Abstract
Autophagy is a degradative process conserved among eukaryotic cells. It allows the elimination of cytoplasm including aberrant protein aggregates and damaged organelles. Accordingly, it is implicated in normal developmental processes and also serves a protective role in tumor suppression and elimination of invading pathogens, whereas defects in autophagy are associated with various human diseases including cancer and neurodegeneration. Atg proteins mediate the sequestration event that occurs at the preautophagosomal structure (PAS) by catalyzing the formation of double-membrane vesicles, termed autophagosomes. In Saccharomyces cerevisiae, the integral membrane protein Atg9 that is required for autophagy cycles through the PAS. Here, we demonstrate that Atg9 shuttles between this location and mitochondria. These data support a new model where mitochondria may provide at least part of the autophagosomal lipids and suggest a novel cellular function for this well-studied organelle.
Keywords: autophagy, cytoplasm to vacuole targeting, endoplasmic reticulum, lysosome
INTRODUCTION
Eukaryotic cells have developed various protective measures to defend themselves against internal and external stresses. Autophagy is one of the most valuable weapons employed to battle a wide range of menaces such as starvation, pathogens, tumors, aging and accumulation of toxic protein aggregates (reviewed in refs. 1 and 2). In addition, autophagy actively participates in certain physiological processes such as type II programmed cell death, development, cellular differentiation and remodeling (reviewed in ref. 3). The method of induction and the actual contribution of autophagy at the molecular level remain largely unknown in most of these situations. What is evident is that autophagy is able to rapidly eliminate unwanted cellular components such as large portions of the cytoplasm and unnecessary or damaged organelles by delivering its cargo into the lysosome/vacuole lumen where it is consumed by hydrolases. 4, 5 This is accomplished by the sequestration of the cargo by a large cytosolic double-membrane vesicle called an autophagosome that successively docks and fuses with the lysosome/vacuole.
Despite the identification of the Atg proteins, it remains unknown how autophagosomes are created. A major challenge in unveiling this process arises from the fact that the origin and the transport mode of the lipids employed to compose these structures is not known. Atg9 is the only integral membrane protein essential for double-membrane vesicle formation. This factor is probably transported to the site of autophagosome formation, the pre-autophagosomal structure (PAS), 6 together with at least part of the lipids or lipid bilayers required to create these vesicles. Interestingly, we have recently discovered that Atg9 cycles between the PAS and several unknown punctate structures dispersed in the cytosol. 7 Atg9 retrieval transport from the PAS is regulated by the Atg1-Atg13 signaling complex and requires Atg2, Atg18 and the phosphatidylinisitol-3-phosphate generated by the Atg14-containing PtdIns 3-kinase complex. Here, we examine the nature of the structures involved in Atg9 cycling.
MATERIALS AND METHODS
Strains and plasmids.
The S. cerevisiae strains used in this study are listed in Table 1. PCR-based integration of GFP at the 3' end of ATG9 was used to generate a strain expressing this fusion protein under the control of its native promoters. The template for the integration was pFA6a-GFP-HIS3. 8 PCR verification and prApe1 processing were used to confirm the functionality of this genomic fusion. The Spo7-RFP (promSPO7RFP424) and RFP-Sso1 (pRFPSSO1416) fluorescent chimeras were generated by excising GFP with only Bam HI or with HindIII and EcoRI from the YEplac122-TRP1-SPO7-GFP and pSSO1416 vectors respectively and replacing it with RFP. 9, 10 The BFP gene was obtained by PCR from the pQBIT7-BFP vector (Qbiogene, Irvine, CA) and cloned into pCu415 11 as a BamHI-SalI fragment, creating pCuBFPSKL415. The 3' primer introduced the coding sequence for SKL before the stop codon. Two-hybrid selection plasmids were created as follows: The pRS416-ProtA-APG9 plasmid 12 was cut with HindIII, blunted with the Klenow fragment and successively digested with SalI to release the ATG9 ORF. This fragment was then cloned into the pGBDU-C3 vector 24 cut with BamHI, blunted with the Klenow fragment and finally digested with SalI to create pGBDU-APG9. pGAD-APG9 was made by releasing ATG9 from pGBDU-APG9 with XmaI and PstI treatment and then subcloning into the pGAD-C3 vector. 13 The pRS416, pCuATG9-GFP(416), pTS470 and pYES-mtBFP plasmids have been described elsewhere. 14-17 The atg1ts allele 6 was cloned into pR415 16 creating pATG1ts415.
Table 1.
S. cerevisiae strains used in this study
| Name | Genotype | Reference |
|---|---|---|
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel GAL | 40 |
| FRY138 | SEY6210 ATG9-YFP::HIS3 S.k. atg1Δ::URA3 | 7 |
| FRY162 | SEY6210 ATG9-GFP:: HIS3 S.k. | This study |
| FRY172 | SEY6210 ATG9-PA::TRP1 pep4Δ::LEU2 | 7 |
| TVY1 | SEY6210 pep4Δ::LEU2 | 41 |
| PSY278 | SEY6210 ATG9-3HA:: HIS3 S.k. | This study |
| PJ69-4A | MATα leu2-3,112 trp1-901 ura3-52 his3-200 gal4 gal80 LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ | 13 |
The PpATG9 gene was amplified by PCR from P. pastoris genomic DNA and first cloned as a HindIII-AgeI fragment into the pSNA3416 plasmid. 18 This procedure generated a PpAtg9-GFP fusion under the control of the TPI1 promoter. The TPI1-PpATG9-GFP fragment was then excised by digestion with XhoI and KpnI and transferred into the P. pastoris pUC19-ARG4 integration vector 19 creating pPpATG9GFP-ARG4. This plasmid was linearized by digestion with NheI and transformed by electroporation into the P. pastoris PPY12 strain (arg4 his4) following a protocol previously described. 20 The new strain was called FRPPY002. To visualize P. pastoris ER in cells expressing PpATG9-GFP, FRPPY002 was transformed with the pIB2-DsRed-HDEL integration plasmid 21 creating the FRPPY003 strain.
Fluorescence microscopy.
