Abstract
We describe here experiments designed to characterize the secondary structure of amyloid fibrils of the Alzheimer's amyloid plaque peptide Aβ, using hydrogen-deuterium exchange measurements evaluated by mass spectrometry. The results show that ≈50% of the amide protons of the polypeptide backbone of Aβ(1–40) resist exchange in aqueous, neutral pH buffer even after more than 1,000 h of incubation at room temperature. We attribute this extensive, strong protection to H-bonding by residues in core regions of β-sheet structure within the fibril. The backbone amide hydrogens exchange at variable rates, suggesting different degrees of protection within the fibril. These data suggest that it is unlikely that the entire Aβ sequence is involved in H-bonded secondary structure within the amyloid fibril. Future studies using the methods described here should reveal further details of Aβ fibril structure and assembly. These methods also should be amenable to studies of other amyloid fibrils and protein aggregates.
Tissue deposits of highly insoluble amyloid fibrils are associated with a variety of human diseases, including Alzheimer's disease (AD) (1) and noninsulin-dependent diabetes (2). More than 15 different proteins are known to form amyloid fibrils. In AD, the main components of amyloid fibrils are the peptides collectively known as Aβ, molecules ranging from 39 to 43 residues in length derived from proteolytic cleavage of the amyloid precursor protein APP (3). The realization that each of the four genes associated with familial forms of AD have an impact on the in vivo concentrations or amyloidogenicity of Aβ has provided strong support for the role of Aβ amyloid assembly in the disease mechanism (4).
Proteins capable of forming amyloid in human diseases have no significant similarities to one another in amino acid sequence, molecular weight, or native folding patterns (5). Despite the dissimilarity of the precursor proteins, amyloid fibrils derived from these different proteins share a number of structural features. Electron microscopy and atomic force microscopy show that fibrils produced by many proteins are 80–100 Å in diameter and several μm in length and are composed of a number of smaller (30–40 Å diameter) protofilaments twisted about each other along the fibril axis (6, 7). Most amyloid fibrils exhibit similar spectroscopic response to the heteroaromatic dyes Congo red and thioflavin T (5), suggesting binding sites for these dyes common to all amyloid fibrils. Both x-ray diffraction (8) and IR spectra (9) show that most of the definable secondary structure of all amyloid fibrils is some form of β-sheet. Solid-state NMR data on the fibrils also have been interpreted to indicate that the Aβ fibril structure includes a parallel β-sheet (10). Unfortunately, amyloid fibrils have proved resistant to conventional methods of higher-order protein structure determination such as x-ray crystallography (fibrils are noncrystalline) and multidimensional NMR (fibrils have short relaxation times and therefore provide broad, unresolved resonances). The lack of high-resolution structural information limits our ability to understand how peptides and proteins with dissimilar native folds are capable of accessing a common amyloid folding motif. Higher-resolution structural data also would facilitate drug design targeting amyloid fibrils.
Significant insight into fibril structure could be derived from knowledge of the pattern and extent of hydrogen bonding within the fibril. For example, we know that amyloid fibrils formed by the Alzheimer's disease peptide Aβ(1–40) are rich in H-bonded β-sheet (8–10), but we do not know which of the 39 backbone amide protons are engaged in H-bonds or even the proportion of total backbone amides so engaged. Defining the secondary structural roles of various primary sequence elements along the fibril-incorporated Aβ peptide would provide important new information that could, for example, be used to build and test models of amyloid fibril structure (8, 11–15).
Hydrogen-deuterium exchange (HX) has proven very useful in defining structural perturbations within globular proteins (16–22). Protons attached to heteroatoms, which normally undergo rapid exchange, experience much slower exchange when involved in H-bonded structures like α-helices and β-sheets and/or when protons are sterically inaccessible to the solvent (16, 17). Analysis normally involves exposing the protein to exchange conditions in D2O, followed by determination of the incorporated deuterium content by methods such as NMR (16–18), MS (19–22), or Fourier transform IR spectroscopy (23, 24).
In this paper we present a protocol enabling rapid fibril dissolution and MS evaluation of the hydrogen bonding of fibrillar Aβ(1–40) by HX. This method provides structural insights for improving our understanding of fibril assembly and offers the prospect for even more detailed information in the future.
