Abstract
HIV-1 subtype C is the most common HIV-1 group M subtype in Africa and many parts of Asia. However, to date HIV-1 vaccine candidate immunogens have not induced potent and broadly neutralizing antibodies against subtype C primary isolates. We have used a centralized gene strategy to address HIV-1 diversity, and generated a group M consensus envelope gene with shortened consensus variable loops (CON-S) for comparative studies with wildtype (WT) Env immunogens. Our results indicate that the consensus HIV-1 group M CON-S Env elicited cross-subtype neutralizing antibodies of similar or greater breadth and titer than the WT Envs tested, indicating the utility of a centralized gene strategy. Our study also shows the feasibility of iterative improvements in Env immunogenicity by rational design of centralized genes.
Keywords: HIV-1, vaccine, neutralization antibody, consensus immunogens
Introduction
Design of an immunogen that can induce broadly reactive neutralizing antibodies against HIV-1 primary isolates is a major goal of HIV-1 vaccine development. To address the problem of HIV-1 diversity for antibody induction, we have begun to design, synthesize, and test group M consensus envelope genes as centralized immunogens using a variety of different vector systems. The first generation group M consensus env gene, termed CON6, was derived from the 1999 Los Alamos HIV env Sequence Database. CON6 env encoded an Env protein that was biologically functional, but contained the variable loops of a contemporary subtype C isolate (Gao et al., 2005). CON-6 Env mediated infection of CD4/CCR5 positive target cells with reduced efficiency compared to wild-type (WT) Env and induced cross-subtype T cell responses in mice (Gao et al., 2005). However, the neutralizing antibody responses induced by CON6 Env were limited to only a subset of subtype B HIV-1 primary isolates (Gao et al., 2005).
Several studies have indicated that mutations or deletions of variable loops in the envelope glycoproteins of HIV-1 and simian immunodeficiency virus (SIV) may increase the number and/or exposure of available neutralization epitopes within the virion-associated viral envelope (Kang et al. 2005; Gzyl et al., 2004; Yang et al. 2004; Kim et al 2003; Barnett et al. 2001; Wyatt et al. 1993). Our second generation group M consensus env gene, termed CON-S, is not only based on a more comprehensive collection of HIV-1 env sequences from the 2001 Los Alamos HIV-1 sequence database, but also contains minimal lengths of consensus rather than wild-type variable loop sequences. In this paper, we describe the biological properties of the CON-S Env glycoprotein and compare its ability to induce neutralizing antibodies with that of CON6 and five WT Env glycoproteins.
Materials and Methods
Design of the CON-S Consensus Env Gene
CON-S was derived by aligning the consensus env sequences of group M subtypes A (including A, A1, and A2), B, C, D, F (including F1 and F2) and G from the 2001 HIV sequence database as described (http://hiv-web.lanl.gov/content/hiv-db/CONSENSUS/M_GROUP/Consensus.html) (Table 1). Only full length proteins and one sequence per individual were included in the alignment. In regions outside of the variable loops, this consensus is virtually identical to a model of the ancestral sequence of the M group based on maximum likelihood phylogenies (Gaschen et al. 2002, Korber et al. 2000). V3 was readily aligned and was treated the same as the more conserved Env regions. The highly variable loop regions evolve by insertion and deletion and are particularly difficult to align. To design the hypervariable loop regions for CON-S, hand alignments were made that first brought potential N-linked glycosyation sites and cysteines into alignment, and then brought repeated sequence motifs within loops into alignment. Amino acids present in the majority of positions were retained for each subtype, and a consensus of consensuses was generated (Table 1). This procedure, by design, generated hypervariable loop sequences (V1, V2, V4 and V5) that tended to comprise a shorter range of lengths found among natural strains. This result was intended since shorter length V1, V2, V4 and V5 loops might expose regions around the variable loops that might conserve neutralizing determinants.
Table 1.
Amino Acid and Nucleotide Sequences of CON-S Env gp160*
| MRVRGIQRNCQHLWRWGTLILGMLMICSAAENLWVTVYYGVPVWKEANTTLFCASDAKAYDTEVHNVWATHACVPTDPNPQEIVLENVTENFNMWKNNMVEQMHEDII
SLWDQSLKPCVKLTPLCVTLNCTNVNVTNTTNNTEEKGEIKNCSFNITTEIRDKKQKVYALFYRLDVVPIDDNNNNSSNYRLINCNTSAITQACPKVSFEPIPIHYCA PAGFAILKCNDKKFNGTGPCKNVSTVQCTHGIKPVVSTQLLLNGSLAEEEIIIRSENITNNAKTIIVQLNESVEINCTRPNNNTRKSIRIGPGQAFYATGDIIGDIRQ AHCNISGTKWNKTLQQVAKKLREHFNNKTIIFKPSSGGDLEITTHSFNCRGEFFYCNTSGLFNSTWIGNGTKNNNNTNDTITLPCRIKQIINMWQGVGQAMYAPPIEG KITCKSNITGLLLTRDGGNNNTNETEIFRPGGGDMRDNWRSELYKYKVVKIEPLGVAPTKAKRRVVEREKRAVGIGAVFLGFLGAAGSTMGAASITLTVQARQLLSGI VQQQSNLLRAIEAQQHLLQLTVWGIKQLQARVLAVERYLKDQQLLGIWGCSGKLICTTTVPWNSSWSNKSQDEIWDNMTWMEWEREINNYTDIIYSLIEESQNQQEKN EQELLALDKWASLWNWFDITNWLWYIKIFIMIVGGLIGLRIVFAVLSIVNRVRQGYSPLSFQTLIPNPRGPDRPEGIEEEGGEQDRDRSIRLVNGFLALAWDDLRSLC LFSYHRLRDFILIAARTVELLGRKGLRRGWEALKYLWNLLQYWGQELKNSAISLLDTTAIAVAEGTDRVIEVVQRACRAILNIPRRIRQGLERALL |
| Nucleotide sequence: |
| ATGCGCGTGCGCGGCATCCAGCGCAACTGCCAGCACCTGTGGCGCTGGGGCACCCTGATCCTGGGCATGCTGATGATCTGCTCCGCCGCCGAGAACCTGTGGGTGACC
GTGTACTACGGCGTGCCCGTGTGGAAGGAGGCCAACACCACCCTGTTCTGCGCCTCCGACGCCAAGGCCTACGACACCGAGGTGCACAACGTGTGGGCCACCCACGCC TGCGTGCCCACCGACCCCAACCCCCAGGAGATCGTGCTGGAGAACGTGACCGAGAACTTCAACATGTGGAAGAACAACATGGTGGAGCAGATGCACGAGGACATCATC TCCCTGTGGGACCAGTCCCTGAAGCCCTGCGTGAAGCTGACCCCCCTGTGCGTGACCCTGAACTGCACCAACGTGAACGTGACCAACACCACCAACAACACCGAGGAG AAGGGCGAGATCAAGAACTGCTCCTTCAACATCACCACCGAGATCCGCGACAAGAAGCAGAAGGTGTACGCCCTGTTCTACCGCCTGGACGTGGTGCCCATCGACGAC AACAACAACAACTCCTCCAACTACCGCCTGATCAACTGCAACACCTCCGCCATCACCCAGGCCTGCCCCAAGGTGTCCTTCGAGCCCATCCCCATCCACTACTGCGCC CCCGCCGGCTTCGCCATCCTGAAGTGCAACGACAAGAAGTTCAACGGCACCGGCCCCTGCAAGAACGTGTCCACCGTGCAGTGCACCCACGGCATCAAGCCCGTGGTG TCCACCCAGCTGCTGCTGAACGGCTCCCTGGCCGAGGAGGAGATCATCATCCGCTCCGAGAACATCACCAACAACGCCAAGACCATCATCGTGCAGCTGAACGAGTCC GTGGAGATCAACTGCACCCGCCCCAACAACAACACCCGCAAGTCCATCCGCATCGGCCCCGGCCAGGCCTTCTACGCCACCGGCGACATCATCGGCGACATCCGCCAG GCCCACTGCAACATCTCCGGCACCAAGTGGAACAAGACCCTGCAGCAGGTGGCCAAGAAGCTGCGCGAGCACTTCAACAACAAGACCATCATCTTCAAGCCCTCCTCC GGCGGCGACCTGGAGATCACCACCCACTCCTTCAACTGCCGCGGCGAGTTCTTCTACTGCAACACCTCCGGCCTGTTCAACTCCACCTGGATCGGCAACGGCACCAAG AACAACAACAACACCAACGACACCATCACCCTGCCCTGCCGCATCAAGCAGATCATCAACATGTGGCAGGGCGTGGGCCAGGCCATGTACGCCCCCCCCATCGAGGGC AAGATCACCTGCAAGTCCAACATCACCGGCCTGCTGCTGACCCGCGACGGCGGCAACAACAACACCAACGAGACCGAGATCTTCCGCCCCGGCGGCGGCGACATGCGC GACAACTGGCGCTCCGAGCTGTACAAGTACAAGGTGGTGAAGATCGAGCCCCTGGGCGTGGCCCCCACCAAGGCCAAGCGCCGCGTGGTGGAGCGCGAGAAGCGCGCC GTGGGCATCGGCGCCGTGTTCCTGGGCTTCCTGGGCGCCGCCGGCTCCACCATGGGCGCCGCCTCCATCACCCTGACCGTGCAGGCCCGCCAGCTGCTGTCCGGCATC GTGCAGCAGCAGTCCAACCTGCTGCGCGCCATCGAGGCCCAGCAGCACCTGCTGCAGCTGACCGTGTGGGGCATCAAGCAGCTGCAGGCCCGCGTGCTGGCCGTGGAG CGCTACCTGAAGGACCAGCAGCTGCTGGGCATCTGGGGCTGCTCCGGCAAGCTGATCTGCACCACCACCGTGCCCTGGAACTCCTCCTGGTCCAACAAGTCCCAGGAC GAGATCTGGGACAACATGACCTGGATGGAGTGGGAGCGCGAGATCAACAACTACACCGACATCATCTACTCCCTGATCGAGGAGTCCCAGAACCAGCAGGAGAAGAAC GAGCAGGAGCTGCTGGCCCTGGACAAGTGGGCCTCCCTGTGGAACTGGTTCGACATCACCAACTGGCTGTGGTACATCAAGATCTTCATCATGATCGTGGGCGGCCTG ATCGGCCTGCGCATCGTGTTCGCCGTGCTGTCCATCGTGAACCGCGTGCGCCAGGGCTACTCCCCCCTGTCCTTCCAGACCCTGATCCCCAACCCCCGCGGCCCCGAC CGCCCCGAGGGCATCGAGGAGGAGGGCGGCGAGCAGGACCGCGACCGCTCCATCCGCCTGGTGAACGGCTTCCTGGCCCTGGCCTGGGACGACCTGCGCTCCCTGTGC CTGTTCTCCTACCACCGCCTGCGCGACTTCATCCTGATCGCCGCCCGCACCGTGGAGCTGCTGGGCCGCAAGGGCCTGCGCCGCGGCTGGGAGGCCCTGAAGTACCTG TGGAACCTGCTGCAGTACTGGGGCCAGGAGCTGAAGAACTCCGCCATCTCCCTGCTGGACACCACCGCCATCGCCGTGGCCGAGGGCACCGACCGCGTGATCGAGGTG GTGCAGCGCGCCTGCCGCGCCATCCTGAACATCCCCCGCCGCATCCGCCAGGGCCTGGAGCGCGCCCTGCTGTAA |
Cleavage site and fusion domain are in italic font and underlined; the immunodominant region is highlighted in bold. The transmembrane and cytoplsmic domains are underlined
Expression of Recombinant HIV-1 Envelopes
The CON-S env gene was generated by converting amino acid sequences of CON-S to nucleotide sequences employing the codon usage of highly expressed human housekeeping genes (André et al., 1998) and de novo synthesized (Table 1). HIV-1 gp140 Envs with the deletion of the cleavage (C) site, fusion (F) and immunodominant (I) region in gp41 were named as gp140ΔCFI, and HIV-1 gp140 Envs with the deletion of only the cleavage (C) site and fusion (F) domain were named as gp140ΔCF. CON-S gp140ΔCFI and CON6 gp140ΔCFI were generated by PCR by introducing a stop codon before the membrane-spanning domain (YIKIFIMIVGGLIGLRIVFAVLSIVN) (Table 1) (Chakrabarti et al., 2002; Gao et al., 2005). CON6 gp140ΔCF and wild-type (WT) subtype B (B.) JRFL gp140ΔCF env genes were constructed as described (Gao et al., 2005). HIV-1 WT subtype A (A.) 92RW020) and subtype C (C.) 97ZA012 env gp140ΔCFI constructs have previously been reported (Seaman et al., 2005). HIV-1 primary isolate B.6101 env gp120 was constructed by introducing a stop codon at the cleavage site using the methods as described (Gao et al., 2005). Recombinant HIV-1 B.BaL gp120 was obtained from the AIDS Reagent repository (Liao et al., 2004). Recombinant vaccinia viruses (rVVs) expressing CON6 gp140ΔCF, CON6 gp140ΔCFI, CON-S gp140ΔCFI, B.JRFL gp140ΔCF, A.92RW020 gp140ΔCFI, C.97ZA012 gp140ΔCFI and B.6101 gp120 genes were generated as previously described ( Earl, 1997; Moss, and Earl, 1998). Identities of the individual rVVs were confirmed by PCR and nucleotide sequence analysis. Recombinant Env glycoproteins were purified from supernatants of 293T cell cultures infected with rVVs using Galanthus nivalis lectin-agarose (Vector Labs, Burlingame, Calif.) column chromatography and stored at −70°C until use.