Yeast cells were grown or starved in the appropriate medium before imaging. For fluorescent labeling of S. cerevisiae mitochondria, 1 ml of growing culture was incubated for 30 min in the presence of 1 μM MitoFluor Red 589 (Molecular Probes, Eugene, OR). Cells were then washed with the same culture medium before imaging in order to remove the excess dye. When necessary, mild conditions were used to fix the cells without destroying the different fluorescent proteins. Thus 2.5 ml of early log cultures were centrifuged, cells resuspended in 1 ml of freshly prepared fixation buffer (50 mM KH2PO4, pH 8.0, 1.5% formaldehyde, 1 μM MgCl2) and incubated at room temperature with occasional mixing for 30 min. The suspension was then centrifuged as above and cells washed twice in 1 ml of fixation buffer without formaldehyde before resuspension in 50-100 μl of the same solution for imaging. For P. pastoris imaging, 10 ml of culture between 0.25-0.8 OD600 were incubated in the presence of 0.05 μM MitoTracker Red CMXRos (Molecular Probes) at 30°C for 30 min. Cells were next centrifuged at 2,000 xg for 4 min, resuspended in 5 ml of fixation buffer and then treated as described above.
Fluorescence signals were visualized with the use of an IX71 fluorescent microscope (Olympus, Melville, NY). The images were captured with an IX-HLSH100 Olympus camera and deconvolved using the DeltaVision software (Applied Precision, Issaquah, WA). Time-lapse experiments were performed by collecting images of the same cells every 5 s for 4 min 10 s (50 pictures). Images were then organized without deconvolution in a movie using the same software.
Subcellular fractionations.
Overnight precultures were diluted in 40 ml of YPD medium and grown for 3-4 h to 1 OD600, harvested and resuspended in 4 ml of 100 mM Tris-HCl, pH 9.6, 10 mM dithiotreitol. After incubation at 30°C for 10 min, cells were harvested, resuspended in 5 ml YPD medium containing 1.2 M sorbitol supplemented with 10 mg lytic enzyme (ICN Biomedicals, Irvine, CA), incubated at 30°C for 40 min and washed in the same solution. Spheroplasts were lysed in 5 ml ice-cold hypo-osmotic buffer (50 mM K2HPO4, pH 7.5, 200 mM sorbitol, 1 mM EDTA) containing freshly added Complete EDTA-free protease inhibitors (Roche Molecular Biochemicals, Indianapolis, IN), 2 mM PMSF, 20 μg/ml α2-macroglobulin, 10 μg/ml pepstatin and 10 μg/ml antipain using 15 strokes in a Dounce homogenizer. The cell lysate was centrifuged twice at 500 xg for 5 min and 500 μl aliquots of the supernatant (TOT) fraction were subjected to 13,000 xg for 12 min. The pellet (P13) fractions were resuspended in 1 ml of hypo-osmotic buffer or the same solution plus 1 M KCl or 1% TX-100, whereas supernatants (S13) were mixed with 500 μl of the same buffer containing nothing, 2 M KCl or 2% TX-100. All samples were then centrifuged at 100,000 xg for 30 min. In order to isolate the proteins after the final centrifugations, supernatants were adjusted to 1 ml and pellets resuspended in the same volume of hypo-osmotic buffer before adding 50 μl of both 10% deoxycholate and 10 mg/ml bovine serum albumin. Tubes were kept on ice for 10 min prior to the addition of 120 μl of trichloroacetic acid and the incubation for other 30 min. After spinning the samples for 5 min, pellets were washed with ice-cold acetone and dried. Precipitates were resuspended in 80 μl of sample buffer with extensive sonication and vortexing in the presence of glass beads. Samples were then heated at 37°C for 15 min, proteins separated by SDS-PAGE and analyzed by immunoblot using antibodies or serum against PA (Dako, Glostrup, Denmark), Pgk1 (a generous gift of Dr. Jeremy Thorner, University of California, Berkeley), Por1 and Pho8 (Molecular Probes).
For membrane separation on sucrose gradients, 500 ml of mid log culture of wild type cells were treated as above maintaining the same cells/buffer ratios. The only differences were that spheroplasts were lysed in 5 ml of hypo-osmotic buffer and only 2 ml of the supernatant fraction (TOT) were subjected to the centrifugations at 13,000 xg. The final P13 pellet fraction was resuspended in 2 ml of hypo-osmotic buffer before being loaded on top of a 22-60% sucrose step gradient (1 ml 22%, 1.5 ml 27%, 2 ml each of 29 and 32%, 1.5 ml 34%, 1ml each of 37 and 60% sucrose in hypo-osmotic buffer). After centrifugation using a TH-641 swinging bucket rotor (Beckman Instruments, Palo Alto, CA) at 170,000 xg for 18 h, fourteen 0.84 ml fractions were collected from the top of the gradient. All fractions were mixed with 4X sample buffer, boiled for 5 min and resolved by SDS-PAGE followed by Western blot using antibodies or serum against Atg9, 22 Por1, Dpm1 (Molecular Probes), Fox3, 23 Vam3 24 and Sso2. 25 Percent sucrose in the final gradient was determined with a refractometer.
Immunoelectron microscopy.
Immunoelectron microscopy was performed as described previously. 26 Antibody dilutions were the following: 1:100 for the monoclonal anti-porin, 1:25 for the polyclonal anti-HA (Santa Cruz Biotechnology, Santa Cruz, CA) preabsorbed with yeast acetone powder, 1:30 for the 12-nm colloidal gold-Affinipure goat anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) and 6-nm colloidal gold-Affinipure goat anti-rabbit IgG (Electron Microscopy Sciences, Washington, PA).
Miscellaneous procedures.
Protein A affinity isolation, growth of yeast cultures and two-hybrid selection were performed as described previously. 13, 27
RESULTS
Atg9 partially localizes to mitochondria.
Atg9 is an integral membrane protein essential for double-membrane vesicle formation. In its absence, Atg components fail to be recruited to the PAS indicating that this factor plays a key role in organizing this specialized site. 6 Interestingly, Atg9 has a unique subcellular distribution; it localizes to several cytoplasmic punctate sites, one of which is the PAS. 7, 22 The peripheral, Atg9-containing punctate structures were located in proximity to the plasma membrane; in yeast this suggests the mitochondria and/or ER. 28 In order to identify the non-PAS sites where Atg9 is confined, we examined cells expressing a functional chromosomally-tagged Atg9-green fluorescent protein (GFP) fusion. Cells in early logarithmic (log) phase were stained with MitoFluor Red 589, a fluorescent dye that allows visualization of yeast mitochondria, which appeared as elongated organelles that formed a reticulum around the cell periphery ( Fig. 1A). 28 Atg9-GFP largely localized to these structures despite not having a known mitochondrial targeting sequence; however, its distribution was not homogeneous but was constrained to certain areas. Some of the Atg9-GFP-containing punctate structures were adjacent to the mitochondria without displaying an obvious overlap and others were found in the cytoplasm. Nitrogen starvation did not significantly affect the Atg9 localization pattern indicating that the distribution of this factor was the same during the formation of both Cvt vesicles and autophagosomes (Fig. 1A).