Materials and Methods
Fibril Synthesis.
Chemically synthesized Aβ(1–40) monomer (Keck Biotechnology Center, Yale University) (NH2-DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVV-COOH) was used in the experiments described here. To eliminate aggregates from the starting peptide, the peptide was treated with trifluoroacetic acid (Pierce) and hexafluoroisopropanol (Sigma) as described by Zagorski et al. (25). To grow fibrils, this disaggregated Aβ was dissolved in H2O to a concentration of 0.5 mg/ml (concentration determined by HPLC analysis against a standard quantified by amino acid composition analysis). An equal volume of PBS (20 mM phosphate, 276 mM sodium chloride, 5.4 mM potassium chloride, pH 7.4) was added and this solution was further cleansed of potential Aβ aggregates by extended ultracentrifugation for at least 15 h. Fibril formation then was initiated by addition of a small quantity (0.2% by weight compared with monomer) of fibrillar Aβ aggregates from a previous fibril synthesis. The mixture was incubated without agitation at 37°C, and fibril growth was monitored by thioflavin T fluorescence (26) until complete (4–6 days). The quality of fibrils was assessed by electron microscopy.
Sample Preparation.
For HX experiments on the monomer, Aβ(1–40) was processed as described above, except that dried peptide after organic solvent treatment was dissolved in 2.5 mM Tris⋅HCl buffer, pH 7.5 (in H2O or D2O depending on the experiment). For HX experiments on fibrillar aggregates of Aβ(1–40), fibrils were first collected by centrifugation, washed once with water, and then resuspended in 2.5 mM Tris⋅HCl or deuterated Tris⋅HCl buffer at pH 7.5.
Electrospray Ionization-MS.
A Micromass (Manchester, U.K.) Quattro II triple quadrupole electrospray ionization mass spectrometer was used in the experiments presented here. Experiments were performed in the positive ion mode with a capillary voltage of 3.0 kV and a cone voltage of 20 V. A 0.25-mm diameter T union (Valco Instruments, Houston) was connected to the electrospray sampling capillary. Two 100-μm i.d. fused silica capillaries (Polymicro, Phoenix) were used for infusing sample (2 μl/min) and processing solvent (18 μl/min) into the T. Sample solutions and processing solvents were delivered with Harvard Bioscience model 22 and model 11 syringe pumps (South Natick, MA), respectively. Source temperature was maintained at 80°C. The flow rates of nebulizing gas and drying gas were set at 20 liter/h and 400 liter/h, respectively.
All spectra were acquired in the multichannel accumulation mode with mass range from 700 to 1,200 Da at a scan rate of 250 Da/s for 1 min. The spectra were smoothed by averaging over a moving three-point window. Centroids of unresolved isotopic envelopes were used to calculate the average molecular mass (corrected for excess protons or deuterons depending on the sample). Charge states +5 and +6 were consistently observed with good signal-to-noise ratio, and therefore molecular masses obtained from these charge states were averaged to obtain the molecular masses in all of the experiments presented here.
Results
Methodological Considerations.
Intact fibrils are far too heavy and heterogeneous for direct mass analysis. MS evaluation of HX in amyloid fibrils therefore requires two steps. In the first step, fibrils are incubated in deuterated buffer to allow forward exchange of exposed, labile hydrogens for deuteriums. In the second step, the resulting partially deuterated fibrils must be rapidly dissolved and disaggregated into monomers before data collection. Using a D2O-based processing solvent for dissolution and disaggregation can result in artifactual additional forward exchange, whereas processing with an H2O-based solvent induces back-exchange. In either case, the desired exchange information is compromised to the extent of the exchange occuring during sample work-up. A 50:50:0.2 (vol/vol/vol) water/acetonitrile/formic acid processing solvent was found to minimize these effects. As described below, this mixture was compatible with MS detection of Aβ and provided rapid fibril dissolution. Moreover, it provided a pH in the range (2.5–3) known to greatly reduce the rate of H/D exchange of many labile protons (27). Although it is in principle possible to correct for the inevitable sample work-up exchange (28), it is preferable to keep the needed corrections (and resulting uncertainties) small by minimizing not only the rate of these exchanges, but also the time for dissolution of fibrils and injection into the MS. This was accomplished by loading fibrillar material into the MS through a mixing T in which fibrils encounter the sample processing solvent, dissolve, and are immediately injected into the MS, all within 10 s. All of the experiments described below were conducted in an identical manner with this on-line mixing T sample port by using either protonated or deuterated forms of the water/acetonitrile/formic acid solvent mixture.