Mabs
Human monoclonal antibodies (MAbs) known to bind epitopes on gp120 (A32), the gp120 V3 loop (19b, F2A3, 39F), and the CCR5 binding site (17b) were kindly provided by James Robinson (Tulane Medical School, New Orleans, La.) (Rizzuto and Sodroski, 2000; Wyatt et al., 1998; Wyatt et al., 1995; Xiang et al., 2002). MAbs 2F5, 4E10 and 2G12 were gifts from Hermann Katinger at the University of Natural Resources and Applied Life Sciences, Vienna, Austria. MAb IgG1b12 was the gift of Dennis Burton, Scripps Research Institute, La Jolla, CA. MAb 447-52D and soluble CD4 (sCD4) were obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program (Bethesda, MD). T8 is a murine MAb that binds to the gp120 C1 region (a gift from P. Earl, NIH).
SDS-PAGE and BN-PAGE
WT B.6101 and B.BaL gp120 proteins were analyzed by SDS-PAGE. Blue native polyacrylamide gel electrophoresis (BN-PAGE) analysis of CON-S, CON6 and WT HIV-1 envelope gp140 CFI or CF proteins was carried out according to the methods as described (Gao et al., 2005; Schagger et al.,1994; Schulke et al., 2002) with modifications due to the highly basic pIs of HIV-1 Env proteins. Briefly, purified HIV-1 Env glycoproteins were diluted in a buffer containing 50 mM MOPS (morpholinepropanesulfonic acid), 50 mM Tris-HCl (pH 7.7), 20% glycerol, and 0.05% Coomassie blue. Protein samples were loaded onto a 3 to 8% Tris-acetate NuPAGE gel (Invitrogen, Carlsbad, Calif.) and electrophoresis was carried out for 1.5 h at 150 V with 50 mM MOPS–50 mM Tris-HCl (pH 7.7)−0.03% Coomassie blue as the cathode running buffer and 50 mM MOPS–50 mM Tris HCl (pH 7.7) as the anode buffer. Thyroglobulin, ferritin, catalase, lactate dehydrogenase and albumin (Amersham Biosciences) with molecular weights of 669, 440, 232, 140 and 66 kDa respectively, were included in BN-PAGE as molecular weight markers.
Surface Plasmon Resonance
Surface plasmon resonance assays were performed on a BIAcore 3000 instrument and data analysis was performed with BIAevaluation 3.0 software (BIAcore Inc, Upsaala, Sweden). Anti-gp120 MAbs (T8, A32, 17b, and 2G12) or sCD4 in 10 mM Na-acetate buffer (pH 4.5) were directly immobilized to CM5 sensor chips using a standard amine coupling protocol for protein immobilization. Purified HIV-1 Env glycoproteins were flowed over CM5 sensor chips at concentrations of 100 and 300 ug/ml, respectively. Binding of HIV-1 envelope proteins was monitored in real time at 25°C with a continuous flow of phosphate-buffered saline (150 mM NaCl, 0.005% surfactant P20 [pH 7.4]) at 10 to 30 ul/min. (Alam et al., 2004; Gao et al., 2005).
Immunizations
Out-bred guinea pigs were purchased from Jackson Laboratory and housed in the Vivarium at Duke University Animal Research Facility. Animals were studied under a protocol approved by the Animal Use and Care Committee of Duke University. Guinea pigs (4 animals per immunogen group) were immunized subcutaneously on day 0, week 3, 6, 9 and 12 with 100 μg/injection of recombinant HIV-1 Env gp140 or gp120 proteins formulated in Ribi-CWS adjuvant (Corixa-Sigma, St. Louis, MO). When combinations of two HIV-1 Env proteins as immunogens were used, 100ug of each Env protein for a total of 200ug/injection were administered. Serum samples were collected 10 days after each immunization and stored at −30°C until use.
Generation of Pseudotyped Viruses
CON-S and CON6 Env-pseudotyped viruses were generated by co-transfection of CON-S gp160 or CON6 gp160 clones (in pcDNA3.1) with pSG3ΔEnv (an env-minus backbone vector) in 293T cells using the method as described (Derdeyn et al., 2000; Wei et al., 2003; Wei et al., 2002). The same approach was used to generate stocks of pseudotyped HIV-1 viruses containing subtype A, B and C Envs from primary HIV-1 isolates for neutralization assays. Infectious titers of pseudotyped viruses were determined in TZM-bl (JC53-BL) cells based on luciferase (Luc) reporter gene expression using the method as described (Montefiori, 2004). Briefly, TZM-bl cells in 96-well plates were infected with 11 serial 5-fold dilutions of pseudotyped viruses in quadruplicates for each dilution and incubated for 48 hours. Uninfected TZM-bl cells were used as negative controls. At the end of incubation, Bright GloTM luceiferase substract was added. The relative luminescence units (RLU) of each well in 96-well microtiter tissue culture plates were measured by a luminometer. Wells with RLU < 2.5 times over the negative controls were considered as negative for the calculation of the tissue culture infection doses (TCID50). TCID50 was calculated using the Reed-Muench accumulative method as described (Poli and Fauci, 2004).
Neutralization Assays
Serum neutralizing antibody levels were measured against a panel of pseudotyped HIV-1 viruses containing Envs from subtype B (B.QH0692, B.SF162, B.SS1196, B.BaL, B.BG1168, B.3988 and B.6101), subtype C (C.TV-1, C.DU123, C.DU172, C.92ZM233M, C.92BR025, C.92ZM197M, C.96ZM651, C.DU151, C.97ZA012, C.DU422, C.DU156, C.02ZM233M and C.02ZM197M), and subtype A (A.92RW010, A.92UG37, A.93TH976.01, A.Q23, A.Q168, A.Q259, A.Q461, A.Q769, and A.Q842). HIV-1 env genes were amplified from DNAs derived from uncultured PBMC of HIV-1 patients or derived from short term PBMC cultures. Neutralization assays were based on reductions in luciferase (Luc) reporter gene expression after a single round of virus infection with pseudotyped HIV-1 viruses in TZM-bl cells (Derdeyn et al., 2000; Wei et al., 2003; Wei et al., 2002; Montefiori, 2004; Li, et al. 2005). Briefly, 200 TCID50 of virus were incubated with serial 3-fold dilutions of serum samples in triplicate in a total volume of 150 μl for 1 hr at 37°C in 96-well flat-bottom culture plates. Freshly trypsinized cells (10,000 cells in 100 μl of growth medium containing 75 μg/ml DEAE dextran) were added to each well. One set of control wells received cells plus virus (virus control) and another set received cells only (background control). After a 48-hour incubation, 100 μl of cells were transferred to a 96-well black solid plate (Costar) for measurements of luminescence using Bright Glo substrate solution as described by the supplier (Promega). Neutralization titer was the dilution at which RLUs were reduced by 50% compared to virus control wells after subtraction of background RLUs. Neutralization was considered positive if the titer of post-immune serum minus the titer of pre-immune bleed titer was >30. For HIV-1 BX08 neutralization assays, PBMC-derived replication competent virus was used in the Luc M.7 assay as described (Montefiori, 2004; Gao et al., 2005). For neutralization of JRFL, neutralization assays were performed in the TZM-bl cells using JRFL virus that was generated in 293T cells by transfection using a infectious molecular clone as described (Derdeyn et al., 2000; Wei et al., 2003; Wei et al., 2002). In CON6 and CON-S infectivity assays, TZM-bl cells were pretreated with ADM3100 (CXCR4 inhibition), TAK-799 (CCR5 inhibitors), or both before infection with Env containing pseudovirions. Neutralization assays of HIV-1 primary isolates, SF162 and TV-1 were also performed in PBMC-based neutralization assays by measuring reduction in HIV-1 p24 antigen in PHA-activated PBMCs (Montefiori et al 1996; Bures et al. 2000).
Peptides
The CON-S and CON6 V1, V2, V3, V4 and V5 peptides (Table 2) were synthesized by SynPep, Inc (Dublin, CA) using F-MOC chemistry, purified by HPLC and confirmed to be correct by mass spectrometry.
Table 2.
CON-S and CON6 Env Variable Loop Peptides Used in Absorption Assay
| Variable Region | Amino Acid Sequence of CON-S | Amino Acid Sequence of CON6 |
|---|---|---|
| V1 | TNVNVTNTTNNTEEKGEIKN | TNVRNVSSNGTETDNEEIKN |
| V2 | TEIRDKKQKVYALFYRLDVVPIDDNNNNSSNYR | TELRDKKQKVYALFYRLDVVPIDDKNSSEISGKNSSEYYR |
| V3 | TRPNNNTRKSIRIGPGQAFYATGDIIGDIRQAH | TRPNNNTRKSIHIGPGQAFYATGEIIGDIRQAH |
| V4 | NTSGLFNSTWIGNGTKNNNNTNDTITLP | NTSGLFNSTWMFNGTYMFNGTKDNSETITLP |
| V5 | RDGGNNNTNETEIFRPGGGD | RDGGNNSNKNKTETFRPGGGD |
For peptide absorption of neutralizing antibodies, serum samples were diluted a minimum of 2-fold in PBS to optimize peptide blocking and to yield approximate equal concentrations of neutralizing antibodies in each sample based on measured neutralizing antibody titers. Separate portions of each diluted serum sample were incubated for 1 hour with the V1, V2, V3, V4 or V5 loop peptides or scrambled V3 control peptide (RGYFTRRNAPSNTARGRPILRRN), each at a final concentration of 50 μg/ml. As an additional control, a portion of each diluted serum sample was incubated with a volume of PBS equal to the volume of peptide added to the other samples. Adsorbed and non-adsorbed samples were then assayed at multiple 3-fold dilutions for neutralizing activity against B.SS1196, C.TV-1, C.DU123, C.DU172, C.02ZM233M and A.92RW020 as described above.
For immunization of guinea pigs with CON-S V3, we used the CON-S V3 peptide (Table 2) and a second peptide, GTH1-V3 that is comprised of HIV-1 gag p24 Th epitope (GTH1) (YKRWIILGLNKIVRM) synthesized N-terminal to the CON-S V3 (TRPNNNTRKSIHIGPGQAFYAT) for enhancement of peptide immunogenicity.