Figure 1.
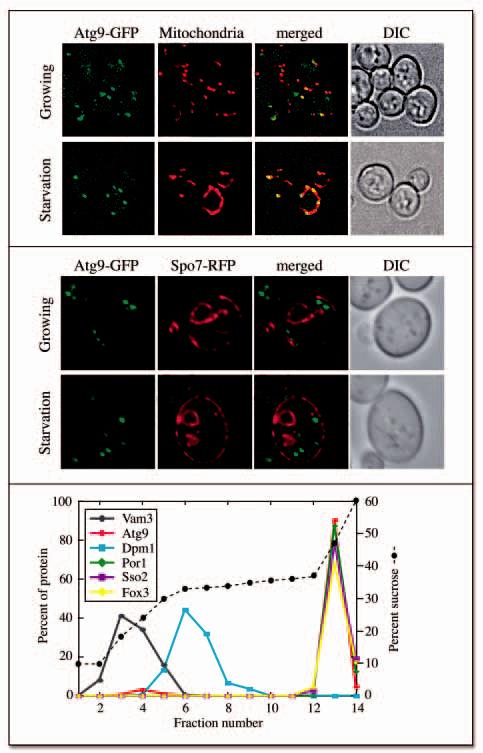
Atg9 localizes to mitochondria. (A) Atg9 distributes to mitochondria. The Atg9-GFP (FRY162) strain was grown to OD600 = 1.0 in YPD or starved in SD-N for 1 h. MitoFluor Red was added prior to imaging. DIC, differential interference contrast. (B) Atg9 does not localize to the ER. The Atg9-GFP strain carrying the SPO7RFP plasmid was grown and imaged as in (A). (C) A population of Atg9 fractionates with mitochondria. A P13 pellet fraction was separated on a sucrose density gradient as described in Materials and Methods. In total, 14 fractions were collected from the top of the gradient, resolved by SDS-PAGE, and membranes were probed with antisera to porin, Vam3, Sso2, Dpm1, Fox3 and Atg9.
We examined if Atg9-GFP was in part distributed to the ER by colocalizing it with the Spo7 marker. 10 As expected, a Spo7-red fluorescent protein (RFP) fusion showed a distribution typical of yeast ER (Fig. 1B). 10, 28 Atg9-GFP was often in close proximity to the ER, but did not colocalize with this organelle in either growing or starvation conditions.
To confirm the fluorescence microscopy observations we separated intracellular membranes on a sucrose density gradient. A total cell extract was prepared from the wild type strain and separated into 13,000 xg pellet and supernatant fractions. In agreement with previous studies, the PAS and most of Atg9 were in the supernatant fraction (Fig. 4A). 26, 29 Mitochondria were exclusively found in the pellet as monitored by a mitochondrial outer membrane protein, porin, along with a small portion of Atg9. To explore if this pool of Atg9 was associated with mitochondria, the membranes present in this fraction were resolved on a density gradient as described in Materials and Methods. Porin was concentrated in fraction 13 together with the plasma membrane protein Sso2 and the peroxisomal marker Fox3 (Fig. 1C). In contrast, the vacuolar SNARE Vam3 and the endoplasmic reticulum integral membrane protein Dpm1 were found in fractions 3-4 and 6-7, respectively, well separated from mitochondria. Atg9 was concentrated in the same fraction as the mitochondria.
Figure 4.
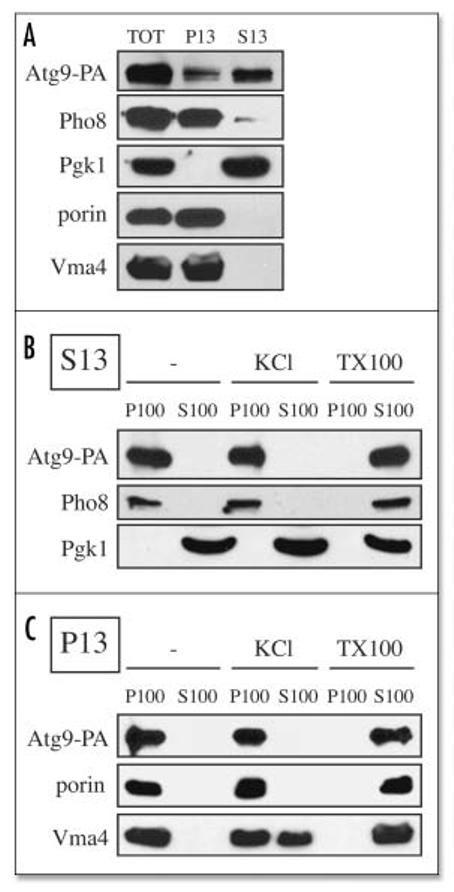
Atg9 is associated with membranous structures. (A) Most of the Atg9 is in a low speed supernatant fraction. Spheroplasts from Atg9-PA pep4Δ (FRY172) cells were lysed osmotically and fractionated at 13,000 xg for 12 min into total (TOT), supernatant (S13), and pellet (P13) fractions. After trichloroacetic acid precipitation, proteins were separated by SDS-PAGE and revealed by Western-blot using antibodies and antisera against protein A (PA), Pgk1, porin, Pho8 and Vma4. (B) Atg9 is always associated with membranes. S13 and P13 fractions prepared as in (A) were subjected to the following treatments: lysis buffer (-), 1 M KCl (KCl) and 1% Triton X-100 (TX-100) before being centrifuged at 100,000 xg for 30 min. Proteins from the supernatant (S100) and pellet (P100) fractions were analyzed as in (A).
To verify that Atg9 does not localize to peroxisomes and the plasma membrane, the Atg9-GFP-expressing strain was transformed with either a plasmid carrying the plasma membrane-localized RFP-Sso1 fusion or one expressing blue fluorescent protein (BFP) C-terminally tagged with the peroxisome targeting sequence SKL. Atg9-GFP did not localize to either of these organelles (Supplemental Fig. S1) suggesting that the Atg9 distribution observed on the gradient was due to its association with mitochondria.
