Abstract
Background
Mycobacteriophage Ms6 integrates into Mycobacterium smegmatis and M. bovis BCG chromosome at the 3' end of tRNAala genes. Homologous recombination occurs between the phage attP core and the attB site located in the T-loop. Integration-proficient vectors derived from Ms6 are useful genetic tools, but their insertion sites in the BCG chromosome remain poorly defined. The primary objective of this study was to identify Ms6 target genes in M. smegmatis and BCG. We then aimed to modify the attP site in Ms6-derived vectors, to switch integration to other tRNAala loci. This provided the basis for the development of recombinant M. bovis BCG strains expressing several reporter genes inserted into different tRNAala genes.
Results
The three tRNAala genes are highly conserved in M. smegmatis and BCG. However, in the T-loop of tRNAalaU and tRNAalaV containing the attB site, a single base difference was observed between the two species. We observed that the tRNAalaU gene was the only site into which Ms6-derived integration-proficient vectors integrated in M. smegmatis, whereas in BCG, the tRNAalaV gene was used as the target. No integration occurred in the BCG tRNAalaU T-loop, despite a difference of only one base from the 26-base Ms6 attP core. We mutated the attP core to give a perfect match with the other tRNAala T-loops from M. smegmatis and BCG. Modification of the seven-base T-loop decreased integration efficiency, identifying this site as a possible site of strand exchange. Finally, two Ms6 vectors were constructed to integrate two reporter genes into the tRNAalaU and tRNAalaV T-loops of the same BCG chromosome.
Conclusion
Small changes in the 7 bp T-loop attP site of Ms6 made it possible to use another attB site, albeit with a lower integration efficiency. These molecular studies on BCG tRNAala genes made it possible to create valuable tools for the site-directed insertion of several genes in the same BCG strain. These tools will be useful for the development of novel multivalent vaccines and genetic studies.
Background
Temperate phages integrate into the bacterial chromosome through a site-specific recombination event catalyzed by a phage-encoded recombinase. This process involves a common core present in the phage attP and the bacterial attB genomic DNA sequences, which are identical [1]. Genetic tools based on phage systems have furthered research into the biology of Mycobacterium tuberculosis, a pathogen responsible for about two million deaths each year [2]. L5 [3] and Ms6 [4], both temperate mycobacteriophages, integrate into genes encoding tRNAs. L5 integrates into a tRNAgly gene in the genome of the fast-growing species M. smegmatis or the slow-growing species M. bovis Bacillus Calmette Guérin (BCG), which is used as a vaccine against tuberculosis [5,6]. Integration-proficient vectors containing the attP site and either a tyrosine-integrase [5,7] or a serine-integrase [8] system integrate into the attB site of the mycobacterial chromosome. Most of these integrative vectors do not retain recombination directionality factors from the phage that mediate excision [9] and are generally stably maintained in the mycobacterial genome, even in the absence of antibiotic selection [10]. However, plasmid loss can occur through low level integrase-mediated excision [11]. These vectors are of considerable use both for the development of recombinant BCG (rBCG) strains with long-term heterologous gene expression for vaccine development [10] and for overcoming the problems associated with the use of multicopy extrachromosomal plasmids in genetic studies. Ms6-derived vectors integrate efficiently into the genomes of both slow- and fast-growing mycobacteria. In M. smegmatis, the attB core site overlaps the 3' end of a tRNAala gene containing the 7 bp T-loop [7]. The precise point of strand exchange between attP and attB is unknown. In silico scanning of the genomes of M. smegmatis and M. tuberculosis showed that there were three tRNAala genes containing 3' end attB sequences either identical or similar to the Ms6 attP core site. We show here that Ms6 integrates into different tRNAala genes in M. smegmatis and BCG, according to the presence of identical attB and attP cores. We also used site-directed mutagenesis of the attP core to construct vectors integrating into tRNAala genes other than that targeted by the wild-type attP. This led to the development of a method for integrating two vectors, carrying different heterologous genes, into two different loci of the same BCG chromosome.
