). GFP targeting was carried out by cloning GFP in frame to the N or C terminus of the cDNA clone and transformation of Arabidopsis suspension cells by biolistic transformation (Thirkettle-Watts et al., 2003; Lee and Whelan, 2004). RFP was fused to the targeting signal of soybean (Glycine max) Aox-RFP and pea rbcS-RFP as mitochondrial and plastidic controls, respectively (Carrie et al., 2007). Fluorescence patterns were obtained 48 h after transformation by visualization under an Olympus BX61 fluorescence microscope and imaged using Cell imaging software.
Antibodies were raised in rabbit against recombinant protein encoded by At4g26670 and At3g49560, respectively (Pineda Antikörper-Service). Proteins with a 6x-His tag located on the C terminus were expressed and purified on a nickel nitrilotriacetic acid column (Qiagen). Chloroplasts from 14-d-old Arabidopsis plants grown on one-half-strength Murashige and Skoog medium were isolated as described by Aronsson and Jarvis (2002). Mitochondria from 4-week-old plants were isolated according to Kruft et al. (2001). Specificity of the antibodies was tested on different recombinant proteins of the PRAT protein family of Arabidopsis.
The yeast strain expressing ScTIM22 under the control of the GAL promoter (GAL-TIM22) has been described previously (Sirrenberg et al., 1996). Because TIM22 is a protein essential for yeast cell viability, this strain requires Gal in the medium for growth. For complementation analysis, Arabidopsis open reading frames At3g25120, At2g42210, and At1g18320/At3g10110 were cloned into the yeast vector pVT-U, which enables the expression of cloned genes under the constitutive alcohol dehydrogenase promoter (Vernet et al., 1987). Obtained plasmids were transformed into the GAL-TIM22 yeast strain using the lithium acetate method (Gietz et al., 1992). Empty plasmid and the plasmid containing ScTIM22 were transformed as controls. The ability of various proteins to substitute for ScTIM22 was assessed on the selective medium lacking or containing 0.5% (w/v) Gal (Sambrook et al., 1989). Two independent transformants were analyzed for each transformation and gave identical results.
QRT-PCR was carried out on RNA isolated from second rosette leaves at different times; the initial time point, labeled 0, was when leaves just emerged and then 1, 2, 3, 4, 5, and 6 weeks after this time. At 6 weeks, the leaves were pale green or yellowing and in an advanced stage of senescence. QRT-PCR was carried out as previously described with the gene listed (Lister et al., 2004).
The nucleic acid sequences for AtTIM23 and AtTIM17 have been deposited previously in GenBank with the accession numbers At1g20350:AY463969 (AtTIM17-1); At2g37410:AY463970 (AtTIM17-2); At5g11690:AY463971 (AtTIM17-3); At1g17350:AY463972 (AtTIM23-1); At1g72750:AY463973 (AtTIM23-1); and At3g04800:AY463974 (AtTIM23-3). The GenBank accession numbers for the remaining cDNA sequences used in this study are as follows: At1g18320: DQ405269; At3g10110: DQ405268; At2g28900 (OEP16-like): AAC79594; At2g42210: DQ386643; At3g25120: DQ405272; At3g49560: DQ405266; At3g62880 (OEP16-like): CAB83138; At4g16160 (OEP16-like): CAB10395; At4g26670: DQ405270; At5g24650: DQ405267; and At5g55510: DQ405271.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Multiple sequence alignment of the predicted proteins of the PRAT family from Arabidopsis and rice.
Supplemental Figure S2. Phylogenetic analysis and gene structure of genes encoding PRAT proteins from Arabidopsis and rice.
Supplemental Figure S3. Matrix indicating the percentage identity (gray) and similarity (white) of proteins encoded by the PRAT family of genes in Arabidopsis and rice.
Supplemental Table S1. Abbreviations and full names for genes whose expression patterns were analyzed in this study. The loci, subcellular location of encoded protein, and reference for expression analysis are listed.