To gain more information about the dynamic relationship between Atg9 and mitochondria, we designed a time-lapse experiment using fluorescence microscopy. The Atg9-GFP-expressing strain was imaged over a 2.5 min time course. In most cells, the Atg9 punctate structures moved rapidly but their trafficking was limited to the mitochondrial boundaries (Fig. 2A and Supplemental Video S2). In addition, the fluorescent signal of this chimera overlapped either totally or partially that generated by the mitochondrial dye, indicating that Atg9 was on the surface of the mitochondria. In some cases, we could see the Atg9-GFP separate from the mitochondria possibly representing a putative budding or fission event (Fig. 2B and Supplemental Video S3). Together, the microscopy and subcellular fractionation data demonstrate that a population of Atg9 associates with mitochondria.
Figure 2.

Clusters of Atg9 undergo dynamic movement on mitochondria. The Atg9-GFP (FRY162) strain was grown to an early log phase and stained with MitoFluor Red. Images of the same cells were collected every 5 s for 2.5 min. (A) The movement of three different Atg9-GFP-containing punctate structures within the mitochondrial boundaries is illustrated, and (B) a putative budding or fission event of an Atg9 cluster from mitochondria is shown. The white arrows mark a constant point for reference. For the complete movies see the supplemental data (Supplemental Video S2 and S3).
Atg9 cycles between mitochondria and the PAS in association with lipid bilayers.
Atg9 trafficking has been dissected with the use of a thermosensitive atg1 allele (atg1ts). 7 Inactivation of this kinase blocks the Atg9 recycling from the PAS causing the accumulation of this protein at this site. Successive Atg1 activation, however, allows Atg9 to redisperse in several punctate structures. In order to reveal whether Atg9 is shuttling between mitochondria and the PAS, we cotransformed an atg1Δ strain expressing Atg9-YFP with a vector carrying the atg1ts allele and a plasmid encoding for CFP-Ape1. Transformants were first grown at the permissive temperature of 24°C and incubated with MitoFluor Red prior to imaging. As expected, at the permissive temperature Atg9-YFP was distributed in several punctate structures some of which were adjacent to or overlapping with mitochondria (Fig. 3). In addition, and in agreement with previous work, only one Atg9-YFP puncta colocalized with the PAS identified by CFP-Ape1. 7, 22 The same cells were then transferred to 35°C for 1 h before being photographed again. As reported, 7 Atg1 inactivation induced Atg9 accumulation at the PAS accompanied by the concomitant disappearance of its pool on the mitochondria (Fig. 3). We concluded that Atg9 is transported from this organelle to the PAS. Importantly, the PAS is not part of the mitochondrial reticulum because it clearly did not localize to this compartment at the nonpermissive temperature.
Figure 3.
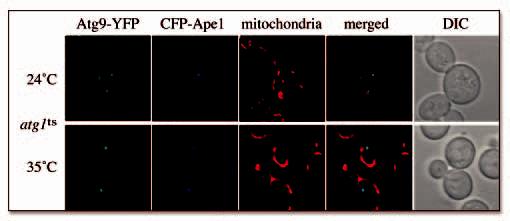
Atg9 cycles between mitochondria and the PAS. The ATG9-YFP atg1Δ (FRY138) strain was cotransformed with the pTS470 (CFP-Ape1) and pATG1ts415 plasmids. Transformants were grown at 24°C to an early log phase, incubated in the presence of MitoFluor Red for 40 min and photographed. The same cells were then transferred to 35°C for 1 h and imaged again.
From these experiments, the mode of Atg9 trafficking to and from the PAS could not be determined. In particular, it was unclear if this factor was transported together with membranes or if its multiple punctate localization patterns were achieved by association and dissociation reactions. Although the latter possibility seemed unlikely because Atg9 is an integral membrane protein, 22 we decided to explore whether the different pools of this protein were associated with lipid bilayers. Spheroplasts derived from wild type cells expressing Atg9-protein A (PA) were lysed and centrifuged at 13,000 xg for 12 min. In agreement with previous reports, the majority of Atg9 was detected in the S13 supernatant fraction that also contained the PAS (Fig. 4A). 26, 29 The complete absence of the cytosolic marker Pgk1 in the P13 pellet fraction indicated efficient lysis (Fig. 4A), suggesting that the Atg9 in the P13 fraction was not present within unlysed spheroplasts. Larger organelles such as the vacuole (Pho8 and Vma4) and mitochondria (porin), along with the Atg9 pool associated with this latter organelle, were in the P13 pellet fraction (Fig. 4A).
To gain insight into the nature of the Atg9 interaction with the lipid bilayers, both P13 and S13 fractions were treated with either 1 M KCl or 1% TX-100 before collecting membranes by centrifugation at 100,000 xg for 30 min. In general, Atg9 distribution was identical to that of two other integral membrane proteins, Por1 and Pho8, and clearly different from cytosolic Pgk1 and the peripheral membrane protein Vma4 (Fig. 4B). Basically, only the detergent was able to extract Atg9 from lipid bilayers. Thus we concluded that both the mitochondrial and the cytosolic/PAS Atg9 populations are associated with membranes. Our membrane separation at 13,000 xg indicated that a small fraction of Atg9, approximately 10-15%, was associated with mitochondria. This result appeared to contrast with our fluorescence data where the majority of this protein localized to this organelle (Figs. 1A and 4A). We suggest that this difference can be explained by the release of a pool of Atg9 from mitochondria during spheroplast lysis.
Atg9 is in a complex with itself.
The intensity of the fluorescent Atg9 signal suggested that Atg9 is organized in clusters (Figs. 1A and 2). To our knowledge, there are no reports describing the presence of either microdomains or lipid rafts on the mitochondrial outer membrane. One possibility that explains the discrete localization of Atg9 is the formation of homo- or hetero-oligomers. To test this hypothesis, we examined Atg9 self-interaction through a yeast two-hybrid analysis. Growth on medium lacking histidine after 2 days indicated that Atg9 was able to interact with itself (Fig. 5A).
Figure 5.