Results
Infectivity and co-receptor usage of the CON-S envelope glycoprotein
We previously demonstrated that the CON6 Env (which contains Group M consensus core regions but contemporary subtype C variable loops) was capable of mediating virus entry into CD4 and CCR5 bearing target cells (Gao et al., 2005) when tested as a full-length (gp160) envelope in the form of pseudovirions. Using this same approach, we determined the infectivity profile and co-receptor usage of the CON-S Env glycoprotein. CON-S gp160 conferred infectivity to HIV-1/SG3Δenv in TZM-bl cells utilizing the CCR5 (but not the CXCR4) co-receptor (Figure 1A). However, compared to WT HIV-1 envelopes (NL4-3 and YU-2), its infectivity was reduced by two orders of magnitude. Consistent with previous observations, CON6 gp160 analyzed in parallel yielded a similar infectivity profile (Figure 1A). Thus, “centralization” seems to confer some functional impairment, at least in the context of the group M consensus envelope glycoproteins. Figure 1B shows that both CON6 and CON-S Env require CCR5 and not CXCR4 for co-receptor.
Figure 1.
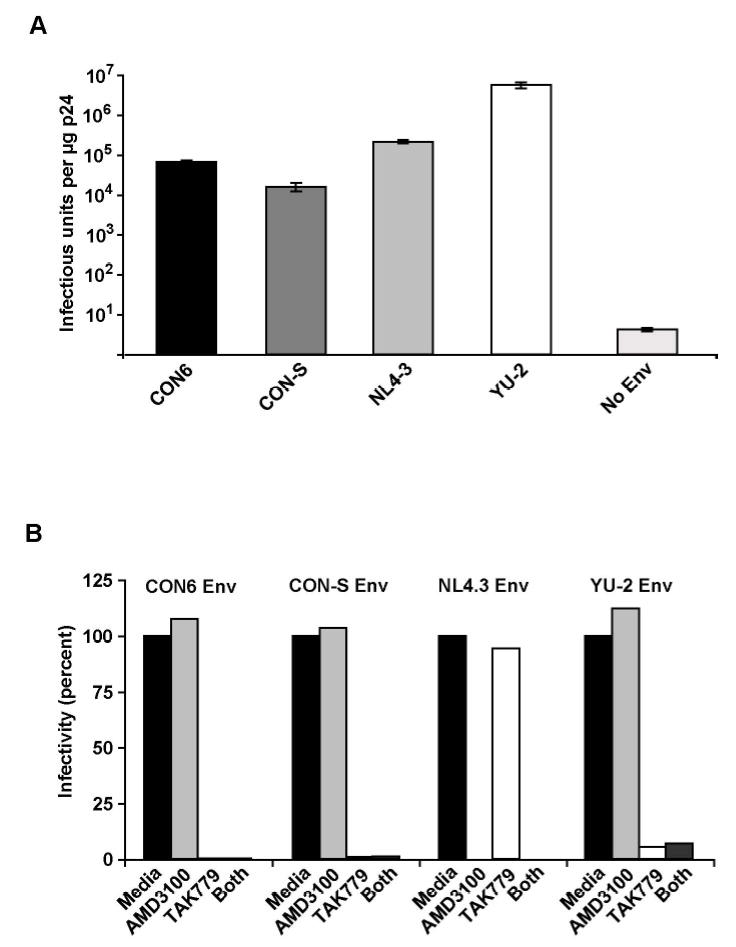
Infectivity and coreceptor usage of CON-S gp160-pseudotyped virus. A. Infectivity in TZM-bl cells. Virus stocks were generated in 293T cells by cotransfection of consensus (CON6 and CON-S) and wildtype Envs (NL4-3, YU-2) with the HIV-1/SG3Δenv backbone (No Env) . Infectivity was determined by counting the number of blue cells (infectious units, IU) per microgram of p24 of pseudovirion stock (IU/μg p24) after staining the infected cells for β-galactosidase expression. Bars indicate standard errors (values are averaged from three independent experiments). B. Coreceptor usage of CON-S Env (gp160) containing pseudovirions. TZM-bl (JC53-BL) cells were pretreated with ADM3100 (CXCR4 inhibitor), TAK-779 (CCR5 inhibitor), both or neither (media) before being infected with CON6, CON-S, NL4-3 and YU-2 Env containing pseudovirions.
We also characterized the neutralization phenotype of both CON6 and CON-S pseudoviruses with sCD4, MAbs, serum and plasma pools from HIV-1 infected patients (Li et al, 2005). We found that both CON6 and CON-S were sensitive to sCD4 neutralization (both ~ 12.0ug/ml ID50) and to MAbs IgG1b12, 2F5 and 4E10 (ID50 range for MAbs for both viruses, 2.9 to 0.5 ug/ml). The analysis of the rank order of the neutralization sensitivity showed that CON-S has a similar sensitivity with B.SS1196.1, and was slightly more sensitive than CON6 to neutralization by plasma and serum pools from HIV-1 infected patients (Li et al, 2005). Both viruses were ~20x less sensitive than the TCLA virus SF162 and MN (Li et al, 2005).
Expression and Characterization of Env gp120 Monomers and gp140 Oligomers
To test the group M consensus and WT HIV-1 subtype A, B and C Env glycoproteins as immunogens, secreted versions (gp140) were expressed as Δ CFI with deletions of the cleavage site, fusion domain and immunodomaint region (CON-S, CON6, A.92RW020, C.97ZA012) or ΔCF with deletions of the cleavage site and fusion domain (CON6, B.JRFL) constructs in recombinant vaccinia virus (rVV) and purified by lectin column chromatography. The ΔCFI or ΔCF forms of the protein were used for these experiments because of their improved immunogenicity and ability to assemble into trimers (Chakrabarti et al., 2002). Two HIV-1 gp120 proteins, B.6101 and B.BaL, were used as controls. Figure 2A shows a SDS-PAGE analysis of the two gp120 proteins, while Figure 2B depicts a BN-PAGE of CON-S gp140ΔCFI, CON6 gp140ΔCF and CON6 gp140ΔCFI as well as WT HIV-1 Env gp140 proteins. As expected, both gp120 proteins migrated as monomers, while CON-S and CON6 gp140ΔCFI Envs migrated as dimers, trimers and tetramers (Gao et al., 2005). B.JRFL gp140ΔCF protein migrated as monomers, dimers and trimers, while C.97ZA012 gp140ΔCFI migrated predominantly as trimers . The A.92RW020 gp140ΔCFI protein preparation contained monomers, dimers, and approximately 40% cleaved Env products in spite of our efforts to eliminate enzymatic degradation of Env during the protein purification process. Figure 2C shows Coomassie strained SDS-PAGE gel under reducing conditions. Approximately 40% of the A.92RW020 Env is cleaved resulting in a 96kd fragment, a pattern compatible with cleavage of gp120 near the V3 loop (Schultz et al. 1993). Analysis of A.92RW020 gp140CF Env after rVV expression and prior to Env purification demonstrated identical results (not shown). Thus, partial cleavage of A.92RW020 Env gp120 is a property of this particular Env when expressed in 293 T cells.
Figure 2.
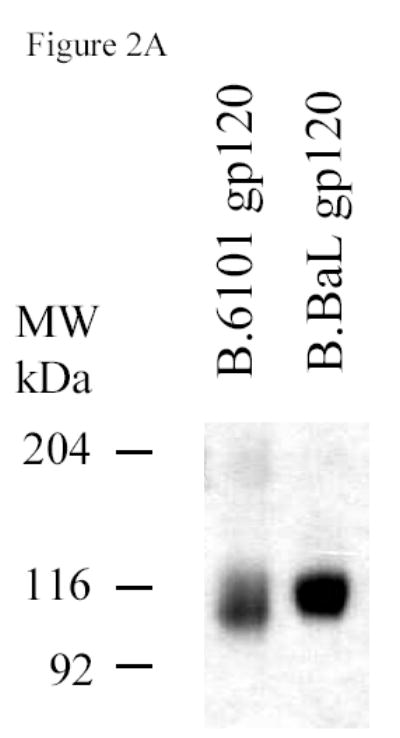
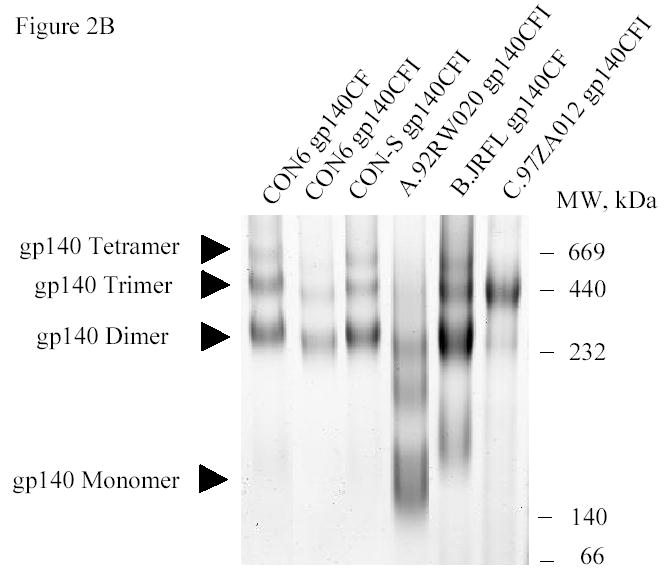
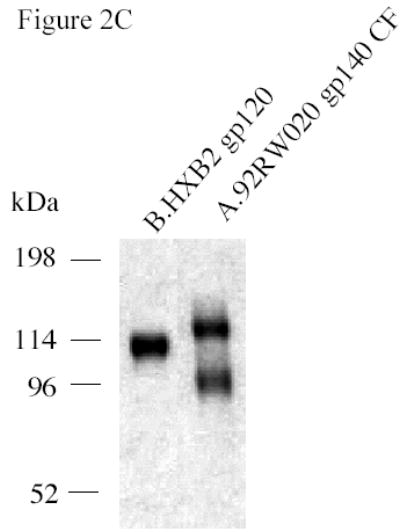
SDS-PAGE and blue native gel analysis of HIV-1 envelope proteins. Panel A shows the results of SDS-PAGE analysis of HIV-1 B.6101 and B.BaL gp120 proteins. HIV-1 gp120 proteins, as indicated on the top of the gel, were fractionated on 4–20% gradient SDS-PAGE under reducing conditions and stained by Coomassie blue. Panel B shows the results of blue native gel analysis of HIV-1 Env gp140 proteins with HIV-1 gp140 proteins indicated on the top of the 3–8% polyacrylamide gels and protein bands of molecular weight markers of 669 (Thyroglobulin), 440 (ferritin), 232 (catalase), 140 (lactate dehydrogenase) and 66 (albumin) kDa indicated on the right hand side of the gel. Arrowheads indicate HIV-1 protein forms (Panel B). Panel C shows SDS-PAGE analysis of the uncleaved HXB2 gp120 Env as a control versus A.92RW020 gp140ΔCF that is approximately 40% cleaved. Gel shows the results of HXB2 gp120 and A.92RW020 gp140ΔCF analyzed under reducing conditions in SDS-PAGE followed by Coomassie blue staining. In this gel, 96kDa band of A.92RW020 Env represents approximately 40% of this preparation. Analysis of 3 separate lots of purified A.92RW020 Env revealed the 96kDa fragment to be 39± 13% of the Env protein preparation.