Results
Genetic organisation of tRNAala genes from M. smegmatis and BCG
A BLASTn search of the M. tuberculosis genome [12] for sequences similar to the Ms6 attP core region [13] revealed three possible attB sites at the 3' ends of three tRNAala genes, named after their anti codon: GGC for tRNAalaU, CGC for tRNAalaV, and TGC for tRNAalaT. In tRNAalaV, the attB core sequence was identical to the 26 bases of the attP core region [7]. In tRNAalaU and tRNAalaT, one and three mismatches, respectively, were observed between the 26 bp Ms6 attP and attB core sequences. Alignment of the attP sequence with the genome of M. bovis [14] revealed the presence of the same three attB sites (data not shown). None of the tRNAalagenes mapped to deletion regions described in BCG [15,16].
Alignment of the Ms6 attP core sequence with the genome of M. smegmatis [17] revealed three possible attB sites displaying sequence similarities. As for BCG, the three possible attB sites were located in tRNAalaU (anticodon GGC), tRNAalaV (anticodon CGC) and tRNAalaT (anticodon TGC). The three M. smegmatis and BCG tRNAala structures were analysed with tRNA-scan SE [18]. A sequence identity block common to attP-attB was identified that encompassed the tRNAala T-loop located at the 3' end, without extending to the variable region (see additional file 2).
In both species, the tRNAala-encoding genes mapped to distant regions of the chromosome. The three tRNAala genes of M. smegmatis and BCG were highly similar, but not identical. Interestingly, the tRNAalaV T-loop from BCG was identical to the tRNAalaU T-loop from M. smegmatis. In both species, the T-loop from two other tRNAala genes differed from the 26 bp core attP site from Ms6 by one to four bases (Fig. 1A)
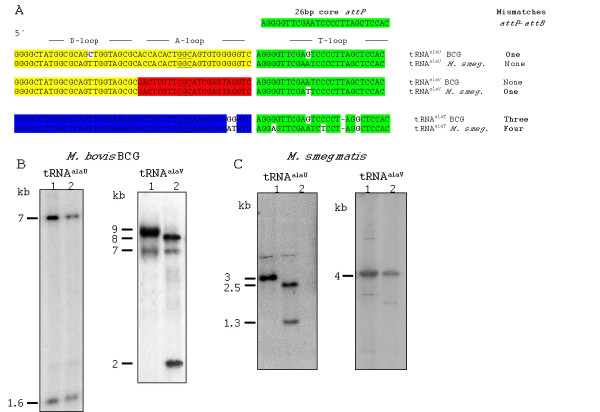
Figure 1.
Southern blot of genomic DNA from wild-type and pAV6950-transfected M. bovis BCG and M. smegmatis integrants. A: Alignment between the three tRNAala gene sequences from M. bovis BCG or M. smegmatis and the Ms6 attP 26-base core sequence. The coloured blocks represent identical sequence stretches, base differences are indicated in bold and the anticodon is underlined. The position of anticodon and T-loops in the tRNAala sequence is indicated. B: Genomic DNA from wild-type BCG (lane 1) and a pAV-SIV integrant (lane 2) was probed with a PCR product based on the tRNAalaU (left panel) or tRNAalaV (right panel) sequence from the M. tuberculosis genome. C: Genomic DNA from wild-type M. smegmatis (lane 1) and a pAV-SIV integrant (lane 2) was probed with a PCR product based on the tRNAalaU (left panel) or tRNAalaV (right panel) sequence from the M. smegmatis genome.