Supplementary Material
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: James Whelan (seamus@cyllene.uwa.edu.au).
Some figures in this article are displayed in color online but in black and white in the print edition.
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Ahting U, Thieffry M, Engelhardt H, Hegerl R, Neupert W, Nussberger S (2001) Tom40, the pore-forming component of the protein-conducting TOM channel in the outer membrane of mitochondria. J Cell Biol 153 1151–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Smith SM (1986) Synthesis of the small subunit of ribulose bisphosphate carboxylase from genes cloned into plasmids containing the SP6 promoter. Biochem J 240 709–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408 796–815 [DOI] [PubMed] [Google Scholar]
- Aronsson H, Jarvis P (2002) A simple method for isolating import-competent Arabidopsis chloroplasts. FEBS Lett 529 215–220 [DOI] [PubMed] [Google Scholar]
- Bathgate B, Baker A, Leaver CJ (1989) Two genes encode the adenine nucleotide translocator of maize mitochondria: isolation, characterisation and expression of the structural genes. Eur J Biochem 183 303–310 [DOI] [PubMed] [Google Scholar]
- Bauer MF, Hofmann S, Neupert W, Brunner M (2000) Protein translocation into mitochondria: the role of TIM complexes. Trends Cell Biol 10 25–31 [DOI] [PubMed] [Google Scholar]
- Beardslee TA, Roy-Chowdhury S, Jaiswal P, Buhot L, Lerbs-Mache S, Stern DB, Allison LA (2002) A nucleus-encoded maize protein with sigma factor activity accumulates in mitochondria and chloroplasts. Plant J 31 199–209 [DOI] [PubMed] [Google Scholar]
- Bedard J, Jarvis P (2005) Recognition and envelope translocation of chloroplast preproteins. J Exp Bot 56 2287–2320 [DOI] [PubMed] [Google Scholar]
- Bhushan S, Lefebvre B, Stahl A, Wright SJ, Bruce BD, Boutry M, Glaser E (2003) Dual targeting and function of a protease in mitochondria and chloroplasts. EMBO Rep 4 1073–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugiere S, Kowalski S, Ferro M, Seigneurin-Berny D, Miras S, Salvi D, Ravanel S, d'Herin P, Garin J, Bourguignon J, et al (2004) The hydrophobic proteome of mitochondrial membranes from Arabidopsis cell suspensions. Phytochemistry 65 1693–1707 [DOI] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Millar AH, Smith SM, Whelan J (2007) Nine 3-ketoacyl-CoA thiolases (KATs) and acetoacetyl-CoA thiolases (ACATs) encoded by five genes in Arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria. Plant Mol Biol (in press) [DOI] [PubMed]
- Carroll J, Shannon RJ, Fearnley IM, Walker JE, Hirst J (2002) Definition of the nuclear encoded protein composition of bovine heart mitochondrial complex I: identification of two new subunits. J Biol Chem 277 50311–50317 [DOI] [PubMed] [Google Scholar]
- Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, Sickmann A, Meyer HE, Truscott KN, Guiard B, et al (2005) Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell 120 817–829 [DOI] [PubMed] [Google Scholar]
- Chew O, Lister R, Qbadou S, Heazlewood JL, Soll J, Schleiff E, Millar AH, Whelan J (2004) A plant outer mitochondrial membrane protein with high amino acid sequence identity to a chloroplast protein import receptor. FEBS Lett 557 109–114 [DOI] [PubMed] [Google Scholar]
- Chew O, Rudhe C, Glaser E, Whelan J (2003. a) Characterization of the targeting signal of dual-targeted pea glutathione reductase. Plant Mol Biol 53 341–356 [DOI] [PubMed] [Google Scholar]
- Chew O, Whelan J (2004) Just read the message: a model for sorting of proteins between mitochondria and chloroplasts. Trends Plant Sci 9 318–319 [DOI] [PubMed] [Google Scholar]