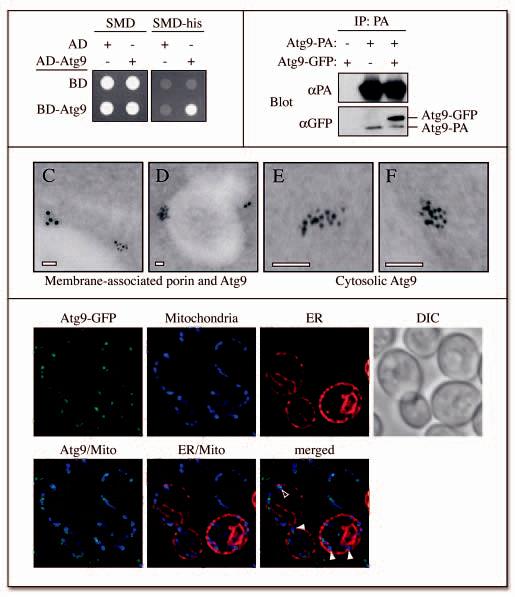
Atg9 is in a complex with itself. (A) A yeast two-hybrid analysis reveals that Atg9 can bind itself. pGAD-ATG9, pGDBU-ATG9 and the vectors without inserts were cotransformed in the appropriate combinations into the PJ69-4A strain. Interactions were monitored by the ability of cells to grow on plates lacking histidine (-his). (B) Atg9 affinity isolation. The Atg9-PA pep4Δ (FRY172) strain carrying either an empty plasmid (pRS416) or one expressing Atg9-GFP (pCuGFPATG9(416)), and the pep4Δ (TVY1) strain expressing Atg9-GFP were used to prepare detergent-solubilized extracts. Protein A fusions together with the associated proteins were affinity purified (IP) with IgG sepharose beads. Eluted polypeptides were separated by SDS-PAGE and visualized by immunoblotting with antibodies to PA (Dako, Glostrup, Denmark) and GFP (Covance Research Products, Berkeley, CA). (C-F) The Atg9-3HA (PSY278) strain expressing the ATG9-triple HA fusion under the control of the authentic ATG9 promoter was grown at 30°C in YPD medium to mid log phase and cells prepared for immuno-electron microscopy as described in Materials and Methods. (C) and (D) represent images of Atg9 (6 nm gold particles) colocalizing with porin (12 nm gold particles) around the mitochondrial surface, whereas (E) and (F) illustrate examples of cytosolic Atg9 clusters. White bars, 0.1 μm. (G) Atg9 is not concentrated to the MAMs. The ATG9-GFP (FRY162) strain carrying both the promSPO7RFP424 and pYES-mtBFP plasmids was grown overnight to an early log phase in a selective medium with galactose as the sole carbon source before being transferred for 3 h to medium containing glucose. Cells were then fixed as described in Materials and Methods and imaged. Mito, mitochondria. Open and closed arrowheads mark the locations of Atg9 colocalizing or not colocalizing with MAMs, respectively.
We next decided to examine if this interaction also occurred in physiological conditions. A strain expressing a genomic Atg9-PA fusion was transformed with either a plasmid encoding Atg9-GFP or an empty vector. Cells carrying the plasmid with Atg9-GFP but also expressing Atg9 not fused to protein A were used as a second control. Spheroplasts obtained from these three strains were lysed, the protein A chimera isolated using IgG-Sepharose beads, and the presence of Atg9-GFP tested by immunoblotting. The fluorescent chimera was selectively pulled down by the Atg9-PA fusion (Fig. 5B) indicating that Atg9 is in a complex with itself in the cytosol or on the mitochondria.
We extended our analysis with immuno-electron microscopy (IEM). A strain expressing a genomic Atg9-triple hemagglutinin (HA) fusion was prepared for IEM and sections were incubated with a polyclonal antiserum against HA (see Materials and Methods). Because the embedding technique employed alters the mitochondrial morphology, we used monoclonal antibody to porin to facilitate the identification of these organelles. The LR White resin gave a negative cell staining with white regions representing membranes (Fig. 5C, D), which we could identify as mitochondria based on staining with the anti-porin antibody (large gold particles). Small gold particles were always observed in clusters further suggesting that Atg9 self-assembles. In certain cases, Atg9 aggregates were observed in the cytosol (Fig. 5E and F) or in close proximity to the vacuole surface.
Finally, we decided to explore if the Atg9 aggregates were segregated in special domains of the mitochondrial network. The mitochondria-associated membranes (MAMs) are regions of the ER that are in close juxtaposition to the mitochondrial outer membrane and act as a conduit for the transfer of lipids but also seem to participate in calcium signaling. 30,31 It is completely unknown how these connections are maintained or the dynamics of the interactions. Furthermore, analysis of protein localization at the MAMs is problematic because no marker proteins have been characterized that exclusively localize to these sites. For this reason, we decided to attempt to visualize the MAMs by simultaneously transforming cells with a plasmid encoding BFP C-terminally tagged with a mitochondrial matrix targeting sequence under the control of a GAL promoter, and a plasmid expressing the ER marker Spo7-RFP. To explore whether Atg9 was concentrated to any sites of coincident ER-mitochondrial colocalization (putative MAMs), we integrated Atg9-GFP into this strain. Transformants were grown overnight in a selective medium with galactose as a sole carbon source in order to induce the expression of the fluorescent mitochondrial marker. Glucose was then added to the culture for 2-3 h before fixing and imaging the cells as described in Materials and Methods. As expected, the mitochondrial network was positioned in proximity to the peripheral ER, which in turn was adjacent to the plasma membrane (Fig. 5G). Contact regions between ER and mitochondria were also observed. Atg9 was sporadically found in these interconnected domains but in most cases it was clearly separated away from these sites. We conclude that Atg9 does not predominantly localize to the MAM sites.
The Pichia pastoris orthologue also localizes to mitochondria.