Method Validation.
To assess the ability of our MS system to observe Aβ peptides, ≈30 μM Aβ(1–40) monomer in 2.5 mM aqueous Tris⋅HCl buffer (pH 7.5) was continuously infused through one arm of the T, where it was mixed with aqueous processing solvent (H2O/CH3CN/HCOOH) being pumped through the second arm of the T. The resulting mixture then was directed to the electrospray emitter through the third arm of the T. Fig. 1a presents a typical result showing the +6 charge state from the mass spectrum of Aβ(1–40). The measured mass (4329.5 Da; Table 1) for Aβ(1–40) is in good agreement with its calculated mass (4329.9 Da). To test the ability of the processing solvent to rapidly dissolve fibrils in the T, we injected a sample of protonated fibrils (equivalent to 11 μM monomer concentration) suspended in 2.5 mM Tris⋅HCl. The resulting spectrum (Fig. 1b) was virtually identical to that of Fig. 1a, resulting in a molecular mass of 4329.5 Da.
Figure 1.
Electrospray ionization-MS of (a) protonated Aβ monomer, (b) protonated Aβ fibrils, (c) deuterated Aβ monomer in deuterated solvent, and (d) deuterated Aβ monomer in protonated solvent. The protein sample and processing solution were pumped through two arms of the T at flow rates of 2 μl/min and 18 μl/min, respectively. The final concentration of Aβ monomer was 2.9 μM, and the equivalent concentration of monomers from fibrils was ≈1.1 μM. Several charge states were obtained but only the +6 charge state is shown here for clarity. The average molecular mass (corrected by subtracting the mass of ionizing protons or deuterons) calculated by using data from +6 and +5 charge states is indicated for each sample.
Table 1.
Summary of molecular masses observed for Aβ (1–40) peptide under the indicated conditions
| Sample | Number of runs | Observed molecular mass, Da* | Mass increase from protonated Aβ Da* | Mass increase after side-chain correction, Da*† |
|---|---|---|---|---|
| Protonated Aβ monomer or fibrils with protonated processing solvent | 6 | 4,329.4 ± 0.1 | ||
| Deuterated Aβ monomer with deuterated processing solvent | 10 | 4,395.1 ± 0.2 | 65.8 ± 0.2 | 38.8 ± 0.2 |
| Deuterated Aβ monomer with protonated processing solvent | 41 | 4,361.7 ± 1.1 | 32.3 ± 1.1 | 27.3 ± 1.1 |
| Deuterated Aβ fibrils with protonated processing solvent‡ (incubation time ∼15 h for deuteration) | 34 | 4,345.7 ± 0.7 | 16.3 ± 0.7 | 11.3 ± 0.7 |
| Deuterated Aβ fibrils with protonated processing solvent (incubation time ∼384 h for deuteration) | 4 | 4,348.0 ± 0.2 | 18.6 ± 0.2 | 13.6 ± 0.2 |
Uncertainties represent the standard deviations of the mean of the indicated number of replicate runs. Data in this table may differ marginally from some data shown in the figures and text, which show individual, representative runs.
Corrected for rapid back-exchange of labile protons (see text).
Multiple deuterated samples prepared from four different fibril preparations were run multiple times.
To determine the extent to which fibrils dissolve in the T, the peak areas from on-line dissolution of a fibril suspension were compared with those obtained from a known quantity of monomer. We determined that under the flow conditions used here at least 50% of the fibrils are dissolved on-line in the T. Although more complete fibril dissolution might be obtained by using slower flow rates, this also would lead to increased exposure to back-exchange and loss of information. It is likely that the dissolved portion of Aβ fibrils analyzed in the MS is representative of fibril structure, because the protofilament-based substructure of fibrils (6–8) does not seem compatible with the existence of a refractory core. Rather, it is more likely that fibrils dissaggregate in the processing solvent through release of Aβ from protofilament termini.