- Chew O, Whelan J, Millar AH (2003. b) Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual targeting of antioxidant defenses in plants. J Biol Chem 278 46869–46877 [DOI] [PubMed] [Google Scholar]
- Christensen AC, Lyznik A, Mohammed S, Elowsky CG, Elo A, Yule R, Mackenzie SA (2005) Dual-domain, dual-targeting organellar protein presequences in Arabidopsis can use non-AUG start codons. Plant Cell 17 2805–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary SP, Tan FC, Nakrieko KA, Thompson SJ, Mullineaux PM, Creissen GP, von Stedingk E, Glaser E, Smith AG, Robinson C (2002) Isolated plant mitochondria import chloroplast precursor proteins in vitro with the same efficiency as chloroplasts. J Biol Chem 277 5562–5569 [DOI] [PubMed] [Google Scholar]
- Clifton R, Lister R, Parker KL, Sappl P, Elhafez D, Millar AH, Day DA, Whelan J (2005) Stress-induced co-expression of alternative respiratory chain components in Arabidopsis thaliana. Plant Mol Biol 58 193–212 [DOI] [PubMed] [Google Scholar]
- Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10 673–676 [DOI] [PubMed] [Google Scholar]
- Drea SC, Lao NT, Wolfe KH, Kavanagh TA (2006) Gene duplication, exon gain and neofunctionalization of OEP16-related genes in land plants. Plant J 46 723–735 [DOI] [PubMed] [Google Scholar]
- Duchene AM, Giritch A, Hoffmann B, Cognat V, Lancelin D, Peeters NM, Zaepfel M, Marechal-Drouard L, Small ID (2005) Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad Sci USA 102 16484–16489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Salvi D, Brugiere S, Miras S, Kowalski S, Louwagie M, Garin J, Joyard J, Rolland N (2003) Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol Cell Proteomics 2 325–345 [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Wilkerson CG, Ray WK, McAndrew RS, Osteryoung KW, Gage DA, Phinney BS (2003) Proteomic study of Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J Proteome Res 2 413–425 [DOI] [PubMed] [Google Scholar]
- Gietz D, St Jean A, Woods RA, Schiestl RH (1992) Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 20 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al (1999) Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286 531–537 [DOI] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini JS, Gout AM, Day DA, Whelan J, Millar AH (2004) Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 16 241–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood JL, Tonti-Filippini J, Verboom RE, Millar AH (2005) Combining experimental and predicted datasets for determination of the subcellular location of proteins in Arabidopsis. Plant Physiol 139 598–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heins L, Schmitz UK (1996) A receptor for protein import into potato mitochondria. Plant J 9 829–839 [DOI] [PubMed] [Google Scholar]
- Hinnah SC, Wagner R, Sveshnikova N, Harrer R, Soll J (2002) The chloroplast protein import channel Toc75: pore properties and interaction with transit peptides. Biophys J 83 899–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansch L, Kruft V, Schmitz UK, Braun HP (1998) Unique composition of the preprotein translocase of the outer mitochondrial membrane from plants. J Biol Chem 273 17251–17257 [DOI] [PubMed] [Google Scholar]
- Kruft V, Eubel H, Jansch L, Werhahn W, Braun HP (2001) Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol 127 1694–1710 [PMC free article] [PubMed] [Google Scholar]
- Lee MN, Whelan J (2004) Identification of signals required for import of the soybean F(A)d subunit of ATP synthase into mitochondria. Plant Mol Biol 54 193–203 [DOI] [PubMed] [Google Scholar]