Like most of the Atg proteins, Atg9 is conserved among eukaryotic organisms (Supplemental Fig. S4). 3,5,32 To determine if Atg9 localization to mitochondria was unique for S. cerevisiae, we extended our analysis of its subcellular distribution to the P. pastoris Atg9 orthologue (PpAtg9; listed as Gsa14 in GenBank®) that is essential for pexophagy, a degradative pathway similar to autophagy that allows the elimination of excess peroxisomes. 33,34 A PpAtg9-GFP fusion was integrated into the P. pastoris PPY12 strain (see Materials and Methods). Fluorescent microscopy showed the presence of several punctate structures that were rapidly moving, making it difficult to determine whether there was colocalization with mitochondria (data not shown). In order to stop this movement, mild conditions were used to fix the cells, which were also incubated in the presence of 0.05 μM MitoTracker Red CMXRos. PpAtg9-GFP was distributed to several punctate structures, some of which colocalized with mitochondria (Fig. 6A). The portion of these structures not colocalizing with mitochondria was higher compared to S. cerevisiae. One possible explanation is that a larger Atg9 fraction was trafficking between the mitochondria and the PAS in P. pastoris. Starving the cells for nitrogen for 1 h did not alter PpAtg9-GFP distribution (Fig. 6A). We then examined if PpAtg9 was in part distributed to the ER by integrating DsRed-HDEL into the PpATG9-GFP-expressing strain. 21 As with S. cerevisiae, PpATG9 was not found in the ER (Fig. 6B). We concluded that Atg9 had a similar subcellular distribution in both S. cerevisiae and P. pastoris.
Figure 6.
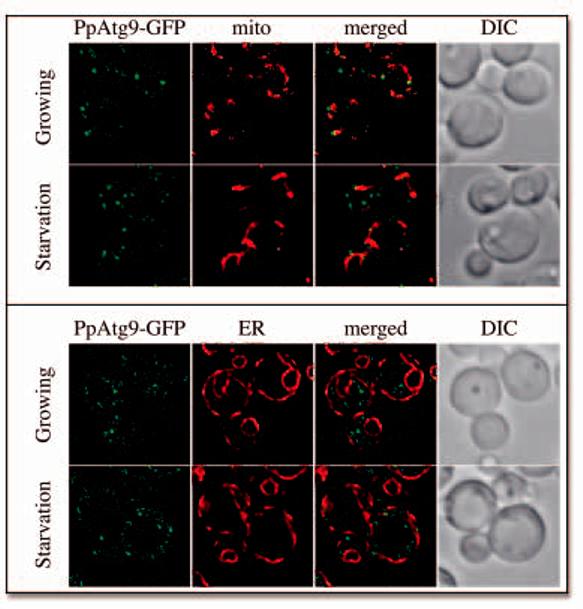
The P. pastoris Atg9 orthologue also localizes to mitochondria. (A) PpAtg9 is partially distributed to mitochondria. The PpATG9-GFP (FRPPY002) strain was grown to early log phase or starved in SD-N medium for 1 h, incubated in the presence of 0.05 μM MitoTracker Red CMXRos for 30 min and fixed as described in Materials and Methods before imaging. (B) PpAtg9 does not localize to the ER. The DsRedHDEL PpATG9-GFP (FRPPY003) strain was grown, fixed and imaged as in (A).
DISCUSSION
In recent years, the degradative pathway of autophagy has attracted great interest because of its involvement in many physiological processes and its induction in numerous pathological situations; however, several important questions are still unanswered. One of the most intriguing concerns the origin of the lipid bilayer used to create the double-membrane vesicles, the hallmark of this transport route. Localization studies of most Atg proteins in different model organisms have furnished no clues because these factors are principally cytosolic and only transiently associate with autophagosomes and the PAS. Accordingly, we have focused on Atg9, the only identified integral membrane protein essential for autophagosome formation. 22 We have recently shown that in S. cerevisiae this protein shuttles between the PAS and several cytoplasmic punctate sites. 7 In the present study we discovered that this punctate localization corresponds in part to mitochondria in both S. cerevisiae and P. pastoris revealing an undiscovered function for this organelle (Fig. 1A and C; Figs. 2 and 3; Fig. 5C, D and G; and Fig. 6A). It should be noted however, that the PAS is not part of the mitochondrial network (Fig. 3). Connections between autophagy and mitochondria have been previously made only in the yeast Hansenula poly-morpha undergoing pexophagy and in macrophages infected by the bacterium Legionella pneumophila. 35, 36 In both cases, the cargo being enwrapped within the double-membrane vesicle is surrounded by mitochondria prior to sequestration. Our data, however, cannot rule out the possibility that Atg9 is in a novel membranous structure closely attached to the outside of the mitochondria.
Atg9 was observed both on the mito-chondria and on the PAS supporting the model that this factor shuttles between these two locations (Fig. 3); 7 however, we cannot exclude the possibility that a population of Atg9 localizes to another structure or organelle for which we do not have adequate marker proteins. The function of Atg9 cycling is not clear, but one possibility is that it is required to supply the forming autophagosomes, at least in part, with membrane. This idea is supported by the fact that Atg9 was always associated with lipid bilayers and some of the Atg9 aggregates were cytosolic (Fig. 1A; Figs. 3 and 4; and Fig. 5C-F). The ER is the principal lipid factory in the cell but mitochondria actively participate in the synthesis and modification of some lipids. The mitochondria-associated membranes (MAM) represent this close relationship between these two organelles in regard to lipid processing. MAM are regions of the ER that are in close juxtaposition to the mitochondrial outer membrane and act as a conduit for the transfer of lipids. 31 Thus, mitochondria are a potential source membrane for the autophagosome. Atg9 however, did not appear to localize to the MAM (Fig. 5G). Even if the “lipid supply” model is correct, we cannot exclude that Atg9 carries out other functions. Under certain conditions, autophagy becomes the principal source of energy for the cell. Because the mitochondria provide the other primary supply of energy, one could imagine that Atg9 is used to coordinate the two sources.
Another aspect of Atg9 localization revealed by our work is that this protein was not homogeneously distributed along the entire mitochondrial surface but was concentrated to discrete areas that have a high mobility in between the organelle boundaries (Fig. 1A; Fig. 2, 5G and 6A). The observation that Atg9 forms clusters is consistent with our IEM analysis of this protein as well as a previous study (Fig. 5C-F). 22 So far, there are no reports describing the presence of microdomains or specialized regions on the mitochondrial outer membrane. Our biochemical data indicate that one explanation for Atg9 clustering is its ability to self-associate (Fig. 5A and B). However, our two-hybrid analysis in yeast and the coimmunoprecipitation experiment cannot exclude the possibility that one or more proteins that are present in these aggregates bridge Atg9 homo-association.