To assess our ability to achieve and detect H→D exchanges into Aβ, fully deuterated Aβ(1–40) monomer was examined. A 30-μM sample was prepared by incubating Aβ monomer in deuterated Tris buffer in D2O at pD 7.5 (pD values were read directly from a pH meter and uncorrected for any isotope effects; ref. 29) at room temperature for ≈15 h. This solution then was infused through one arm of the mixing T, while deuterated processing solvent was pumped through the other arm of the T. This protocol provided essentially no opportunity for back-exchange. Fig. 1c presents the +6 charge state of a representative mass spectrum. The molecular mass of the completely deuterated peptide was calculated to be 4395.1 Da (Table 1), ≈66 Da higher than that found for protonated Aβ(1–40) monomer in a protonated solvent (Fig. 1a). Because every replacement of H by D produces a mass increase of 1 Da, this finding suggests that a total of 66 protons exchanged for deuterons under these conditions. This is in excellent agreement with the 66 total exchangeable protons predicted for Aβ(1–40), which in its nonionized state contains 39 backbone amides, two N-terminal amine protons, one C-terminal carboxylate proton, and 24 labile side-chain protons. These data suggest that Aβ monomer contains no highly protective secondary structure and confirm the ability of our protocol to count H→D conversions in Aβ samples.
Exchange Experiments.
To test the ability of the sample processing protocol to preserve exchange information during MS analysis, we next exposed the fully deuterated Aβ monomer to analysis using the protonated processing solvent. The +6 charge state of a representative mass spectrum is shown in Fig. 1d. The peaks obtained here are ≈1.5 times wider than for the corresponding protonated sample (Fig. 1a) because of heterogeneity in the deuterium content. Using the centroid of the isotopic envelope to calculate molecular mass, we obtain a measured mass of 4361.9 Da (Fig. 1d, Table 1) for this sample, representing the net incorporation of ≈32 deuteriums into the monomer. Relative to the fully deuterated material (Fig. 1c), this indicates back-exchange of 34 of the 66 sites, slightly more than expected if only the 27 labile side-chain and terminal sites are vulnerable to rapid back-exchange (see the next section). Evidently, some of the 39 backbone amide protons back-exchange on the time scale of mixing and sampling using the T.
The kinetics of monomer HX was studied in more detail by quickly dissolving dried protonated monomer with deuterated Tris buffer (to make a 30-μM solution), then immediately and with periodic repetitions infusing aliquots of this solution into the T, along with protonated processing solvent. The data (Fig. 2) indicate very fast kinetics; all hydrogens are exchanged within 5 min. The spectrum after 5 min of room-temperature incubation was indistinguishable from that after 15 h (i.e., Fig. 1d). The rapid, single-phase exchange kinetics further supports the absence of significant protective secondary structure within the monomer.
Figure 2.
Deuterium content vs. time for deuterated monomer (●), corrected deuterated monomer (⧫), deuterated fibrils (■), and corrected deuterated fibrils (▴). The data presented in corrected deuterated monomer and fibril curves have been adjusted to remove contributions from side-chain protons (see text). (a) Magnified view of the first 42-min segment from c. (b) Magnified view of the 0- to 21-h segment from c. (c) HX kinetics data for monomer (792 h) and fibrils (1135 h). The kinetic plots presented here are averaged from data on three monomer samples and four fibril samples. The error bars in b and c represent the standard deviation of the mean and in some cases are equal to or smaller than the size of the symbol. a represents single kinetic runs at each time point.