The integration of Ms6-derived vectors requires 100% identity of the 26 base cores of attP and attB
We have previously described the construction of pAV-SIV, an Ms6-derived integration-proficient vector, for the production of rBCG candidate vaccines genetically stable in vivo and expressing genes from the simian immunodeficiency virus SIVmac251 [10]. Here, we analysed the insertion locus of pAV-SIV in the BCG chromosome. We carried out Southern blot analysis on genomic DNA using a probe (see additional file 1 for all primers plasmids and strains used in this study) designed from the tRNAalaU gene from M. tuberculosis, as we previously showed that the target insertion sequence corresponded to tRNAalaU [7]. However, we did observe no band disruption when the genomic DNA hybridisation profiles of pAV-SIV integrants and wild-type BCG were compared, indicating that the integration-proficient plasmids had not inserted into the tRNAalaU gene (Fig. 1B, left panel). We then amplified another probe derived from the tRNAalaV sequence and observed, on a Southern blot of DNA from wild-type BCG, two specific bands : a 9 kb band showing a strong positive signal and a weakly hybridizing 7 kb band (Fig. 1B, right panel). Following transformation with pAV-SIV, the 9 kb band disappeared, giving rise to two bands of 8 and 2 kb. The 7 kb band, due to cross-hybridisation of tRNAalaU attB with the tRNAalaV probe, remained intact. This confirmed the specific integration of pAV-SIV into the tRNAalaV gene displaying identity with the attP rather than into the attB site from tRNAalaU, which has a base mismatch. We then investigated whether Ms6-derived vectors also targeted the M. smegmatis attB site displaying 100% identity with the Ms6 attP core – the tRNAalaU gene. A Southern blot analysis with two probes containing either the tRNAalaU or the tRNAalaV gene sequence revealed that the 3 kb band hybridising with the tRNAalaU probe in genomic DNA from wild-type M. smegmatis was disrupted in DNA from pAV-SIV integrants, giving 2.5 kb and 1.3 kb bands (Fig. 1C, left panel). When DNA was probed with tRNAalaV, no difference in hybridisation profile was observed between the wild type and integrants, demonstrating an absence of integration into the tRNAalaV locus (Fig. 1C, right panel).
We investigated whether integration into an attB site carrying mismatches with the attP core site was possible, as reported for L5 [5], by designing three primer pairs to amplify tRNAalaU, tRNAalaV or tRNAalaT regions both from M. smegmatis and BCG genome sequences. The PCR amplification of genomic DNA from fifty pAV-SIV BCG integrants identified tRNAalaV as the only gene target for Ms6 integration. By contrast, in the 50 M. smegmatis integrants analysed, pAV-SIV had inserted into the tRNAalaU gene (data not shown).
Modification of the attP core makes integration into the T-loop of other tRNAala genes possible
The tRNAalaU gene (anticodon GGC) of BCG and the tRNAalaV gene of M. smegmatis (anticodon CGC) differ from the attP core by one base in the T-loop, whereas the tRNAalaT genes differ from this core sequence by three and four bases, respectively (Fig. 1A). We then investigated whether modification of the attP site to match these T-loops would allow us to target Ms6-derived integrative vectors to other tRNAala genes. Site-directed mutagenesis was performed on pAV6950, carrying the natural Ms6 attP core sequence, to construct four vectors carrying modified 26 bp attP cores identical to the various tRNAala T-loops.
In M. smegmatis, the integration efficiency of pSV, carrying an attP core identical to the tRNAalaV T-loop, was only 12% that of the non-mutated pAV6950 targeting tRNAalaU (Table 1). PCR amplification of M. smegmatis pSV integrants confirmed that the vector had targeted tRNAalaV in all clones, as expected. When four base changes were introduced, to match the tRNAalaT T-loop sequence (plasmid pST), integration efficiency was only 0.05% that with the non-mutated pAV6950.
Table 1.