The majority of the Atg9 complexes were on the mitochondrial network (Fig. 1A). After subcellular fractionation, however, only a relatively small amount of this protein was found bound to this organelle (Fig. 4A). Thus cell lysis seems to provoke Atg9 dissociation from mitochondria. This loss of binding could be caused by the stimulation of the Atg9 sorting event. Alternatively, release of Atg9 could be due to the peripheral association of certain Atg9 aggregates. Importantly, the released material is associated with lipid (Fig. 4B) and further stripping of the mitochondria with high salt released additional membrane bound Atg9 (our unpublished data). All together, these findings support the idea that Atg9 leaves mitochondria in association with membranous structures, possibly vesicles or small cisternae. It is interesting to notice that the Atg9-containing clusters released from mitochondria or present in the cytosol (S13 supernatant fraction) have a very similar density as that of the PAS because they cofractionate on density gradients. 26
During the preparation of this manuscript, a report was published that demonstrates that the two human proteins with high homology to Atg9, HsAtg9L1 and HsAtg9L2, are its functional orthologues. 32 The yeast P. pastoris is evolutionarily close to S. cerevisiae and not surprisingly Atg9 localization in this organism was almost identical. In contrast, our preliminary results with mammalian cells indicate that HsAtg9L1 32 was not distributed on the mitochondrial network indicating that higher eukaryotes could supply autophagy with membranes by extracting lipids from a different reservoir (data not shown). This observation could also explain why HsAtg9 cannot substitute for the yeast Atg9 (data not shown). 32 In human adult tissues, HsATG9L1 is ubiquitously expressed whereas HsATG9L2 is highly expressed in placenta and pituitary gland. Importantly, the authors showed that these two factors are not distributed on mitochondria but to a perinuclear region in complete support of our preliminary observations. It is interesting to note, however, that HsAtg9L2 possesses a non-functional mitochondrial targeting sequence that is also present in its closest higher eukaryote homologues. 32 This characteristic raises the possibility that this is an ancient localization signal.
What is the role of Atg9-containing membranes during autophagosome biogenesis? In the current model, the elongation of a small template membrane, termed the isolation membrane or phagophore, leads to autophagosome formation in mammalian cells (Fig. 7). 22, 37 The surface of this small cisterna is decorated with Atg16 and Atg5. 37, 38 Conjugation of the ubiquitin-like Atg12 to Atg5 seems to be the induction step that triggers the elongation of this compartment. It is not known how the Atg12-Atg5 complex recruits additional lipid bilayers but the crescent autophagosome acquires more Atg12-Atg5 as well as a second ubiquitin-like molecule, Atg8/LC3, that is unconventionally linked to phosphatidylethanolamine. In yeast, the PAS very likely represents this growing cisterna. In the absence of Atg9, Atg5 is not targeted to the PAS and accordingly Atg8/LC3-containing membranes cannot be recruited to this location, thus double-membrane vesicles are not formed. 6, 22 Atg8-containing membranes seem to have a different origin than the mitochondria, possibly the Golgi apparatus. 26, 39
Figure 7.
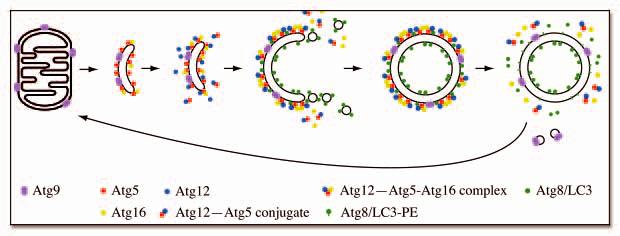
Putative model for Atg9 trafficking in yeast S. cerevisiae. Atg9-containing membranes are derived from mitochondria and become an isolation membrane or phagophore by acquiring Atg16 and Atg5. The covalent linkage of Atg12 to Atg5, and other events that follow autophagic induction trigger autophagosome formation. Additional cytosolic Atg12-Atg5 conjugate are recruited to this compartment and start to redistribute, becoming concentrated mostly on the external lipid bilayer. The Atg12-Atg5-Atg16 complex then forms larger oligomers that dictate the elongation of the phagophore by conveying Atg8-containing membranes and possibly additional Atg9 clusters to the crescent structure. Once the autophagosome is completed, the Atg12-Atg5 conjugate and Atg16 dissociate from this structure whereas Atg4 releases the Atg8 on the external lipid bilayer into the cytosol by proteolytically cleaving this molecule from the lipid moiety. At the same time, Atg9 is recycled back to mitochondria for reuse. These final uncoating events allow the autophagosome to fuse with the lysosome/vacuole.
Our work has revealed Atg9 to be the first Atg component that localizes to a distinct subcellular compartment making it the first marker that allows monitoring at least part of the lipid bilayers used during autophagosome assembly. Future work on this factor will help to unveil the mode of membrane transport to and from the PAS. In addition, identification of proteins involved in the sorting of Atg9 from the mitochondria, or present in the Atg9 aggregates, will help to explain how autophagosomes are formed and potentially also reveal how their lipid bilayers are derived from the membrane origin, providing a model for the study of the same process in higher eukaryotes.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank R. Tsien for providing the mRFP1 plasmid, and B. Glick, Y. Ohsumi, S. Siniossoglou, P. Stromhaug and W. Wickner for reagents. We are also grateful to Drs. J. Komduur and W.-P. Huang for providing plasmids and H. Chong for providing technical assistance. This work was supported by the National Institutes of Health Public Health Service grant GM53396 (to D.J.K.). F.R. is supported by a Swiss National Foundation Fellowship for advanced researchers.