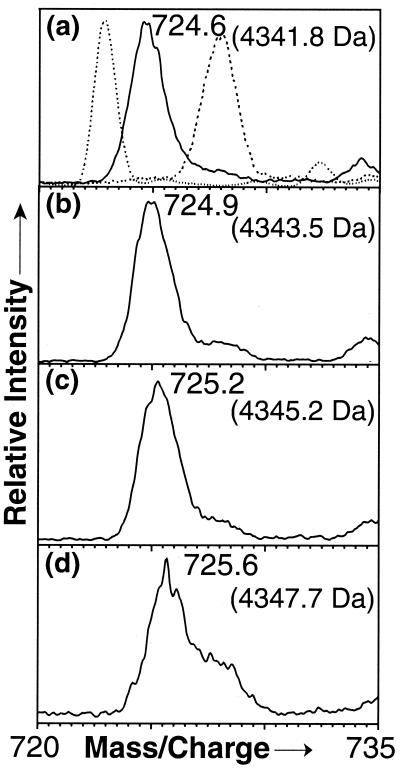
Identical protocols were followed to study the kinetics of deuterium exchange into Aβ fibrils. Protonated fibrils were collected by centrifugation, washed once with water to remove any remaining phosphate buffer, then rapidly suspended in deuterated Tris buffer and incubated at room temperature. At various times, samples from this suspension were mixed with protonated processing solvent in the T and analyzed in the MS. Fig. 3a, collected 3.5 min after mixing with deuterated buffer, shows a single mass distribution centered at 4341.8, corresponding to a total gain of 12 deuteriums, significantly fewer than the 32 incorporated into the monomer in under 5 min. When the fibril suspension was evaluated again after 21 min of incubation, the Aβ(1–40) peak center had shifted to 4343.5 (Fig. 3b). Analysis after 4.8 h showed a total gain of only ≈16 deuteriums (Fig. 3c), and even after incubation at room temperature for 384 h, the spectrum reflects only ≈18 H→D exchanges (Fig. 3d).¶ This sample of partially deuterated fibrils analyzed after 1,135 h contained ≈19 deuteriums.
Figure 3.
Electrospray ionization-MS of deuterated fibrils in protonated solvent after (a) 3.5 min, (b) 21 min, (c) 4.8 h, or (d) 384 h. Mass spectra of protonated and partially deuterated Aβ(1–40) monomer from Fig. 1 a and d are reproduced for comparison as dashed lines in a. The average molecular mass (calculated as in Fig. 1) is indicated for each sample.
The full kinetics curve for deuterium exchange into fibrils also is shown in Fig. 2. The data representing only the exchange into the backbone amides (see below) also are presented in Fig. 2. In contrast to the monomer exchange experiments, these data suggest a number of kinetic phases. Seven deuteriums exchange into fibrils within a few minutes, whereas another seven exchange over a period of several hours to days. The remaining backbone amide hydrogens do not exchange even after 47 days. These different classes presumably represent backbone amide protons residing in increasingly protective environments in the amyloid fibrils.
Hydrogen Exchange During Sample Processing.
As discussed above, there are generally two major contributors to the HX process: very fast exchange of terminal and side-chain hydrogens, and relatively slow exchange of the main-chain, backbone amide hydrogens. Both modes of exchange operate in the mixing T against a solvent that is a mixture of D2O (from the Aβ incubation) and H2O (from the processing solvent). The fraction of water that is D2O in the T (determined by the relative flow rates of the two streams) is 18.2% in these experiments. Thus, if back exchange were complete in any of these experiments (due, for example, to excessively long processing times), then only ≈18% of the maximum 66 deuterons (≈12) would be detected in the MS. The data clearly show that this is not the case for either monomeric or fibrillar Aβ and that significant exchange information therefore has been recovered in the MS analysis. Nonetheless, the measured amounts of deuterium in both the monomer and fibril experiments have been influenced by some degree of back/forward exchange during sample work-up. As discussed next, correction for rapid exchange into side chains is straightforward, but correction for backbone amide exchange is not.
For unaggregated monomers, the labile terminal and side-chain hydrogens exchange very rapidly even at pH 2–3 (27), so that the final measured deuterium content should include an equilibrium distribution of deuterium into these sites. Because there are 27 such hydrogens in Aβ, a total of ≈5 (18.2% of 27) deuteriums should be incorporated at these sites for both monomeric and fibrillar Aβ.