Integration efficiency of vectors carrying attP core modifications
| Vector | 26 bp core attPa | Integration efficiency/pAV6950b | % integration into the tRNAala T-loopc | ||
| M. smegmatis | M. bovis BCG | M. smegmatis | M. bovis BCG | ||
| pAV6950 | AGGGGTTCGAATCCCCTTAGCTCCAC | 100% | 100% | 100% in tRNAalaU | 100% in tRNAalaV |
| pBU | AGGGGTTCGAGTCCCCTTAGCTCCAC | 1.1% | 34% | ND | 100% in alaU |
| pBT | AGGGGTTCGAGTCCCCTAGGCTCCAC | 0.7% | 18% | ND | 100% in tRNAalaT |
| pSV | AGGGGTTCGATTCCCCTTAGCTCCAC | 12% | 0.8% | 100% tRNAalaV | ND |
| pST | AGGAGTTCGAATCTCCTAGGCTCCAC | 0.05% | ND | ND | ND |
a: Changes introduced by site-directed mutagenesis, with respect to the original attP site from pAV6950 are indicated in bold
b: Identical amounts of plasmid DNA were used to electroporate competent M. smegmatis or BCG. Integrants were numbered and the percentage of [integrants with modified attP/integrants with pAV6950] was calculated.
c: The insertion locus was determined by PCR amplification and the percentage of clones in the given tRNAala T-loop was calculated
In BCG, pBU, in which the sequence TTCGAA was mutated to TTCGAG to match the tRNAalaU T-loop, integration efficiency was 34% that for the non-mutated pAV6950 targeting tRNAalaV (Table 1). In all integrants, pBU targeted the tRNAalaU T-loop, as expected. When three base changes were introduced, to match the tRNAalaT T-loop, integration efficiency was 18% with respect to the non-mutated attP. In all clones tested, integration into tRNAalaT had occurred.
Ms6-derived vectors targeting two different tRNAala T-loops in BCG can be used to construct bivalent recombinant BCG strains
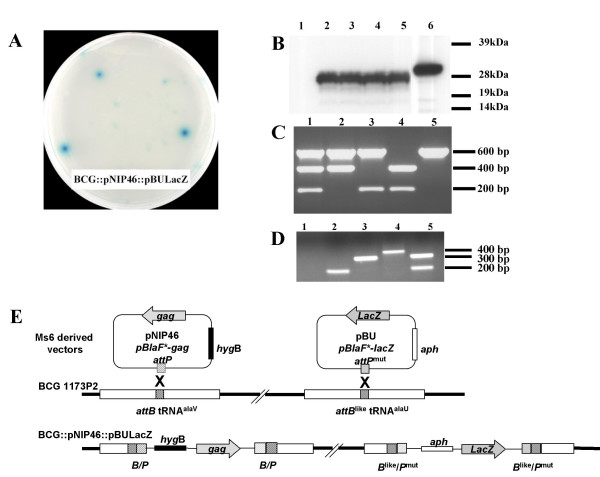
As pBU made possible integration into tRNAalaU, we investigated whether both tRNAalaU and tRNAalaV could be targeted in the same BCG strain. BCG was electroporated with pNIP46, a pAV6950-derived vector (targeting the tRNAalaV T-loop in BCG) containing the SIVmac251 gagp26 gene and a gene conferring resistance to hygromycin. The recombinant BCG::pNIP46 strain expressing gagp26 was then transformed with pBU-lacZ, containing the Escherichia coli lacZ and a kanamycin resistance gene. Recombinant clones, selected on medium supplemented with kanamycin and X-gal, displayed β-galactosidase activity (Fig. 2A). The expression of gagp26 was also detected in these clones by western blotting (Fig. 2B). PCR was used to determine the site of integration of the various plasmids in the BCG integrants (Fig. 2C). It was found that pNIP46 had inserted into tRNAalaV and pBU-lacZ into tRNAalaU. In the double integrant, BCG::pNIP46::pBU-lacZ, both tRNAalaU and tRNAalaV were disrupted. A diagram of this double integration of an Ms6-derived vector carrying natural attP in BCG tRNAalaV and an Ms6-derived vector with modified attP in BCG tRNAalaU is provided in Figure 2D.
Figure 2.