- Linke D, Frank J, Pope MS, Soll J, Ilkavets I, Fromme P, Burstein EA, Reshetnyak YK, Emelyanenko VI (2004) Folding kinetics and structure of OEP16. Biophys J 86 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Chew O, Lee MN, Heazlewood JL, Clifton R, Parker KL, Millar AH, Whelan J (2004) A transcriptomic and proteomic characterization of the Arabidopsis mitochondrial protein import apparatus and its response to mitochondrial dysfunction. Plant Physiol 134 777–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister R, Hulett JM, Lithgow T, Whelan J (2005) Protein import into mitochondria: origins and functions today (review). Mol Membr Biol 22 87–100 [DOI] [PubMed] [Google Scholar]
- Marmagne A, Rouet MA, Ferro M, Rolland N, Alcon C, Joyard J, Garin J, Barbier-Brygoo H, Ephritikhine G (2004) Identification of new intrinsic proteins in Arabidopsis plasma membrane proteome. Mol Cell Proteomics 3 675–691 [DOI] [PubMed] [Google Scholar]
- Martin W, Herrmann RG (1998) Gene transfer from organelles to the nucleus: how much, what happens, and why? Plant Physiol 118 9–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AH, Heazlewood JL, Kristensen BK, Braun HP, Moller IM (2005) The plant mitochondrial proteome. Trends Plant Sci 10 36–43 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Elhafez D, Millar AH, Whelan J (2005) The C-terminal region of TIM17 links the outer and inner mitochondrial membranes in Arabidopsis and is essential for protein import. J Biol Chem 280 16476–16483 [DOI] [PubMed] [Google Scholar]
- Murcha MW, Lister R, Ho AY, Whelan J (2003) Identification, expression, and import of components 17 and 23 of the inner mitochondrial membrane translocase from Arabidopsis. Plant Physiol 131 1737–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehls U, Hemmer S, Rohlen DA, Van der Pas JC, Preis D, Sackmann U, Weiss H (1991) cDNA and genomic DNA sequence of the 21.3 kDa subunit of NADH:ubiquinone reductase (complex I) from Neurospora crassa. Biochim Biophys Acta 1088 325–326 [DOI] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66 863–917 [DOI] [PubMed] [Google Scholar]
- Peeters N, Small I (2001) Dual targeting to mitochondria and chloroplasts. Biochim Biophys Acta 1541 54–63 [DOI] [PubMed] [Google Scholar]
- Pfanner N, Geissler A (2001) Versatility of the mitochondrial protein import machinery. Nat Rev Mol Cell Biol 2 339–349 [DOI] [PubMed] [Google Scholar]
- Pohlmeyer K, Soll J, Steinkamp T, Hinnah S, Wagner R (1997) Isolation and characterization of an amino acid-selective channel protein present in the chloroplastic outer envelope membrane. Proc Natl Acad Sci USA 94 9504–9509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM (2005) Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rassow J, Dekker PJ, van Wilpe S, Meijer M, Soll J (1999) The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol 286 105–120 [DOI] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N (2004) Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol 5 519–530 [DOI] [PubMed] [Google Scholar]
- Rehling P, Wiedemann N, Pfanner N, Truscott KN (2001) The mitochondrial import machinery for preproteins. Crit Rev Biochem Mol Biol 36 291–336 [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Pollmann S, Springer A, James RJ, Tichtinsky G, Reinbothe C (2005) A role of Toc33 in the protochlorophyllide-dependent plastid import pathway of NADPH:protochlorophyllide oxidoreductase (POR) A. Plant J 42 1–12 [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Quigley F, Gray J, Schemenewitz A, Reinbothe C (2004. a) Identification of plastid envelope proteins required for import of protochlorophyllide oxidoreductase A into the chloroplast of barley. Proc Natl Acad Sci USA 101 2197–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe S, Quigley F, Springer A, Schemenewitz A, Reinbothe C (2004. b) The outer plastid envelope protein Oep16: role as precursor translocase in import of protochlorophyllide