As noted above, there is evidence for some back-exchange into the backbone amide hydrogens in the analysis of the fully deuterated monomer in protic processing solvent (Fig. 1d). By subtracting the five deuteriums contributed by rapid equilibration of the labile sites, from the final measured deuterium content of 32, we obtain a total of 27 deuteriums distributed over the 39 backbone amide hydrogens; that is, 12 have back-exchanged. If the 39 backbone amide hydrogens were completely exchanged and equilibrated with the 18.2% deuterium in the final processing mixture, only seven (not 27) deuteriums would be retained.
In principle, it should be possible to use this kind of information to correct the fibril data and thus determine the absolute number of deuteriums present in fibrillar Aβ before exposing the fibrils to processing. Correction equations for data from soluble proteins have been described and are considered to be accurate to approximately 5% (although deviations of >25% are noted in some instances) (28). Some investigators prefer to use uncorrected data in comparative analyses of different molecules (19, 31, 32). Because no mathematical correction scheme has been developed or validated for treatment of data from experiments involving solubilization before analysis, we have chosen for this discussion (and for Fig. 2) to correct only for the equilibration of the labile sites, leaving for future work the small (but uncertain) correction for back-exchange into backbone amide hydrogens.
As described above, after more than 1,000 h of exchange time, Aβ from fibrils was found to contain ≈19 deuteriums. According to the above analysis, five of these must reside in the labile groups, leaving about 14 backbone amide hydrogen positions occupied by deuteriums. This is in contrast to the 27 backbone amide hydrogens occupied by deuterium in the monomer after only 5 min of exposure to D2O (data of Fig. 2, corrected for side-chain exchange). Our data thus indicate that approximately 50% of the amide backbone hydrogens are involved in some kind of protective structure when Aβ is incorporated into amyloid fibrils. This estimate is in good agreement with exchange protection experiments analyzed by Fourier transform IR spectroscopy (R.W., I.K., Yongsung Kim, and John Carperter, unpublished results).
Discussion
Backbone amide protons in globular proteins occupy a variety of structures that exhibit varying degrees of protection against hydrogen exchange that span at least 6 orders of magnitude of protection factors (17). Core portions of β-sheet are more highly protected than edge regions of sheets and α-helix, which in turn are more protected than loop and random coil regions (17). In this paper we show that all of the backbone amide hydrogens of monomeric Aβ undergo rapid HX consistent with the absence of protective structure within the molecule. On the contrary, Aβ incorporated into fibrils undergoes much slower exchange with more complex kinetics. Fig. 2 suggests at least three classes of backbone amides in fibrils: those that exchange as rapidly as the backbone amide hydrogens of the monomer, those that exchange at intermediate rates, and those that do not exchange even after 1,000 h of exposure to D2O. At least 50% of the Aβ peptide backbone resides in this highly protected, rigid core structure in the fibrils. Such high extents of protection over extended exchange times generally are not observed in HX studies on globular proteins.
Global exchange data such as that described here should assist in elucidating the structure of amyloid fibrils by allowing the testing and refinement of structural models. For example, our data suggest that a significant portion of the Aβ peptide is not involved in protective β-sheet structure in the amyloid fibrils. Our data are thus inconsistent with any model of fibril structure invoking strong H-bonding along the entire length of the Aβ peptide. HX-MS also should prove useful in characterizing the secondary structure contents of amyloid fibrils generated from biologically relevant forms of Aβ, such as the 1–42 peptide (33) and the “Dutch” (34) mutant. It also can be used to characterize fibril assembly intermediates (35, 36) as well as other Aβ aggregates not on the assembly pathway (37).
An important future direction of hydrogen exchange experiments is to move beyond global analysis of the entire Aβ molecule and to localize exchange protection to particular molecular regions, and ultimately particular residues of the amino acid sequence. In the MS approach, this might be accomplished by fragmentation of the solubilized Aβ, either by proteolysis before MS analysis (21, 28) or by high-energy decomposition during MS-MS analysis (22), to determine exchange protection in individual fragments.
Progress in our understanding of amyloid structure and its impact on disease pathology will depend to a large extent on the development of novel approaches to the structural analysis of protein aggregates. In this paper we show that HX-MS studies of Aβ fibrils provide global information on the extent of the β-sheet network and protection within the fibrils. In principle, these methods also should be useful in analyzing the structures of other amyloid fibrils and protein aggregates.