Bivalent recombinant BCG strain containing two pAV6950-derived vectors inserted into the tRNAalaU and tRNAalaV T-loops. BCG was transformed with the two vectors – pNIP46, targeting tRNAalaV, and pBU-lacZ, targeting tRNAalaU – to obtain a strain coexpressing the two foreign genes. A: Expression of the lacZ gene in rBCG::pNIP46::pBU-lacZ was detected as blue coloration on agar plates supplemented with the β-galactosidase substrate X-gal. B: Expression of gagp26 was detected by western blotting and wild-type BCG (lane 1), rBCG::pNIP46, (lane 2) and three different clones of rBCG::pNIP46::pBU-lacZ (lanes 3, 4, 5) were analysed. Lane 6 corresponds to purified recombinant Gagp26 produced in E. coli with an N-terminal His-tag. C: Multiplex PCR using the three sets of primers located upstream and downstream from the three different attB sites in BCG. Absence of the 200 bp, 400 bp or 600 bp amplification products indicates disruption of the tRNAalaU, tRNAalaV and tRNAalaT T-loops, respectively. The three fragments were present in the wild-type BCG chromosome (lane 1). After a single integration of pBU-lacZ into tRNAalaU, only the 600 and 400 bp fragments were present (lane 2), and in BCG::pNIP46, in which tRNAalaV had been disrupted, both the 600 and 200 bp fragments were amplified (lane 3). pBT disrupted the tRNAalaT locus (lane 4). In the double integrant, BCG::pNIP46::pBU-lacZ, only the tRNAalaT locus was intact and gave the 600 bp amplification fragment (lane 5). D Integration of each vector was confirmed by PCR using a mixture of the three primers upstream from tRNAalaU, tRNAalaV and tRNAalaT combined with LM2, annealing to the Ms6 integrase. No amplification was detected in wild type BCG chromosome (lane 1), by contrast, single products were detected in BCG::pBU-lacZ (lane 2), BCG::pNIP46 (lane 3) and BCG::pBT (lane 4). In BCG::pNIP46::pBU-lacZ two bands were obtained confirming the double integration (lane 5). E A diagram of the double integration of pNIP46 and pBU-lacZ is provided
Discussion
Determination of the complete genome sequences of M. tuberculosis [12] and M. smegmatis [17] made it possibe to analyse the possible integration targets of Ms6-derived vectors. In contrast to expectations [7], we observed that Ms6 targeted two different tRNAala genes in the fast-growing species M. smegmatis and the slow-growing species M. bovis BCG. The critical factor defining the exclusive integration site was the presence in the attB region of a core sequence identical to that of Ms6 attP. Thus, whereas in M. smegmatis 100% of the transformants obtained with Ms6-derived integration-proficient vectors displayed insertion into tRNAalaU, in BCG, all transformants carried the vector in tRNAalaV. In the case of L5, another temperate mycobacteriophage widely used for genetic studies [3], integration into the attB site of the BCG chromosome may occur in a tRNAglyV gene carrying one mismatch with the attP minimal core [5]. However, analysis of the three tRNAgly genes of BCG showed that there was no tRNAgly carrying an attB sequence identical to L5 attP in BCG (data not shown). L5-derived vectors therefore target an attB sequence carrying one mismatch with attP. This mismatch does not seem to affect the efficiency of L5 integration in BCG [5]. However, this mismatch maps outside the tRNAglyV 7 bp anticodon loop in which strand exchange beween attP and attB occurs [6].
The three tRNAala gene sequences displayed a high degree of similarity between M. smegmatis and BCG. Only two base differences in tRNAalaU and one base difference in tRNAalaV were observed. Interestingly, whereas the first 25 bases of the tRNAalaU and tRNAalaV anticodon loops were identical, the nucleotides involved in the formation of anticodon loop flanking symmetry differed in the tRNAalaU and tRNAalaV genes. This may be a signature of ancestral recombination between the tRNAala genes during evolution. The T-loops were remarkably conserved between tRNAalaU and tRNAalaV and between the species. However, one base difference occurred in this region containing the attB site. Point mutations occurring separately in the two species during evolution may account for these observations. Alternatively, the Ms6 phage or its ancestors may have introduced some of these mutations, as mycobacteriophages have actively participated in remodelling of the bacterial chromosome [19].