To compare antigenic and functional epitopes expressed on the CON-S gp140ΔCFI protein with those expressed on CON6 gp140 and other WT HIV-1 gp120 and gp140ΔCF/CFI proteins, Envs were assayed for their ability to bind sCD4, as well as well-characterized anti-HIV-1 gp120 and gp41antibodies (Figure 3, Table 3). We found that CON-S gp140ΔCFI bound sCD4 and also bound human MAbs A32, 4E10, 2G12, 2F5 and mouse MAb T8 that recognizes the COOH-terminal region of HIV-1 gp120 (Table 3). To determine whether the CD4i MAb 17b that binds to the CCR5 binding site could be induced on the purified CON-S gp140ΔCFI Env, either sCD4, MAb A32, or MAb T8 was applied to a sensor chip to capture CON-S gp140ΔCFI, and then MAb 17b binding activity was determined. CON-S gp140CΔFI Env was induced to bind MAb17b only following binding to either sCD4 or MAb A32, but not following binding to MAb T8 (Figure 3B). The results of characterization of antigenic epitopes expressed on the various recombinant HIV-1 Env glycoproteins using surface plasmon resonance analysis are summarized in Table 3. All recombinant Envs bound to sCD4 and MAb A32. Most of the Envs expressed epitopes recognized by V3 specific antibodies, CD4-inducible antibodies and other epitopes recognized by MAb 2G12, 2F5 and 4E10 (Table 3). While the protein preparations are heterogeneous, these studies provided a relative assessment of expression of antigenic epitopes by the rVV-expressed proteins. In addition, that A.92RW020 Env was able to bind sCD4 and upregulate MAb17b binding provided additional rationale for testing this Env as an immunogen.
Figure 3.
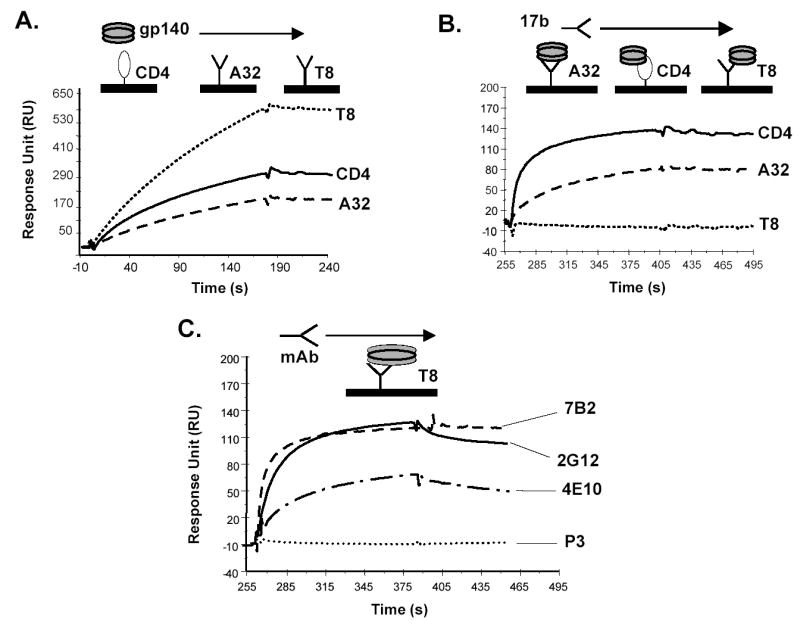
Analysis of antigenic epitopes expressed on CON-S gp140ΔCFI by surface plasmon resonance assays. Surface plasmon resonance assays were performed as described in Materials and Methods. Panel A shows the ability of CON-S gp140 CFI to bind to sCD4, MAbs A32 and T8.. sCD4 or HIV-1 MAbs T8 and A32 were covalently immobilized to a CM5 sensor chip (BIAcore), and CON-S gp140 CFI was injected over each surface (100 and 300 ug/ml, respectively). To determine induction of 17b MAb binding to CON-S gp140ΔCFI, CON-S gp140ΔCFI protein was captured (400 to 580 response units) on individual flow cells immobilized with sCD4 or MAb A32 or T8. Following stabilization of each of the surfaces, MAb 17b was injected and allowed to flow over each of the immobilized flow cells (Panel B). To determine binding of CON-S gp140ΔCFI protein to human HIV-1 MAbs, CON-S gp140ΔCFI protein was captured (400 to 580 response units) on individual flow cells immobilized with MAb T8. Following stabilization of each of the surfaces, the indicated human MAb 7B2, 2G12, 4E10b or irrelevant control MAb P3 was injected to flow over each of the immobilized flow cells (Panel C). Each analysis was performed at least twice.
Table 3.
Characterization of Antigenic Epitopes Expressed on Recombinant HIV-1 Env Immunogens.
| Antigens | CON6gp140ΔCF | CON6gp140ΔCFI | CON-S gp140ΔCFI | B.JRFL gp140ΔCF | A.92RW020gp140ΔCFI | C.97ZA012gp140CΔFI | B.BaL gp120 | B.6101gp120 |
|---|---|---|---|---|---|---|---|---|
| CD4 bs | ||||||||
| sCD4 | +* | +* | +* | +* | +* | +* | +* | +* |
| 1b12 | ++ | ++ | ++ | ++ | + | ND | ++ | − |
| V3 Loop Epitopes | ||||||||
| 19b | ++ | ++ | ++ | ++ | + | + | ++ | ++ |
| F2A3 | − | + | − | − | + | − | − | − |
| 39F | ++ | ++ | ++ | ++ | + | − | ++ | ++ |
| CCR5 binding site (MAb 17b) | ||||||||
| Constitutive | − | − | − | − | − | − | + | − |
| CD4 induced | + | + | + | + | + | − | ++ | − |
| A32 induced | + | + | + | ++ | ++ | − | ++ | |
| Other Epitopes | ++ | |||||||
| 2G12 | +++ | +++ | +++ | − | − | + | +++ | + |
| 2F5 | + | + | ++ | − | − | − | − | |
| 4E10 | ++ | + | + | − | − | − | − | − |
| A32 | +* | +* | +* | +* | +* | − | +* | +* |
For binding of Mab A32 or sCD4 to ENVs, + = specific binding. Binding was assessed by injecting Env proteins at 150ug/ml, over A32, CD4, or irrelevant mAb surfaces. About 150–200 responding units (RU) of specific binding was observed after subtraction of non-specific binding to the control surface.
Binding of Env proteins to all other mAbs were normalized by calculating binding ratios of each mAb to Env proteins captured on either immobilized T8 or A32 sensor surface. Binding ratio = Binding of mAbs over binding of ENVs to A32 or T8 in RU. Binding ratios were scored for this Table as follows: + = or > 0.05; ++ = >0.5; +++ = >1.0; − = no specific binding.
Immunogenicity of Consensus and WT HIV-1 Envs
To determine the ability of gp120, gp140 CFI, and gp140 CF envelope constructs to induce antibodies that neutralize HIV-1 primary isolates, guinea pigs were immunized with 100 ug of glycoprotein in RIBI adjuvant. Sera collected after 4 or 5 immunizations were tested for neutralizing activity using a panel of pseudoviruses containing the full length (gp160) Env proteins of subtype A, B, and C primary isolates. Sera of guinea pigs immunized with B.6101 gp120 induced neutralizing activities against three (B.BaL, B.SS1196 and B.SF162) of ten subtype B viruses and did not significantly neutralize any subtype C and A viruses. As previously reported (Liao et al., 2004), immune sera of animals immunized with B.BaL gp120 induced antibodies that neutralized six (B.BX08, B.QH0692, B.QH0515, B.SS1196, B.SF162 and the autologous BaL) out of ten subtype B viruses tested, but failed to induce antibodies that neutralized subtype C and subtype A viruses (Table 4).
Table 4.
Neutralization Titers of Guinea Pigs Immunized with HIV-1 Subtype B Monomeric gp120 Immunogens
|
B. 6101 gp120 |
B.BaL gp120 |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Guinea Pig Number |
Guinea Pig Number |
|||||||
| HIV-1 Isolate | 858 | 859 | 860 | 861 | 889 | 890 | 891 | 902 |
| B.BX08# | 21 | 20 | <20 | 24 | 368 | 540 | 637 | <20 |
| B.QH0692.42 | <20 | <20 | <20 | <20 | 27 | 38 | 55 | 45 |
| B.QH0515.1 | <20 | <20 | <20 | <20 | <20 | 36 | <20 | 40 |
| B.BaL.26 | 107 | 166 | 201 | 540 | 112 | 1,844 | 1,475 | 507 |
| B.SS1196.1 | 141 | 165 | 210 | 577 | 105 | 2,395 | 2,475 | 1,475 |
| B.SF162.LS | 265 | 180 | 227 | 1,298 | 530 | 130 | 1,397 | 424 |
| B.JRFL-MC ** | <20 | <20 | <20 | <20 | <20 | 23 | 43 | <20 |
| B.BG1168.1 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| B.3988.25 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| B.6101.10 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
50% Neutralization titers of serum after 4th or 5th immunizations as determined in the pseudotyped HIV-1 neutralization assays as described in Methods. Neutralization was considered positive (number in bold) if the titer of post-immune serum minus the titer of pre-immune bleed serum was >30. Guinea pigs anti-sera were tested as negative against 11 subtype C pseudotyped viruses including TV-1.21, DU123.6, DU172.17, 02ZM233.PB6, 92BR025.9, 02ZM197.PB7, 96ZM651.2, DU151.2, 97ZA012.29, DU422.01 and DU156.12, and 2 subtype A pseudotyped viruses, 92RWO20.2 and 92UG037.02. ND = Not done.
Assayed in Luc M7 cells using virus stock grown in PBMC.
A full-length molecular clone, and the assay stock of virus was produced by transfection in 293T cells.
We next compared the ability of the consensus and WT gp140 Envs to induce neutralizing antibodies against a panel of Tier 1 and Tier 2 HIV-1 Env-containing pseudovirions (Mascola et al., 2005) (Table 5). Of the WT proteins, we found that B.JRFL and C.97ZA012 Envs induced the least cross-reactive neutralizing antibodies, with neutralization of only B.SF162, and minimal neutralizationof C.TV-1 and C.92BR025.9 subtype C pseudovirions as well as group M consensus CON-S and CON6 Env-pseudotyped viruses. The subtype A Env protein induced a broader neutralizing antibody response, with neutralizing activity detectable against two subtype B (B.SS1196, B.SF162), two subtype C (C.TV-1 and C.92BR025.9) and the autologous subtype A (A.92RW020) Env-pseudotyped viruses (Table 5). There was minimal neutralization activity against CON6 Env-pseudotyped virus and no reactivity against seven additional subtype A primary viruses (Table 5, Table Legend). In contrast, the broadest neutralization response was observed with the CON-S Env immunogen. Specifically, we found that CON-S gp140 CFI envelope induced neutralizing antibodies that neutralized four subtype B (B.BX08, B.QH0692, B.SS196 and B.SF162), four subtype C (C.TV-1, C.DU123, C.DU172 and C.92BR025.9) and one subtype A (A.92RW020) isolate (Table 5). Moreover, the titers of neutralizing antibodies induced by CON-S gp140 CFI against most pseudoviruses were higher than those induced by WT Envs. In additional repeat experiments summarized in Table 6, four of four CON-S Env-immunized animals also had neutralizing activity to B.QH0692 and B.BaL pseudoviruses.
Table 5.