References
- 1.Levine B. Eating oneself and uninvited guests; autophagy-related pathways in cellular defense. Cell. 2005;120:159–62. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Shintani T, Klionsky DJ. Autophagy in health and disease: A double-edged sword. Science. 2004;306:990–5. doi: 10.1126/science.1099993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–77. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 4.Klionsky DJ, editor. Autophagy. Landes Bioscience; Georgetown, TX: 2004. [Google Scholar]
- 5.Reggiori F, Klionsky DJ. Autophagy in the eukaryotic cell. Eukaryot Cell. 2002;1:11–21. doi: 10.1128/EC.01.1.11-21.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. doi: 10.1093/emboj/20.21.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 8.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–61. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 9.Reggiori F, Black MW, Pelham HRB. Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol Biol Cell. 2000;11:3737–49. doi: 10.1091/mbc.11.11.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siniossoglou S, Santos-Rosa H, Rappsilber J, Mann M, Hurt E. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 1998;17:6449–64. doi: 10.1093/emboj/17.22.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Labbé S, Thiele DJ. Copper ion inducible and repressible promoter systems in yeast. Methods Enzymol. 1999;306:145–53. doi: 10.1016/s0076-6879(99)06010-3. [DOI] [PubMed] [Google Scholar]
- 12.Wang C-W, Kim J, Huang W-P, Abeliovich H, Stromhaug PE, Dunn WA, Jr, Klionsky DJ. Apg2 is a novel protein required for the cytoplasm to vacuole targeting, autophagy, and pexophagy pathways. J Biol Chem. 2001;276:30442–51. doi: 10.1074/jbc.M102342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–36. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Huang W-P, Stromhaug PE, Klionsky DJ. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J Biol Chem. 2002;277:763–73. doi: 10.1074/jbc.M109134200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shintani T, Huang W-P, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. doi: 10.1016/s1534-5807(02)00373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Westermann B, Neupert W. Mitochondria-targeted green fluorescent proteins: Convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421–7. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 18.Reggiori F, Pelham HRB. Sorting of proteins into multivesicular bodies: Ubiquitin-dependent and -independent targeting. EMBO J. 2001;20:5176–86. doi: 10.1093/emboj/20.18.5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rossanese OW, Soderholm J, Bevis BJ, Sears IB, O’Connor J, Williamson EK, Glick BS. Golgi structure correlates with transitional endoplasmic reticulum organization in Pichia pastoris and Saccharomyces cerevisiae. J Cell Biol. 1999;145:69–81. doi: 10.1083/jcb.145.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sears IB, O’Connor J, Rossanese OW, Glick BS. A versatile set of vectors for constitutive and regulated gene expression in Pichia pastoris. Yeast. 1998;14:783–90. doi: 10.1002/(SICI)1097-0061(19980615)14:8<783::AID-YEA272>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 21.Bevis BJ, Hammond AT, Reinke CA, Glick BS. De novo formation of transitional ER sites and Golgi structures in Pichia pastoris. Nat Cell Biol. 2002;4:750–6. doi: 10.1038/ncb852. [DOI] [PubMed] [Google Scholar]
- 22.Noda T, Kim J, Huang W-, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol. 2000;148:465–80. doi: 10.1083/jcb.148.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutchins MU, Veenhuis M, Klionsky DJ. Peroxisome degradation in Saccharomyces cerevisiae is dependent on machinery of macroautophagy and the Cvt pathway. J Cell Sci. 1999;112:4079–87. doi: 10.1242/jcs.112.22.4079. [DOI] [PubMed] [Google Scholar]
- 24.Nichols BJ, Ungermann C, Pelham HRB, Wickner WT, Haas A. Homotypic vacuolar fusion mediated by t- and v-SNAREs. Nature. 1997;387:199–202. doi: 10.1038/387199a0. [DOI] [PubMed] [Google Scholar]
- 25.Aalto MK, Ronne H, Keranen S. Yeast syntaxins Sso1p and Sso2p belong to a family of related membrane proteins that function in vesicular transport. EMBO J. 1993;12:4095–104. doi: 10.1002/j.1460-2075.1993.tb06093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reggiori F, Wang C-W, Nair U, Shintani T, Abeliovich H, Klionsky DJ. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol Biol Cell. 2004;15:2189–204. doi: 10.1091/mbc.E03-07-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reggiori F, Wang C-W, Stromhaug PE, Shintani T, Klionsky DJ. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J Biol Chem. 2003;278:5009–20. doi: 10.1074/jbc.M210436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prinz WA, Grzyb L, Veenhuis M, Kahana JA, Silver PA, Rapoport TA. Mutants affecting the structure of the cortical endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Biol. 2000;150:461–74. doi: 10.1083/jcb.150.3.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tucker KA, Reggiori F, Dunn WA, Jr, Klionsky DJ. Atg23 is essential for the cytoplasm to vacuole targeting pathway and efficient autophagy but not pexophagy. J Biol Chem. 2003;278:48445–52. doi: 10.1074/jbc.M309238200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rizzuto R, Pinton P, Carrington W, Fay FS, Fogarty KE, Lifshitz LM, Tuft RA, Pozzan T. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science. 1998;280:1763–6. doi: 10.1126/science.280.5370.1763. [DOI] [PubMed] [Google Scholar]
- 31.Daum G, Vance JE. Import of lipids into mitochondria. Prog Lipid Res. 1997;36:103–30. doi: 10.1016/s0163-7827(97)00006-4. [DOI] [PubMed] [Google Scholar]
- 32.Yamada T, Carson AR, Caniggia I, Umebayashi K, Yoshimori T, Nakabayashi K, Scherer SW. Endothelial nitric oxide synthase antisense (NOS3AS) gene encodes an autophagy-related protein (APG9-like2) highly expressed in trophoblast. J Biol Chem. 2005;280:18283–90. doi: 10.1074/jbc.M413957200. [DOI] [PubMed] [Google Scholar]
- 33.Stromhaug PE, Bevan A, Dunn WA., Jr GSA11 encodes a unique 208-kDa protein required for pexophagy and autophagy in Pichia pastoris. J Biol Chem. 2001;276:42422–35. doi: 10.1074/jbc.M104087200. [DOI] [PubMed] [Google Scholar]
- 34.Mukaiyama H, Oku M, Baba M, Samizo T, Hammond AT, Glick BS, Kato N, Sakai Y. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells. 2002;7:75–90. doi: 10.1046/j.1356-9597.2001.00499.x. [DOI] [PubMed] [Google Scholar]
- 35.Kiel JAKW, Venhuis M. In: Autophagy. Klionsky DJ, editor. Landes Bioscience; Georgetown, TX: 2004. pp. 126–37. [Google Scholar]
- 36.Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: Implications for conversion of plasma membrane to the ER membrane. J Cell Sci. 2001;114:4637–50. doi: 10.1242/jcs.114.24.4637. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–68. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–88. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 39.Legesse-Miller A, Sagiv Y, Glozman R, Elazar Z. Aut7p, a soluble autophagic factor, participates in multiple membrane trafficking processes. J Biol Chem. 2000;275:32966–73. doi: 10.1074/jbc.M000917200. [DOI] [PubMed] [Google Scholar]
- 40.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: Isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–48. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gerhardt B, Kordas TJ, Thompson CM, Patel P, Vida T. The vesicle transport protein Vps33p is an ATP-binding protein that localizes to the cytosol in an energy-dependent manner. J Biol Chem. 1998;273:15818–29. doi: 10.1074/jbc.273.25.15818. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