We next investigated whether the two "attB-like" sequences located in the two other tRNAala T-loops were potential target sites for modified Ms6. Indeed, with an integrase trained on one gene, a small change in the attP sequence may allow the element to switch to another tRNAala gene. We therefore mutated the 26-base attP core and assessed integration into the other mycobacterial loci. In BCG, one base difference between natural attP (TTCGAA) and pBU attP-like (TTCGAG) sequences reduced integration efficiency by 70%, despite identity with attB (tRNAalaU). The mutation introduced in attP was located in the 7 bp identical to the T-loop of the tRNAala gene but did not directly affect flanking symmetry (for review [1]. This change, although minor in nature, caused a dramatic drop in integration efficiency. Two additional changes in the 26-base sequence (pBT) only slightly reduced integration efficiency further in BCG, from the 70% decrease with pBU (one base change) to an 80% decrease with pBT (three base changes). This identifies the 7 bp T-loop sequence TTCGAA as important for Ms6 integration. Indeed, when M. smegmatis was electroporated with pBU (TTCGAG), despite there being only one base difference between this mutated attP and the three attB sites available in the genome, very few integrants were obtained (99% decrease in efficiency). Phages have been classified according to integration site sublocation within tRNA or tmRNA genes [20]. Class I phage integrase targets the tRNA anticodon loop whereas class II targets the tRNA T-loop. The precise crossover segment has been examined in class I phages, in which it maps to the 7 bp anticodon-encoding loop. This is also the case for mycobacteriophage L5 [6], which belongs to class I [20]. The precise location for crossover in class II phages remains unknown. Our data suggest that strand exchange between the class II phage Ms6 attP and mycobacterial attB occurs precisely in the 7 bp T-loop from tRNAala.
Integration-proficient vectors derived from the temperate mycobacteriophage Ms6 are particularly useful for constructing recombinant BCG strains that are genetically stable in vivo [10]. BCG is also of considerable interest for use in the development of vaccines for simultaneous immunisation against several pathogens. The next step was to use Ms6 integration-proficient vectors targeting different tRNAala T-loops in BCG to construct multivalent vaccine strains. We therefore constructed one vector carrying the natural attP core (TTCGAA/tRNAalaV), a SIVmac251 gene encoding Gagp26 and another carrying an attP site targeting tRNAalaU (TTCGAG) and carrying the lacZ gene from E. coli. In the rBCG::pNIP46::pBU-lacZ strain transformed with the two vectors, we observed expression of both the gagp26 and lacZ genes. Interestingly, the level of expression of gagp26 in the bivalent strain was similar to that observed in the monovalent rBCG::pNIP46 strain. This method may therefore be useful for the future development of new rBCG vaccines carrying multiple heterologous genes that will help in immunisation programmes.
Methods
Bacterial strains and plasmids
The E. coli, M. smegmatis, M. bovis BCG strains and the plasmids and primers used in this study are described in additional file 1. Mycobacterial strains were electroporated as previously described [21]. The plasmid pAV-SIV [10] was derived from pAV6950 [7], containing the attP-int region from mycobacteriophage Ms6 and an origin of replication from E. coli. It contains SIVmac251 genes inserted into the single EcoRI site of pAV6950 located upstream from the attP core and int gene. Similarly, pBU-lacZ contains the gene encoding β-galactosidase from E. coli inserted upstream from the modified attP core. Both pAV-SIV and pBU-lacZ express the aph3' gene from Tn5, conferring resistance to kanamycin (20 μg ml-1). pNIP46 is a pAV6950 derivative containing the hygB gene from Streptomyces hygrospicus conferring resistance to hygromycin (50 μg ml-1) and the gagp26 gene from SIVmac251. BCG transformants were selected on solidified Middlebrook 7H11 medium (Difco) supplemented with OADC (0.05% oleic acid, 5% bovine serum albumin fraction V, 2% dextrose, 0.004% beef catalase, 0.85% NaCl) and the appropriate antibiotic. β-galactosidase activity in BCG transformed with pBU-lacZ was visualised by adding X-gal (80 μg ml-1, MP Biomedicals, Inc.) to the medium.