Neutralization Titers of Guinea pigs Immunized with HIV-1 Subtype A, B, C and Group M Consensus (CON-S) Env Immunogens*
|
92RW020 gp140
ΔCFI (Subtype A) |
JRFL
gp140ΔCF (Subtype B) |
97ZA012
gp140ΔCFI (Subtype C) |
CON-S
gp140ΔCFI |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Guinea Pig Number |
Guinea Pig Number |
Guinea Pig Number |
Guinea Pig Number |
|||||||||||||
| HIV-1 Isolate | 854 | 855 | 856 | 857 | 791 | 793 | 796 | 797 | 862 | 863 | 864 | 865 | 776 | 777 | 778 | 780 |
| B.BX08# | <20 | <20 | <20 | <20 | 23 | 22 | <20 | <20 | <20 | <20 | <20 | <20 | 1,196 | 412 | 4,856 | 1,817 |
| B.QH0692.42 | 34 | <20 | <20 | 36 | 108 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | 109 | <20 | <20 | <20 |
| B.SS1196.1 | 115 | 83 | 100 | 150 | 2,203 | 2,095 | 506 | 489 | 23 | 27 | <20 | <20 | 796 | 296 | 1,339 | 423 |
| B.SF162.LS | 1,546 | 412 | 1,301 | 984 | 1,489 | 1,888 | 92 | 290 | 128 | 421 | 88 | 106 | 31,224 | 8,186 | 41,667 | 13,369 |
| B.JRFL-MC** | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| C.TV-1.21 | 540 | 443 | 449 | 711 | <20 | <20 | <20 | <20 | 93 | 148 | <20 | <20 | 1,339 | 770 | 2,442 | 724 |
| C.DU123.6 | 41 | <20 | 48 | 37 | <20 | <20 | <20 | <20 | <20 | 115 | <20 | <20 | 176 | 329 | 387 | 378 |
| C.DU172.17 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | 235 | <20 | 213 |
| C.02ZM233M.PB6 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | 84 | 61 | 86 | 43 |
| C.92BR025.9 | 403 | 168 | 258 | 311 | <20 | <20 | <20 | <20 | 55 | 50 | <20 | 39 | 1,819 | 1,408 | 3,207 | 1,336 |
| C.02ZM197M.PB7 | <20 | <20 | <20 | 27 | 23 | 22 | <20 | <20 | 21 | 22 | <20 | <20 | <20 | 33 | 30 | <20 |
| A.92RW020.2 | 150 | 71 | 100 | 106 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | 116 | 204 | 95 | 177 |
| A.92UG037.01 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| CON-S | 44 | <20 | 27 | 112 | 22 | 26 | <20 | ND | 58 | 72 | 183 | 24 | 819 | 1,091 | 25 | <20 |
| CON6 | 27 | <20 | 22 | 45 | <20 | <20 | <20 | ND | 53 | 30 | <20 | <20 | 30 | <20 | <20 | 28 |
50% Neutralization titers of serum after 4th or 5th immunizations as determined in the pseudotyped HIV-1 neutralization assays as described in Methods. Neutralization was considered positive (number in bold) if the titer of post-immune serum minus the titer of pre-immune bleed serum was >30. All guinea pig sera were assayed against additional subtype B pseudotyped viruses, BG1168.1, 3988.25 and 6101.10, and subtype C pseudotyped viruses, 02ZM197M.PB7, 96ZM651.2, DU151.2, DU422.01 and DU156.12 and were negative. In addition, anti-CON-S sera, No. 776, 777, 778 and 780, and anti-92RW020 gp140 sera, 854, 855, 856, and 857 were assayed against additional 7 subtype A pseudotyped viruses (93TH976.01, Q23, Q168, Q259, Q461, Q769 and Q842) and were negative. ND= Not done.
Assayed in Luc M7 cells using virus stock grown in PBMC.
A full-length molecular clone, and the assay stock of virus were produced by transfection in 293T cells.
Table 6.
Geometric Means of Neutralization Titers of Guinea pigs Immunized with CON-S Env gp140ΔCFI Combined with Other Envs‡.
| CON-S gp140 ΔCFI |
CON-S gp140 ΔCFI
+C.97ZA012 gp140 ΔCFI |
CON-S gp140 ΔCFI
+B.BaL gp120 |
|||
|---|---|---|---|---|---|
| HIV-1 Isolate | Geometric Mean | Geometric Mean | p value* | Geometric Mean | p value* |
| B.SF162.LS | 17,697 | 5,451 | 0.004 | 16,846 | NS |
| B.BX08 # | 75 | 154 | NS | 152 | NS |
| B.QH0692.42 | 48 | 27 | 0.03 | 29 | NS |
| B.BaL.26 | 1,159 | 85 | 0.001 | 1,028 | NS |
| B.SS1196.1 | 627 | 89 | 0.02 | 880 | NS |
| C.TV-1.21 | 1,165 | 275 | 0.01 | 530 | NS |
| C.DU123.6 | 59 | 21 | NS | 36 | NS |
| C.92BR025.9 | 1,314 | 360 | 0.003 | 708 | NS |
Neutralization geometric means (GMT) of guinea pig sera after the 4th or 5th immunizations as determined in the pseudotyped HIV-1 neutralization assays as described in Methods.
Assayed in Luc M7 cells using virus stock grown in PBMC.
Data for CON-S gp140ΔCFI were from animals additional to those in Table 5. p values were obtained when comparison was made with GMT of guinea pigs immunized with CON-S gpΔ140CFI Env alone. For CON-S gp140ΔCFI as well as CON-S gp140ΔCFI + 97ZA012 gp140ΔCFI, n=4; for CON-S gp140ΔCFI + BaL gp120, n=3. NS = not statistically significant.
Next, we tested anti-CON-S and anti-C.97ZA012 guinea pig sera against B.SF162 and C.TV-1 in PBMC-based HIV-1 neutralization assays to verify the results obtained in the pseudotyped HIV-1 neutralization assays. We found that CON-S guinea pig sera neutralized both B.SF162 (geometric mean titer of 276, n=8) and C.TV-1 (geometric mean titer of 100, n=8), while anti-C.97ZA012 guinea pig did not neutralize either B.SF162 or C.TV-1 (geometric mean titer <20, n=4). We had previously observed that CON6 gp140ΔCF induced antibodies that neutralized selected subtype B but not non-B viruses (Gao et al., 2005). To examine whether enhanced induction of neutralizing antibodies by CON-S was due to the absence of the immunodominant gp41 region in the ΔCFI constructs, we generated the corresponding CON6 gp140ΔCFI construct (Chakrabarti et al., 2002; Gao et al., 2005). Immunization studies in guinea pigs showed that the additional deletion of 28 amino acids of the gp41 ectodomain had no effect on the immunogenicity of CON6 Env (Table 7). Both CON6 gp140 ΔCF and CON6 gp140CΔFI oligomers induced antibodies that neutralized B.BX08, B.QH0692 and B.SS1196 among a total of seven subtype B Env-containing pseudovirions. We next determined if anti-CON-S or CON6 antibodies could neutralize the CON-S or CON6 pseudoviruses.
Table 7.
Neutralization Titers of Guinea Pigs Immunized with CON6 gp140ΔCF and gp140ΔCFI Immunogens
|
CON6
gp140ΔCF |
CON6
gp140ΔCFI |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Guinea Pig Number |
Guinea Pig Number |
|||||||
| HIV-1 Isolate | 770 | 771 | 772 | 775 | 781 | 783 | 784 | 786 |
| B.BX08# | 520 | 257 | 428 | 189 | 1,930 | 390 | 2,979 | 645 |
| B.QH0692.42 | 46 | 55 | 58 | 77 | <20 | 91 | 100 | 76 |
| B.SS1196.1 | 398 | 306 | 284 | 222 | 482 | 323 | 1,151 | 446 |
| B.SF162.LS | 791 | 151 | 34 | 200 | 578 | 2,022 | 249 | 351 |
| B.JRFL-MC ** | <20 | <20 | <20 | <20 | <20 | 169 | <20 | <20 |
| B.BG1168.1 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| B.3988.25 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| B.6101.10 | <20 | <20 | <20 | <20 | <20 | <20 | <20 | <20 |
| CON6 | 65 | 151 | 48 | <20 | <20 | 1,308 | 41 | 260 |
50% Neutralization titers of serum after 4th or 5th immunizations as determined in the pseudotyped HIV-1 neutralization assays as described in Methods. Neutralization was considered positive (number in bold) if the titer of post-immune serum minus the titer of pre-immune bleed serum was >30. Guinea pigs sera were aslo tested as negative against 11 subtype C pseudotyped viruses including TV-1.21, DU123.6, DU172.17, 02ZM233.PB6, 92BR025.9, 02ZM197.PB7, 96ZM651.2, DU151.2, 97ZA012.29, DU422.01, DU156.12, and 2 subtype A pseudotyped viruses, 92RWO20.2 and 92UG037.02.
Assayed in Luc M7 cells using virus stock grown in PBMC.
A full-length molecular clone, and the assay stock of virus were produced by transfection in 293T cells.
We found that two of four anti-CON-S antisera neutralized CONS pseudovirus, but did not significantly neutralize CON6 (Table 5), while six of eight animals immunized with CON6 Envs developed antibodies that neutralized CON6 pseudovirus (Table 7).
Ability of CON-S variable loop peptides to absorb the neutralizing activity of CON-S Env-induced antibodies
Most of the amino acid sequence differences between CON6 and CON-S Envs are localized in the variable loops (Figure 4). To determine if any particular variable loop contributed to the ability of CON-S to induce antibodyes that neutralized non-subtype B isolates, peptides derived from the V1, V2, V3, V4 and V5 loop regions of both CON-S and CON6 Envs were synthesized, purified, and tested for their ability to absorb the neutralizing activity against B.SS1196, C.TV-1, C.DU123, C.DU172 and C.02ZM233M and A.92RW020 Env-pseudotyped viruses.
Figure 4.
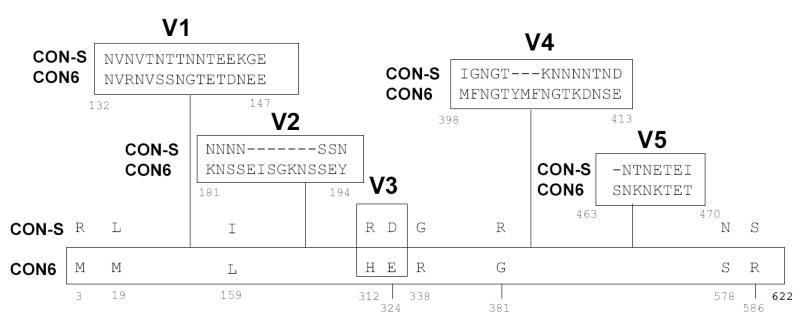
Amino acid differences in CON6 gp140ΔCFI and CON-S gp140ΔCFI. Differences were indicated using single letter code. Positions of a.a. are indicated by numbers beneath the amino acids. The dashed line(s) indicate the deletion of a.a. at the position. Amino acid differences in the Env variable loops are shown in boxes.
While CON-S V1, V2, V4 or V5 peptides did not absorb the neutralizing activity of anti-CON-S sera to any of 6 Env-containing pseudoviruses tested (Figure 5), 86% of the neutralizing activity of anti-CON-S sera for B.SS1196 and 92% of the neutralizing activity for C.TV-1 was absorbed by CON-S V3 peptide. In addition, an average of 48% of the neutralizing activity of anti-CON-S sera for non-subtype B Env-pseudotyped viruses including C.DU123 (67%), C.DU172 (33%), C.02ZM233M (43%) and A.92RW020 (48%), was absorbed by the CON-S V3 peptide (Figure 5A). In comparison, the CON6 V3 peptide had weaker but significant ability to absorb neutralization activity of anti-CON-S sera for subtype B pseudoviruses (upper left panel in Figure 5A). While CON6 V3 peptide absorbed 50% of the neutralizing activity of anti-CON-S sera for B.SS1196 pseudovirus, the CON6 V3 peptide could not absorb any CON-S-induced anti-subtype C neutralizing antibodies (Figure 5A). Thus, the CON6 V3 peptide could assume a conformer structure to partially absorb the CON-S-induced subtype B neutralizing antibodies, but could not assume a conformer sufficiently similar to CON-S V3 peptide to absorb the CON-S-induced subtype C neutralizing antibodies.
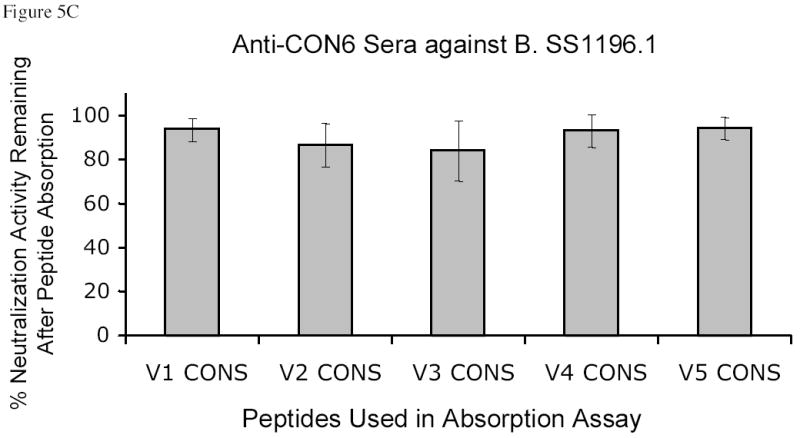
Figure 5.