Acknowledgments
We thank John Wall for his assistance with data analysis and Al Tuinman for helpful discussions. We also acknowledge helpful discussions with Sheena Radford and Andrew Miranker. I.K. is supported by a National Research Service Award from the National Institutes of Health (F32 AG05869), and S.Z. is supported by the University of Tennessee Measurement and Control Engineering Center. R.W. acknowledges support from the Lindsay Young Alzheimer's Disease Research Fund.
Abbreviation
- HX
hydrogen deuterium exchange
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250288897.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250288897
In the MS of samples from the fibril exchange reaction (Fig. 3), we consistently observe a shoulder at a mass-to-charge ratio coinciding with that of the partially deuterated monomer (Fig. 1d) and growing relatively larger at long incubation times. One possible source is monomeric (and hence unprotected) Aβ in equilibrium with the fibrils (30), which would exchange at the rate and to the extent of the monomer. Further work is required to fully characterize the source of this peak.
References
- 1.Selkoe D J. J Am Med Assoc. 2000;283:1615–1617. doi: 10.1001/jama.283.12.1615. [DOI] [PubMed] [Google Scholar]
- 2.Clark A, de Koning E J, Hattersley A T, Hansen B C, Yajnik C S, Poulton J. Diabetes Res Clin Pract. 1995;28,Suppl.:S39–S47. doi: 10.1016/0168-8227(95)01075-o. [DOI] [PubMed] [Google Scholar]
- 3.Selkoe D J. Annu Rev Neurosci. 1994;17:489–517. doi: 10.1146/annurev.ne.17.030194.002421. [DOI] [PubMed] [Google Scholar]
- 4.Selkoe D J. Science. 1997;275:630–631. doi: 10.1126/science.275.5300.630. [DOI] [PubMed] [Google Scholar]
- 5.Sipe J D. Annu Rev Biochem. 1992;61:947–975. doi: 10.1146/annurev.bi.61.070192.004503. [DOI] [PubMed] [Google Scholar]
- 6.Serpell L C, Sunde M, Fraser P E, Luther P K, Morris E P, Sangren O, Lundgren E, Blake C C. J Mol Biol. 1995;254:113–118. doi: 10.1006/jmbi.1995.0604. [DOI] [PubMed] [Google Scholar]
- 7.Fraser P E, Duffy L K, OMalley M B, Nguyen J, Inouye H, Kirschner D A. J Neurosci Res. 1991;28:474–485. doi: 10.1002/jnr.490280404. [DOI] [PubMed] [Google Scholar]
- 8.Sunde M, Blake C. In: Advances in Protein Chemistry. Wetzel R, editor. Vol. 50. New York: Academic; 1997. pp. 123–159. [DOI] [PubMed] [Google Scholar]
- 9.Fraser P E, Nguyen J T, Inouye H, Surewicz W K, Selkoe D J, Podlisny M B, Kirschner D A. Biochemistry. 1992;31:10716–10723. doi: 10.1021/bi00159a011. [DOI] [PubMed] [Google Scholar]
- 10.Benzinger T L S, Gregory D M, Burkoth T S, Miller-Auer H, Lynn D G, Botto R E, Meredith S C. Biochemistry. 2000;39:3491–3499. doi: 10.1021/bi991527v. [DOI] [PubMed] [Google Scholar]
- 11.Kirschner D A, Inouye H, Duffy L K, Sinclair A, Lind M, Selkoe D J. Proc Natl Acad Sci USA. 1987;84:6953–6957. doi: 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chaney M O, Webster S D, Kuo Y-M, Roher A E. Protein Eng. 1998;11:761–767. doi: 10.1093/protein/11.9.761. [DOI] [PubMed] [Google Scholar]