PCR, Southern blotting and site-directed mutagenesis
PCR was performed in a final volume of 50 μl containing chromosomal DNA, 1.5 mM MgCl2, 200 μM of each dNTP, 50 pmoles of each primer and 1 unit of polymerase. Thirty-five cycles of [94°C 30 seconds, 60°C 30 seconds, 72°C 90 seconds] were run. For Southern blot analysis, genomic DNA from BCG and M. smegmatis pAV-SIV integrants was digested with BamHI for 18 hours at 37°C. Probes were generated by PCR amplification (see primers in Table 1), using M. tuberculosis cosmid 22D7 for tRNAalaU or cosmid 237 for tRNAalaV or genomic M. smegmatis DNA. For site-directed mutagenesis of the attP core site, the QuickChange™ Site-Directed Mutagenesis kit from Stratagene (La Jolla, CA) was used according to the manufacturer's instructions. This procedure uses double-stranded DNA and two primers complementary to opposite strands of the vector and each containing the desired mutation. For site-directed mutagenesis, only the sequence primers identical to the attP core site are listed in additional file 1. Both this primer and the complementary primer were used in the reaction.
Western blotting
Recombinant BCG colonies were grown at 37°C in Middlebrook 7H9 (Difco) medium supplemented with 10% ADC (5% bovine serum albumin fraction V, 2% dextrose, 0.003% beef catalase) and 0.05% Tween 80. We evaluated gagp26 expression with total protein extracts prepared as previously described [21]. Gagp26 was detected by incubation with a 1:500 dilution of polyclonal rabbit anti-Gag antibody followed by a 1:10000 dilution of anti-rabbit peroxidase-conjugated IgG (Amersham) and visualisation with an enhanced chemiluminescence kit (Amersham).
Abbreviations
BCG: Bacillus Calmette-Guérin; rBCG: recombinant BCG; SIV: simian immunodeficiency virus.
Authors' contributions
TDV and IM carried out the molecular and genetic studies and wrote the draft manuscript. They contributed equally to the work. VA carried out the southern blots. MP and JMP were involved in designing the studies. BG participated in the design of the study and contributed to the draft version of the manuscript. JMR participated in the design of the study, contributed to data analysis and helped with the writing of the draft manuscript. NW had the initial idea, co-ordinated the study and wrote the final version of the manuscript. All authors have read and approved the final manuscript.
Supplementary Material
Table S1. List of bacterial strains, primers and plasmids used in this study.
tRNAala secondary structures. Cartoon of the tRNAala genes secondary structures of M. smegmatis and M. bovis BCG designed by the tRNA-scan SE program [14].
Acknowledgments
Acknowledgements
We thank Edgar Badell for BCG cultures, Patricia Charles for excellent technical assistance and Jean Rauzier for sequencing. We also thank Alan Murray and Christophe Sola for critical reading of the manuscript. This work was supported by Agence Nationale de Recherche contre le SIDA and European Community contracts IC4A-1999-40006 and QLRT-PL-1999-00228 as well as Marie Curie fellowship for T. Dos Vultos.
Contributor Information
Tiago Dos Vultos, Email: tvultos@pasteur.fr.
Isabelle Méderlé, Email: imederle@wanadoo.fr.
Valérie Abadie, Email: abadie@chups.jussieu.fr.
Madalena Pimentel, Email: mpimentel@ff.ul.pt.
José Moniz-Pereira, Email: jpereira@ff.ul.pt.
Brigitte Gicquel, Email: bgicquel@pasteur.fr.
Jean-Marc Reyrat, Email: reyrat@necker.fr.
Nathalie Winter, Email: nwinter@pasteur.fr.