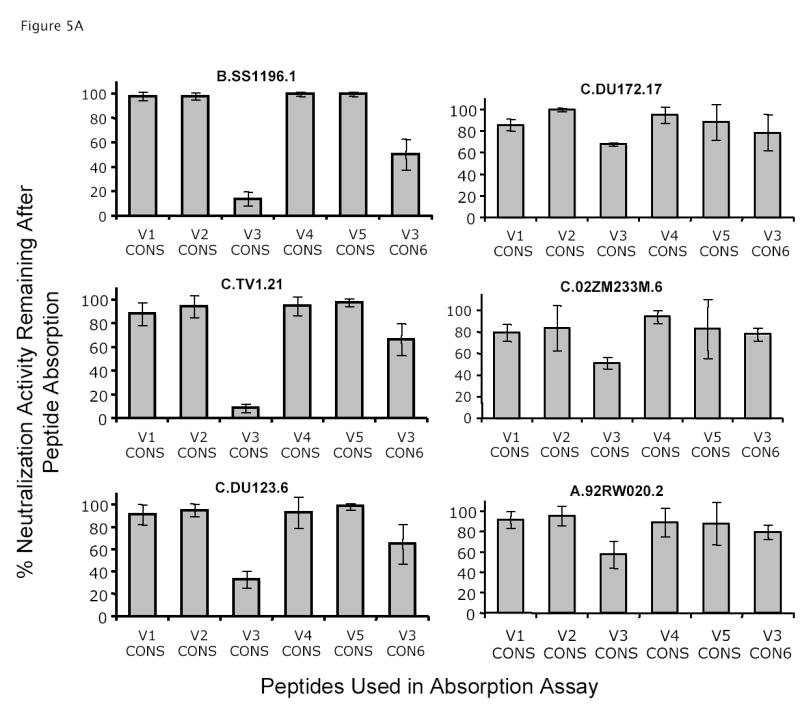
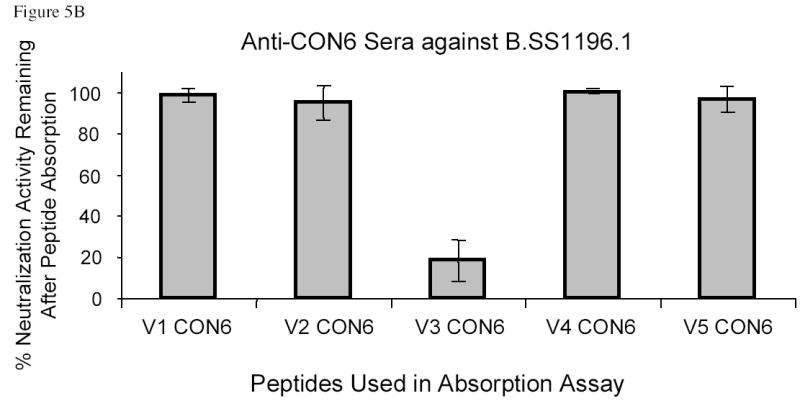
Specificity of neutralization activity of anti-CON-S and anti-CON6 guinea pig sera. Absorption assays were performed as described in the Materials and Methods. Results were plotted on y axis as mean % of neutralization activity remaining after absorption of 4 serum samples with the indicated CON-S or CON6 V1, V2, V3, V4, and V5 peptides (V1–V5 CON-S or CON6). The error bars indicated the standard error deviations. Panel A shows the neutralization activity against six HIV-1 primary isolates by anti-CON-S sera after absorption with CON-S variable loop peptides or CON6 V3 peptide as indicated in the bottom of the column plots. Panel B shows the neutralization activity against B.SS1196 by anti-CON6 sera after absorption with CON6 variable loop peptides. Panel C shows the neutralization activity against B.SS1196 by anti-CON6 sera after absorption with CON-S V3 variable loop peptides.
CON6 V1, V2, V4 and V5 peptides did not absorb neutralizing activity induced by either CON6 (Figure 5B) or CON-S gp140ΔCFI proteins (data not shown), although CON6 V3 peptides did absorb CON6 Env-induced neutralizing activity against B.SS1196. Further, none of CON-S V1, V2, V3, V4 and V5 peptides absorbed the anti-B.SS1196 neutralizing antibodies induced by CON6 gp140ΔCFI (Figure 5C).
Since high titers of ELISA binding antibody were detected in anti-CON-S sera that reacted with the CON-S V2 and V3 peptides (data not shown), we tested if a combination of CON-S V2 and V3 peptides could absorb more neutralization activity of anti-CON-S sera than CON-S V3 peptide alone. We found no additive or synergistic effect of CON-S V2 peptide mixed with CON-S V3 peptide on the absorption of neutralizing activity (data not shown).
Certain HIV-1 V3 peptides have been shown to be potent inducers for the induction of antibodies that neutralize T cell line adapted isolates and selected HIV-1 primary isolates (Palker et al., 1988, 1989; Rusche et al., 1988; LaRosa et al., 1990; Liao et al., 2000; Haynes et al., 2005). Since the CON-S V3 peptide absorbed a major proportion of the neutralizing activity of anti-CON-S gp140ΔCFI sera, we next tested if the same V3 peptide used in the absorption studies or an optimally immunogenic CON-S V3 peptide (GTH1-V3) that contains the CON-S V3 loop sequence with a N-terminal T helper gag p24 epitope (GTH1), could induce a similar spectrum of neutralizing antibodies as the CON-S gp140ΔCFI Env. We found in 4 guinea pigs for each peptide that neither CON-S V3 peptide nor CON-S GTH1-V3 peptide induced antibodies that neutralized subtype B or C HIV-1 Env containing pseudoviruses (data not shown). Thus, CON-S V3 peptides could absorb much of the anti-CON-S neutralizing activity, but when used as immunogens themselves, they could not induce the neutralizing activity, indicating a requirement of the CON-S V3 conformation in the context of the gp140 Env for immunogenicity.
Induction of Neutralizing Antibodies with CON-S gp140ΔCFI Combined with Other Envs
We have previously reported that B.BaL gp120 induced antibodies that neutralized approximately 40% of subtype B isolates tested (Liao et al., 2004). To determine if the ability of CON-S gp140ΔCFI to induce HIV-1 neutralizing antibodies could be enhanced by immunization with a combination of CON-S and other HIV-1 envelope immunogens, we immunized guinea pigs with CON-S gp140ΔCFI combined with either WT C.97ZA012 gp140ΔCFI or B.BaL gp120 (100ug of each Env/injection). We found that immune sera of guinea pigs immunized with the combination immunogens neutralized the same number of HIV-1 isolates with essentially the same antibody titers compared to immune sera of guinea pigs immunized with CON-S gp140ΔCFI alone (Table 6). Interestingly, not only did the combination of CON-S with subtype C gp140ΔCFI protein not enhance the ability to induce neutralizing antibodies, the neutralizing antibody titers induced by the combination of WT C + CON-S gp140ΔCFI were significantly reduced (Table 6). For example, the addition of C.97ZA012 gp140ΔCFI to CON-S gp140ΔCFI reduced the geometric mean titer (GMT) of anti-B.SF162 neutralizing antibodies from 17,697 to 5,451 (p=0.004, n=4) and of anti-C.TV-1 neutralizing antibodies from 1,165 to 275 (p=0.01, n=4). Thus, not only was the subtype C Env not additive in inducing neutralizing antibodies, the WT subtype C Env was in some way suppressing the neutralizing antibody responses induced by CON-S gp140ΔCFI.
Discussion
In this study, we have compared the ability of a group M consensus Env immunogen (CON-S gp140ΔCFI) to induce neutralizing antibodies to that of five WT Env constructs representing subtypes A, B and C. Among the wild-type Env immunogens, the subtype A Envs induced antibodies in guinea pigs that neutralized a subset of subtype B and C Envs. However, the group M consensus Env, CON-S, elicited cross-subtype neutralizing antibodies with similar or greater breadth and titer than WT Envs.
Seaman et al. (2005) have studied the immunogenicity of WT A (A.92RW020), WT B (B.HXB2 with BaL V3 loop), and WT C Envs (C.97ZA012), and showed that when formulated as a DNA prime, recombinant adenovirus (rAd) boost, the WT A was superior to WT B and C Env genes for the induction of neutralizing antibodies. They also showed that the WT C Env gene induced minimal neutralizing antibodies to primary isolates (Seaman et al., 2005). Our studies using the same subtype A and C recombinant protein gp140ΔCFI immunogens confirm these studies, and further to show that the WT C immunogen was suppressive of subtype B and C neutralizing responses induced by CON-S. These studies may in part provide an explanation for the limited magnitude and breadth of neutralization responses that have been induced by subtype C immunogens. In this regard, Seaman et al. studied combinations of C.97ZA012 Env genes with WT subtype B and A Env genes and did not see suppression of antibody responses in Env gene combinations that included WT C. It is possible that this was due to the use of DNA prime, rAd boost or due to specific interactions of subtype C and CON-S Env in the induction of neutralizing antibodies. Nonetheless, additional subtype C Envs should be studied to determine if the subtype C Env in some generic manner inhibits the induction of anti-HIV-1 neutralizing antibodies.
Our year 2001 group M consensus CON-S Env now represents the third centralized Env that has been studied for immunogenicity (Gao et al., 2005) (Doria-Rose et al., 2005). Here and in our previous study of CON6 Env, we show that this immunogen induces neutralizing antibodies only against a subset of subtype B viruses. Similarly, Doria-Rose et al. reported only weak neutralizing responses exclusively against subtype B viruses and no neutralization of the non-subtype B strains (suntype C isolate S080 and clade E isolate CM244) using an ancestral subtype B Env gene in a DNA prime and recombinant protein boost immunization regimen (Doria-Rose et al., 2005). One difference of possible significance between the ancestral B antigen and the M group consensus sequences described here is that the B ancestor sequence had very long hypervariable loops with more glycosylation sites than natural strains.
A critical issue in HIV-1 vaccine development is to determine the criteria for choosing any one particular Env or combination of Envs for inclusion into candidate immunogens (Hurwitz et al., 2005; Rollman et al., 2004; Srivastava et al., 2004; VanCott et al., 1997, 1999; Barnett et al., 2001; Fouts et al., 2002; Hewer and Meyer, 2002; Dong et al., 2003; Seaman et al., 2005; Mascola et al., 2005; Poon et al., 2005; Lian et al., 2005; Pal et al., 2005). Our data indicate that consensus env genes such as CON-S can be designed that express the same antigenic epitopes as WT Envs, but have improved immunogenicity. Our observations demonstrate that iterative improvement of Env immunogenicity, whether using centralized and/or structure based designs, is achievable. The variable loops of CON-S were purposefully shortened to mimic the shorter variable loops observed for env genes of acute subtype A and C infections. Several reports indicate that exposure of certain neutralizing epitopes participating in the envelope-CD4 and -coreceptor binding can be increased by mutations and deletions of the variable domains of HIV-1 envelope glycorprotein (Kang et al. 2005; Gzyl et al., 2004; Kim et al 2003; Barnett et al. 2001; Wyatt et al. 1993). Yang et al. (2004) have demonstrated induction of improved levels of neutralizing antibodies with Envs with shortened V3 loops. Conserved neutralization epitopes of receptor binding sites are located in the recessed core of HIV-1 envelope, partially masked by variable loops and glycosylations. Thus, to elicit antibodies that can broadly neutralize genetically diverse HIV-1 isolates, it may be important to design vaccines in which the conserved neutralizing epitopes are exposed. However, it remains to be determined that if the enhanced ability of CON-S Env for induction of broadly reactive neutralizing antibody was contributed by the shortened variable loops in CON-S Env. In this regards, we have recently determined if anti-CON-S sera have antibodies capable of blocking the binding of MAbs 2F5 and 2G12 as well as sCD4 to CON-S gp140 oligomers. While anti-CON-S sera did not block the binding of MAbs 2F5 and 2G12 to CON-S Env, anti-CON-S sera (n=4) did block >75% binding of sCD4 to CON-S Env at 1:100 dilution (Liao, H-X., Xia, S-M., Park, R. and Haynes, BF. manuscript in preparation). Thus, anti-CD4 binding site antibodies may contributed to the neutralization of non-subtype B isolates by anti-CON-S sera.