- 13.Lazo N D, Downing D T. Biochemistry. 1998;37:1731–1735. doi: 10.1021/bi971016d. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Darden T A, Bartolotti L, Kominos D, Pedersen L G. Biophys J. 1999;76:2871–2878. doi: 10.1016/S0006-3495(99)77442-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jimenez J L, Guijarro J I, Orlova E, Zurdo J, Dobson C M, Sunde M, Saibil H R. EMBO J. 1999;18:815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodward C, Simon I, Tuchsen E. Mol Cell Biochem. 1982;48:135–160. doi: 10.1007/BF00421225. [DOI] [PubMed] [Google Scholar]
- 17.Englander S W, Kallenbach N R. Q Rev Biophys. 1983;16:521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- 18.Li R, Woodward C. Protein Sci. 1999;8:1571–1591. doi: 10.1110/ps.8.8.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robinson C V. In: Mass Spectrometry of Biological Materials. Larsen B S, McEwen C N, editors. New York: Dekker; 1998. pp. 369–387. [Google Scholar]
- 20.Anderegg R J. In: Mass Spectrometry in the Biological Sciences. Burlingame A L, Carr S A, editors. Totowa, NJ: Humana; 1996. pp. 85–104. [Google Scholar]
- 21.Smith D L, Deng Y, Zhang Z. J Mass Spectrom. 1997;32:135–146. doi: 10.1002/(SICI)1096-9888(199702)32:2<135::AID-JMS486>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 22.Miranker A, Robinson C V, Radford S E, Dobson C M. FASEB J. 1996;10:93–101. doi: 10.1096/fasebj.10.1.8566553. [DOI] [PubMed] [Google Scholar]
- 23.Heimburg T, Marsh D. Biophys J. 1993;65:2408–2417. doi: 10.1016/S0006-3495(93)81299-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baello B I, Pancoska P, Keiderling T A. Anal Biochem. 2000;280:46–57. doi: 10.1006/abio.2000.4483. [DOI] [PubMed] [Google Scholar]
- 25.Zagorski M G, Yang J, Shao H, Ma K, Zeng H, Hong A. Methods Enzymol. 1999;309:189–204. doi: 10.1016/s0076-6879(99)09015-1. [DOI] [PubMed] [Google Scholar]
- 26.LeVine H., III Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 27.Wuthrich K, Wagner G. J Mol Biol. 1979;130:1–18. doi: 10.1016/0022-2836(79)90548-5. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Z, Smith D L. Protein Sci. 1993;2:522–531. doi: 10.1002/pro.5560020404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glasoe P F, Long F A. J Phys Chem. 1960;64:188–193. [Google Scholar]
- 30.Harper J D, Lansbury P T., Jr Annu Rev Biochem. 1997;66:385–407. doi: 10.1146/annurev.biochem.66.1.385. [DOI] [PubMed] [Google Scholar]
- 31.Ehring H. Anal Biochem. 1999;267:252–259. doi: 10.1006/abio.1998.3000. [DOI] [PubMed] [Google Scholar]
- 32.Nemirovskiy O, Giblin D E, Gross M L. J Am Soc Mass Spectrom. 1999;10:711–718. doi: 10.1016/S1044-0305(99)00036-7. [DOI] [PubMed] [Google Scholar]
- 33.Naslund J, Haroutunian V, Mohs R, Davis K L, Davies P, Greengard P, Buxbaum J D. J Am Med Assoc. 2000;283:1571–1577. doi: 10.1001/jama.283.12.1571. [DOI] [PubMed] [Google Scholar]
- 34.Levy E, Carman M D, Fernandez-Madrid I J, Power M D, Lieberburg I, Van Duynen S G, Bots G T A M, Luyendijk W, Frangione B. Science. 1990;248:1124–1126. doi: 10.1126/science.2111584. [DOI] [PubMed] [Google Scholar]
- 35.Harper J D, Wong S S, Lieber C M, Lansbury P T. Chem Biol. 1997;4:119–125. doi: 10.1016/s1074-5521(97)90255-6. [DOI] [PubMed] [Google Scholar]
- 36.Walsh D M, Lomakin A, Benedek G B, Condron M M, Teplow D B. J Biol Chem. 1997;272:22364–22372. doi: 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- 37.Wood S J, Maleeff B, Hart T, Wetzel R. J Mol Biol. 1996;256:870–877. doi: 10.1006/jmbi.1996.0133. [DOI] [PubMed] [Google Scholar]