References
- Campbell AM. Chromosomal insertion sites for phages and plasmids. J Bacteriol. 1992;174:7495–7499. doi: 10.1128/jb.174.23.7495-7499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye C. Global epidemiology of tuberculosis. The Lancet. 2006;367:938–940. doi: 10.1016/S0140-6736(06)68384-0. [DOI] [PubMed] [Google Scholar]
- Hatfull GF, Sarkis GJ. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- Anes E, Portugal I, Moniz-Pereira J. Insertion into the Mycobacterium smegmatis genome of the aph gene through lysogenization with the temperate mycobacteriophage Ms6. FEMS Microbiol Lett. 1992;74:21–25. doi: 10.1111/j.1574-6968.1992.tb05337.x. [DOI] [PubMed] [Google Scholar]
- Lee MH, Pascopella L, Jacobs WR, Jr., Hatfull GF. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guerin. Proc Natl Acad Sci U S A. 1991;88:3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena CE, Stoner JE, Hatfull GF. Positions of strand exchange in mycobacteriophage L5 integration and characterization of the attB site. J Bacteriol. 1996;178:5533–5536. doi: 10.1128/jb.178.18.5533-5536.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas-Vieira A, Anes E, Moniz-Pereira J. The site-specific recombination locus of mycobacteriophage Ms6 determines DNA integration at the tRNA(Ala) gene of Mycobacterium spp. Microbiology. 1998;144 :3397–3406. doi: 10.1099/00221287-144-12-3397. [DOI] [PubMed] [Google Scholar]
- Bibb LA, Hatfull GF. Integration and excision of the Mycobacterium tuberculosis prophage-like element, PhiRv1. Molecular Microbiology. 2002;45:1515–1526. doi: 10.1046/j.1365-2958.2002.03130.x. [DOI] [PubMed] [Google Scholar]
- Lewis JA, Hatfull GF. Control of directionality in integrase-mediated recombination: examination of recombination directionality factors (RDFs) including Xis and Cox proteins. Nucl Acids Res. 2001;29:2205–2216. doi: 10.1093/nar/29.11.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méderlé I, Bourguin I, Ensergueix D, Badell E, Moniz-Pereira J, Gicquel B, Winter N. Plasmidic versus insertional cloning of heterologous genes in Mycobacterium bovis BCG : impact on in vivo antigen persistence and immune responses. Infect Immun. 2002;70:303–314. doi: 10.1128/IAI.70.1.303-314.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer B, Sander P, Sedlacek L, Ellrott K, Böttger EC. Instability and site-specific excision of integration-proficient mycobacteriophage L5 plasmids: development of stably maintained integrative vectors. Int J Med Microbiol. 2001;290:669–675. doi: 10.1016/S1438-4221(01)80004-7. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell BG, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- www.sanger.ac.uk
- www.sanger.ac.uk/projects/M.bovis
- Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. doi: 10.1126/science.284.5419.1520. [DOI] [PubMed] [Google Scholar]
- Brosch R, Gordon SV, Buchrieser C, Pym AS, Garnier T, Cole ST. Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast. 2000;17:111–123. doi: 10.1002/1097-0061(20000630)17:2<111::AID-YEA17>3.0.CO;2-G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://cmr.tigr.org
- Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucl Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR. Origins of highly mosaic mycobacteriophage genomes. Cell. 2003;113:171–182. doi: 10.1016/S0092-8674(03)00233-2. [DOI] [PubMed] [Google Scholar]
- Williams KP. Integration dites for genetic elements in prokaryotic tRNA and tmRNA genes : sublocation preference of integrase subfamilies. Nucl Acids Res. 2002;30:866–875. doi: 10.1093/nar/30.4.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter N, Lagranderie M, Rauzier J, Timm J, Leclerc C, Guy B, Kieny MP, Gheorghiu M, Gicquel B. Expression of heterologous genes in Mycobacterium bovis BCG: induction of a cellular response against HIV-1 Nef protein. Gene. 1991;109:47–54. doi: 10.1016/0378-1119(91)90587-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. List of bacterial strains, primers and plasmids used in this study.
tRNAala secondary structures. Cartoon of the tRNAala genes secondary structures of M. smegmatis and M. bovis BCG designed by the tRNA-scan SE program [14].