Given the limited heterogeneity of most transmitted HIV-1 quasispecies (Chohan et al., 2005; Derdeyn et al., 2004), it is reasonable to expect continued improvement in centralized Env design when a large database of transmitted virus Envs of multiple HIV-1 subtypes becomes available. Such a database is currently under development (Korber, B, Haynes, B, Hahn, B et al. unpublished observations). Furthermore, experiments are underway to test the next generation M group consensus Envs based on the 2003 HIV-1 Sequence Database, to include only the minimal conserved elements in the hypervariable loops. It is also interesting to note that while V3 peptides absorbed >85% anti-CON-S neutralizing activity for the Tier 1 B.SS1196 and C.TV-1 viruses, V3 peptides were only able to reduce 48% of the neutralizing activity for the Tier 2 C.DU123, C.DU172, C.92M233M, and A.92RW020 strains. It will be important to construct a V3 deleted CON-S to determine if, in the absence of V3, CON-S can induce enhanced levels of non-V3 neutralizing antibody.
That much of the neutralizing activity induced by CON-S Env was absorbed by the CON-S V3 peptide was of interest, and suggested that the V3 loop of CON-S gp140 CFI was in a conformation required for induction of neutralizing antibodies to subtype C primary viruses. The V3 of subtype C is not particularly variable (Ping et al., 1999; Williamson et al., 2003; Gaschen et al., 2003). As such, the inability of CON-S to induce neutralizing antibodies to six of eleven subtype C strains suggests that like subtype B (Sullivan et al., 1998), the WT C V3 loop may not be accessible on the surface of the virion in a substantial number of WT subtype C Envs. Alternatively, the V3 loop may be accessible, but anti-V3 antibodies may bind with insufficient strength to neutralize HIV-1 (Gorny et al., 2004).
In this regard, it is important to note that the CON6, WT subtype C, and CON-S Envs all have GPGQ at the tips of the V3 loops, yet only CON-S induced any antibodies against subtype C viruses (Table 7). Gorny et al. (2005) have recently demonstrated that human anti-V3 MAbs derived from patients with non-B HIV-1 with GPGQ V3s have greater breadth against HIV-1 primary isolates than human mabs derived from patients with subtype B HIV-1 with GPGR V3s (Zolla-Pazner et al., 2004). This suggests that in patients with chronic HIV-1 infection, the GPGQ V3 in select patients is able to induce broadly neutralizing antibodies against a functional conformation of V3. However, the opposite result is observed in the setting of animals immunized with either V3 immunogens or V3 containing WT Envs, in that subtype B V3s and oligomeric Envs induce neutralizing antibodies with more breadth than do subtype C V3 peptides and subtype C Envs (Haynes et al., 2005; Liao et al., 2004; Haynes, B.F., Korber, B.T., unpublished). Whether this difference is due to the duration of immunizations, adjuvant formulations, or conformations of the Env and/or V3 Env subunits remain to be determined. Whatever the explanation, the gp140 CON-S Env differs from WT subtype C Env in its ability to induce neutralizing antibodies. Study of the structural differences between CON6, CON-S, and WT Envs should provide important information regarding the design of immunogens for induction of neutralizing antibodies with more breadth than is currently attainable.
In summary, the results reported here suggest that with continued iterative improvements in design, centralized Env immunogens may be of value for the induction of cross-reactive neutralizing antibody responses.
Acknowledgments
The authors thank Robert Parks and Christopher Ryan for expert technical assistance, James Robinson, Hermann Katinger, Dennis Burton and the NIAID AIDS Reagent Repository for reagents, and Kim McClammy for expert secretarial assistance. Supported by NIH grants PO1 AI52816, P30 AI51445, HIVRAD PO-1, AI061734, NO1 AI 85338, R21AI55386 and PO1 AI 28147.
References
- Andre S, Seed B, Eberle J, Schraut W, Bultmann A, Haas J. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J Virol. 1998;72:1497–503. doi: 10.1128/jvi.72.2.1497-1503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam S, Paleos C, Liao HX, Scearce R, Robinson J, Haynes B. An inducible HIV type 1 gp41 HR-2 peptide-binding site on HIV type 1 envelope gp120. AIDS Res Hum Retroviruses. 2004;20(8):836–845. doi: 10.1089/0889222041725181. [DOI] [PubMed] [Google Scholar]
- Barnett S, Lu S, Srivastava I, Cherpelis S, Gettie A, Blanchard J, Wang S, Mboudjeka I, Leung L, Lian Y, Fong A, Buckner C, Ly A, Hilt S, Ulmer J, Wild C, Mascola J, Stamatatos L. The Ability of an Oligomeric Human Immunodeficiency Virus Type 1 (HIV-1) Envelope Antigen To Elicit Neutralizing Antibodies against Primary HIV-1 Isolates Is Improved following Partial Deletion of the Second Hypervariable Region. J Virol. 2001;75:5526–5540. doi: 10.1128/JVI.75.12.5526-5540.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bures R, Gaitan A, Zhu T, Graziosi C, McGrath KM, Tartaglia J, Caudrelier P, El Habib R, Klein M, Lazzarin A, Stablein DM, Deers M, Corey L, Greenberg ML, Schwartz DH, Montefiori DC. Immunization with recombinant canarypox vectors expressing membrane-anchored gp120 followed by gp160 protein boosting fails to generate antibodies that neutralize R5 primary isolates of human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 2000;16:2019–2035. doi: 10.1089/088922200750054756. [DOI] [PubMed] [Google Scholar]
- Chakrabarti BK, Kong WP, Wu BY, Yang ZY, Friborg J, Ling X, King SR, Montefiori DC, Nabel GJ. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J Virol. 2002;76(11):5357–68. doi: 10.1128/JVI.76.11.5357-5368.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien PC, Jr, Cohen S, Kleeberger C, Giorgi J, Phair J, Zolla-Pazner S, Hioe CE. High levels of antibodies to the CD4 binding domain of human immunodeficiency virus type 1 glycoprotein 120 are associated with faster disease progression. J Infect Dis. 2002;186(2):205–13. doi: 10.1086/341297. [DOI] [PubMed] [Google Scholar]
- Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1–V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79:6528–66531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derdeyn C, Decker J, Bibollet-Ruche F, Mokili J, Muldoon MSA, SA D, Heil M, Kasolo F, Musonda R, Hahn B, Shaw G, Korber B, Allen S, Hunter E. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O’Brien WA, Ratner L, Kappes JC, Shaw GMEH. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong M, Zhang PF, Grieder F, Lee J, Krishnamurthy G, VanCott T, Broder C, Polonis VR, Yu XF, Shao Y, Faix D, Valente P, Quinnan GVJ. Induction of primary virus-cross-reactive human immunodeficiency virus type 1-neutralizing antibodies in small animals by using an alphavirus-derived in vivo expression system. J Virol. 2003;77:3119–3130. doi: 10.1128/JVI.77.5.3119-3130.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria-Rose N, Learn GH, Rodrigo A, Nickle DC, Li F, Mahalanabis M, Hensel MT, McLaughlin S, Edmonson P, Montefiori D, Barnett S, Haigwood N, JI M. Human immunodeficiency virus type 1 subtype B ancestral envelope protein is functional and elicits neutralizing antibodies in rabbits similar to those elicited by a circulating subtype B envelope. J Virol. 2005;79:11214–11224. doi: 10.1128/JVI.79.17.11214-11224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl P. Short Technical Reports: Compacts, synthetic, vaccinia virus early/late promoter for protein expression. BioTechniques. 1997;23:1094–1097. doi: 10.2144/97236st07. [DOI] [PubMed] [Google Scholar]
- Fouts T, Godfrey K, Bobb K, Montefiori D, Hanson C, Kalyanaraman VS, DeVico A, Pal R. Crosslinked HIV-1 envelope-CD4 receptor complexes elicit broadly cross-reactive neutralizing antibodies in rhesus macaques. Proc Natl Acad Sci (USA) 2002;99:11842–11847. doi: 10.1073/pnas.182412199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Weaver E, Lu Z, Li Y, Liao H-X, Ma B-J, Alam SM, Scearce R, Sutherland LL, Decker J, Shaw G, Montefiori D, Korber B, Hahn B, Haynes B. Antigenicity and Immunogenicity of a Synthetic HIV-1 Group M Consensus Envelope Glyclprotein. J Virol. 2005;79(2):1154–1163. doi: 10.1128/JVI.79.2.1154-1163.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Novitsky V, Haynes B, Hahn B, Bhattacharya T, B K. Diversity considerations in HIV-1 vaccine selection. Science. 2003;299(5612):1515–1518. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- Gorny MK, VanCott TC, Williams C, Revesz K, Zolla-Pazner S. Effects of oligomerization on the epitopes of the human immunodeficiency virus type 1 envelope glycoproteins. Virology. 2000;267:2208. doi: 10.1006/viro.1999.0095. [DOI] [PubMed] [Google Scholar]
- Gorny MK, Revesz K, Williams C, Volsky B, Louder MK, Anyangwe CA, Krachmarov C, Kayman SC, Pinter A, Nadas A, Nyambi PN, Mascola JR, Zolla-Pazner S. The V3 loop is accessible on the surface of most human immunodeficiency virus type 1 primary isolates and serves as a neutralization epitope. J Virol. 2004;78(5):2394–404. doi: 10.1128/JVI.78.5.2394-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorny MK, Williams C, Volsky B, Revesz K, Wang X-H, Burda S, Nyambi P, Krachmarov C, Pinter A, Zolla-Pazner S. Cross-neutralizing activity of human anti-V3 monoclonal antibodies derived from the cells of individuals infected with non-B subtypes of HIV-1. (Abstract), page 23, No. 24; AIDS Vaccine 2005, Sept. 2005. Montreal, Quebec; Canada: [Google Scholar]
- Gzyl J, Bolesta E, Wierzbicki A, Kmieciak D, Naito T, Honda M, Komuro K, Kaneko Y, Kozbor D. Effect of partial and complete variable loop deletions of the human immunodeficiency virus type 1 envelope glycoprotein on the breadth of gp160-specific immune responses. Virology. 2004;318(2):493–506. doi: 10.1016/j.virol.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Haynes BF, Ma B-J, Montefiori DC, Wrin T, Petropoulos CJ, Sutherland LL, Scearce RM, Denton C, Xia S-M, Korber BT, Liao H-X. Analysis of HIV-1 Subtype B Third Variable Region Peptide Motifs For Induction of Neutralizing Antibodies Against HIV-1 Primary Isolates. Virology. 2006;345:44–55. doi: 10.1016/j.virol.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Hewer R, Meyer D. Producing a highly immunogenic synthetic vaccine construct active against HIV-1 subtype C. Vaccine. 2002;20:2680–2683. doi: 10.1016/s0264-410x(02)00213-x. [DOI] [PubMed] [Google Scholar]
- Hurwitz JL, Slobod K, Lockey T, Wang S, Chou T, Lu S. Hurwitz JL., Slobod KS., Lockey TD., Wang S., Chou, TH., Lu S., Application of the polyvalent approach to HIV-1 vaccine development. Current Drug Targets - Infectious Disorders. 2005;5(2):143–156. doi: 10.2174/1568005054201517. [DOI] [PubMed] [Google Scholar]