oxidoreductase A. Proc Natl Acad Sci USA 101 2203–2208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudhe C, Chew O, Whelan J, Glaser E (2002) A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. Plant J 30 213–220 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Schwacke R, Schneider A, van der Graaff E, Fischer K, Catoni E, Desimone M, Frommer WB, Flugge UI, Kunze R (2003) ARAMEMNON, a novel database for Arabidopsis integral membrane proteins. Plant Physiol 131 16–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedow JN, Day DA (2002) Respiration and photosynthesis. In B Buchanan, W Gruissem, RL Jones, eds, Biochemistry and Molecular Biology of Plants. John Wiley and Sons, New York, pp 676–728
- Sirrenberg C, Bauer MF, Guiard B, Neupert W, Brunner M (1996) Import of carrier proteins into the mitochondrial inner membrane mediated by Tim22. Nature 384 582–585 [DOI] [PubMed] [Google Scholar]
- Soll J, Schleiff E (2004) Protein import into chloroplasts. Nat Rev Mol Cell Biol 5 198–208 [DOI] [PubMed] [Google Scholar]
- Stahl A, Nilsson S, Lundberg P, Bhushan S, Biverstahl H, Moberg P, Morisset M, Vener A, Maler L, Langel U, et al (2005) Two novel targeting peptide degrading proteases, PrePs, in mitochondria and chloroplasts, so similar and still different. J Mol Biol 349 847–860 [DOI] [PubMed] [Google Scholar]
- Stojanovski D, Johnston AJ, Streimann I, Hoogenraad NJ, Ryan MT (2003) Import of nuclear-encoded proteins into mitochondria. Exp Physiol 88 57–64 [DOI] [PubMed] [Google Scholar]
- Swofford DL (2002) PAUP: Phylogenetic Analysis Using Parsimony Version 4.0 b10. Sinauer Associates, Sunderland, MA
- Szponarski W, Sommerer N, Boyer JC, Rossignol M, Gibrat R (2004) Large-scale characterization of integral proteins from Arabidopsis vacuolar membrane by two-dimensional liquid chromatography. Proteomics 4 397–406 [DOI] [PubMed] [Google Scholar]
- Tamayo P, Slonim D, Mesirov J, Zhu Q, Kitareewan S, Dmitrovsky E, Lander ES, Golub TR (1999) Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA 96 2907–2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanudji M, Sjoling S, Glaser E, Whelan J (1999) Signals required for the import and processing of the alternative oxidase into mitochondria. J Biol Chem 274 1286–1293 [DOI] [PubMed] [Google Scholar]
- Thirkettle-Watts D, McCabe TC, Clifton R, Moore C, Finnegan PM, Day DA, Whelan J (2003) Analysis of the alternative oxidase promoters from soybean. Plant Physiol 133 1158–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25 4876–4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truscott KN, Brandner K, Pfanner N (2003) Mechanisms of protein import into mitochondria. Curr Biol 13 R326–R337 [DOI] [PubMed] [Google Scholar]
- Truscott KN, Wiedemann N, Rehling P, Muller H, Meisinger C, Pfanner N, Guiard B (2002) Mitochondrial import of the ADP/ATP carrier: the essential TIM complex of the intermembrane space is required for precursor release from the TOM complex. Mol Cell Biol 22 7780–7789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T, Osumi T (1994) Characterization of the signal peptide at the amino terminus of the rat peroxisomal 3-ketoacyl-CoA thiolase precursor. J Biol Chem 269 6001–6010 [PubMed] [Google Scholar]
- van Wijk KJ (2004) Plastid proteomics. Plant Physiol Biochem 42 963–977 [DOI] [PubMed] [Google Scholar]
- Vernet T, Dignard D, Thomas DY (1987) A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52 225–233 [DOI] [PubMed] [Google Scholar]
- Winning BM, Sarah CJ, Purdue PE, Day CD, Leaver CJ (1992) The adenine nucleotide translocator of higher plants is synthesized as a large precursor that is processed upon import into mitochondria. Plant J 2 763–773 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.