- Kang SM, Quan FS, Huang C, Guo L, Ye L, Yang C, Compans RW. Modified HIV envelope proteins with enhanced binding to neutralizing monoclonal antibodies. Virology. 2005;331(1):20–32. doi: 10.1016/j.virol.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Kim YB, Han DP, Cao C, Cho MW. Immunogenicity and Ability of Variable Loop-Deleted Human Immunodeficiency Virus Type 1 Envelope Glycoproteins to Elicit Neutralizing Antibodies. Virology. 2003;305:124–137. doi: 10.1006/viro.2002.1727. [DOI] [PubMed] [Google Scholar]
- LaRosa GJ, Davide JP, Weinhold K, Waterbury JA, Profy AT, Lewis JA, Langlois AJ, Dreesman GR, Boswell RN, Shadduck P, Holley LH, Karplus M, Bolognesi DP, Matthews TJ, Emini EASDP. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science. 1990;249(4971):932–935. doi: 10.1126/science.2392685. [DOI] [PubMed] [Google Scholar]
- Li M, Gao F, Mascola JR, Stamatatos L, Polonis VR, Koutsoukos M, Voss G, Goepfert P, Gilbert P, Greene KM, Bilska M, Kothe DL, Salazar-Gonzalez JF, Wei X, Decker JM, Hahn BH, Montefiori DC. Human immunodeficiency virus type 1 env clones from acute and early subtype B infections for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2005;79:10108–10125. doi: 10.1128/JVI.79.16.10108-10125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian Y, Srivastava I, Gomez-Roman VR, Megede JZ, Sun Y, Kan E, Hilt S, Engelbrecht S, Himathongkham S, Luciw PA, Otten G, Ulmer JB, Donnelly JJ, Rabussay D, Montefiori D, van Rensburg EJ, Barnett SW. Evaluation of envelope vaccines derived from the South African subtype C human immunodeficiency virus type 1 TV1 strain. J Virol. 2005;79(21):13338–49. doi: 10.1128/JVI.79.21.13338-13349.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-X, Alam M, Mascola J, Robsinson J, Ma B-J, Montefiori D, Rhein M, Sutherland L, Scearce M, Haynes B. Immunogenicity of Constrained Monoclonal Antibody A32- HIV Env gp120 Complexes Compared to Recombinant HIV-1 gp 120 Envelope Glycoproteins. J Virol. 2004;78:5270–5278. doi: 10.1128/JVI.78.10.5270-5278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H-X, Etemad-Moghadam B, Montefiori DC, Sun Y, Sodroski J, Scearce RM, Doms RW, Thomasch JR, Robinson S, Letvin NLBFH. Induction of antibodies in guinea pigs and rhesus monkeys against the human immunodeficiency virus type 1 envelope: Neutralization of non-pathogenic and pathogenic primary isolate simian/Human immunodeficiency virus strains. J Virol. 2000;74(1):254–263. doi: 10.1128/jvi.74.1.254-263.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, Sambor A, Beaudry K, Santra S, Welcher B, Louder MK, Vancott TC, Huang Y, Chakrabart iBK, Kong WP, Yang ZY, Xu L, Montefiori DC, Nabel GJ, Letvin NL. Neutralizing antibodies elicited by immunization of monkeys with DNA plasmids and recombinant adenoviral vectors expressing human immunodeficiency virus type 1 proteins. J Virol. 2005;79:771–79. doi: 10.1128/JVI.79.2.771-779.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascola JR, D’Souza P, Gilbert P, Hahn BH, Haigwood NL, Morris L, Petropoulos CJ, Polonis VR, Sarzotti M, Montefiori DC. Recommendations for the design and use of standard virus panels to assess neutralizing antibody responses elicited by candidate human immunodeficiency virus type 1 vaccines. J Virol. 2005;79:10103–10107. doi: 10.1128/JVI.79.16.10103-10107.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montefiori DC, Pantaleo G, Fink LM, Zhou JT, Zhou JY, Bilska M, Miralles GD, Fauci AS. Neutralizing and infection-enhancing antibody responses to human immunodeficiency virus type 1 in long term non-progressors. J Infect Dis. 1996;173:60–67. doi: 10.1093/infdis/173.1.60. [DOI] [PubMed] [Google Scholar]
- Montefiori DC. In: Evaluating neutralizing antibodies against HIV, SIV and SHIV in luciferase reporter gene assays. Current Protocols in Immunology. Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, Coico R, editors. John Wiley & Sons; 2004. pp. 12.11.1–12.11.15. [DOI] [PubMed] [Google Scholar]
- Moss B, Earl P. In: Current Protocols in Molecular Biology. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. John Wiley & Sons, Inc; Indianapolis, IN: 1998. p. 16:15.116.19.9. [Google Scholar]
- Pal R, Wang S, Kalyanaraman VS, Nair BC, Whitney S, Keen T, Hocker L, Hudacik L, Rose N, Cristillo A, Mboudjeka I, Shen S, Wu-Chou T-H, Montefiori D, Mascola J, Lu S, Markham P. Polyvalent DNA prime and envelope protein boost HIV-1 vaccine elicits humoral and cellular responses and controls plasma viremia in rhesus macaques following rectal challenge with an R5 SHIV isolate. J Med Primat. 2005;34:226–236. doi: 10.1111/j.1600-0684.2005.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palker TJ, Clark ME, Langlois AJ, Matthews TJ, Weinhold KJ, Randall RR, Bolognesi DP, Haynes BF. Type-specific neutralization of the human immunodeficiency virus with antibodies to env-encoded synthetic peptides. Proc Natl Acad Sci (USA) 1988;85:1932–1936. doi: 10.1073/pnas.85.6.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palker TJ, Matthews TJ, Langlois AL, Tanner ME, Martin ME, Scearce RM, Kim JE, Berzofsky JA, Bolognesi DP, Haynes BF. Polyvalent human immunodeficiency virus synthetic immunogen comprised of envelope gp120 T helper cell sites and B cell neutralization epitopes. J Immunol. 1989;142(10):3612–3619. [PubMed] [Google Scholar]
- Ping LH, Nelson JA, Hoffman IF, Schock J, Lamers SL, Goodman M, Vernazza P, Kazembe P, Maida M, Zimba D, Goodenow MM, Eron JJJ, Fiscus SA, Cohen MS, Swanstrom R. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J Virol. 1999;73:6271–6281. doi: 10.1128/jvi.73.8.6271-6281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poli G, Fauci A. In: Isolation and Quantitation of HIV in Peripheral Blood. Current Protocols in Immunology. Coligan JE, Kruisbeek AM, Margulies DH, Shevach EM, Strober W, Coico R, editors. John Wiley & Sons; 2004. pp. 12.2.5–12.2.6. [DOI] [PubMed] [Google Scholar]
- Poon B, Hsu JF, Gudeman V, Chen IS, Grovit-Ferbas K. Formaldehyde-treated, heat-inactivated virions with increased human immunodeficiency virus type 1 env can be used to induce high-titer neutralizing antibody responses. J Virol. 2005;79:10210–10707. doi: 10.1128/JVI.79.16.10210-10217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzuto C, Sodroski J. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res Hum Retroviruses. 2000;16(8):741–749. doi: 10.1089/088922200308747. [DOI] [PubMed] [Google Scholar]
- Rollman E, Hinkula J, Arteaga J, Zuber B, Kjerrstrom A, Liu M, Wahren B, Ljungberg K. Multi-subtype gp160 DNA immunization induces broadly neutralizing anti-HIV antibodies. Gene Therapy. 2004;11:1146–1154. doi: 10.1038/sj.gt.3302275. [DOI] [PubMed] [Google Scholar]
- Rusche J, Javaherian K, McDanal C, Petro J, Lynn D, Grimaila R, Langlois A, Gallo R, Arthur L, Fischinger P, Bolognesi DP, Matthews TJ, Putney S. Antibodies that inhibit fusion of human immunodeficiency virus-infected cells bind a 24-amino acid sequence of the viral envelope, gp120. Proc Natl Acad Sci (USA) 1988;89(9):3198–3202. doi: 10.1073/pnas.85.9.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochemistry. 1994;217:220–230. doi: 10.1006/abio.1994.1112. [DOI] [PubMed] [Google Scholar]
- Schulke N, Vesanen MS, Sanders RW, Zhu P, Lu M, Anselma DJ, Villa AR, Parren PW, Binley JM, Roux KH, Maddon PJ, Moore JP, Olson WC. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J Virol. 2002;76:7760–7776. doi: 10.1128/JVI.76.15.7760-7776.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz TF, Reeves JD, Hoad JG, Tailor C, Stephens P, Clements G, Ortlepp S, Page KA, Moore JP, Weiss RA. Effect of mutations in the V3 loop of HIV-1 gp120 on infectivity and susceptibility to proteolytic cleavage. AIDS Res Hum Retroviruses. 1993;9:159–66. doi: 10.1089/aid.1993.9.159. [DOI] [PubMed] [Google Scholar]
- Seaman MS, Xu L, Beaudry K, Martin KL, Beddall MH, Miura A, Sambor A, Chakrabarti BK, Huang Y, Bailer R, Koup RA, Mascola JR, Nabel GJ, Letvin NL. Multiclade human immunodeficiency virus type 1 envelope immunogens elicit broad cellular and humoral immunity in rhesus monkeys. J Virol. 2005;79:2956–2963. doi: 10.1128/JVI.79.5.2956-2963.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava I, Ulmer J, Barnett S. Neutralizing antibody responses to HIV: role in protective immunity and challenges for vaccine design. Expert Review of Vaccines. 2004;3(4 suppl):533–552. doi: 10.1586/14760584.3.4.s33. [DOI] [PubMed] [Google Scholar]
- Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodrosk i. SCD4-Induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. Journal of Virology. 1998;72(6):4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trkola A, Kuster H, Rusert P, Joos B, Fischer M, Leemann C, Manrique A, Huber M, Rehr M, Oxenius A, Weber R, Stiegler G, Vcelar B, Katinger H, Aceto L, Gunthard H. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nature Medicine. 2005;11(6):615–622. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- VanCott TC, Mascola JR, Kaminski RW, Kalyanaraman V, Hallberg PL, Burnett PR, Ulrich JT, Rechtman DJ, Birx DL. Antibodies with specificity to native gp120 and neutralization activity against primary human immunodeficiency virus type 1 isolates elicited by immunization with oligomeric gp160. J Virol. 1997;71:4319–4330. doi: 10.1128/jvi.71.6.4319-4330.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanCott TC, Mascola JR, Loomis-Price LD, Sinangil F, Zitomersky N, McNeil J, Robb ML, Birx DL, Barnett S. Cross-Subtype Neutralizing Antibodies Induced in Baboons by a Subtype E gp120 Immunogen Based on an R5 Primary Human Immunodeficiency Virus Type 1 Envelope. J Virol. 1999;73:4640–4650. doi: 10.1128/jvi.73.6.4640-4650.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Decker J, Wang S, Hui H, Kappes J, Wu X, Salazar-Gonzalez J, Salazar M, Kilby J, Saag M, Komarova N, Nowak M, Hahn B, Kwong P, GM S. Antibody neutralization and escape by HIV-1. Nature. 2003;422(6929):307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother. 2002;46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson C, Morris L, Maughan MF, Ping LH, Dryga SA, Thomas R, Reap EA, Cilliers T, van HarmelenJ, Pascual A, Ramjee G, Gray G, Johnston R, Karim SA, Swanstrom R. Characterization and selection of HIV-1 subtype C isolates for use in vaccine development. AIDS Res Hum Retroviruses. 2003;19(2):133–44. doi: 10.1089/088922203762688649. [DOI] [PubMed] [Google Scholar]
- Wyatt R, Sullivan N, Thali M, Repke H, Ho D, Robinson J, Posner M, Sodroski J. Functional and immunologic characterization of human immunodeficiency virus type 1 envelope glycoproteins containing deletions of the major variable regions. J Virol. 1993;67:4557–4565. doi: 10.1128/jvi.67.8.4557-4565.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69(9):5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt R, Kwong P, Desjardins E, Sweet R, Robinson J, Hendrickson W, Sodroski J. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;39(6686):705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- Xiang SH, Doka N, Choudhary RK, Sodroski J, Robinson JE. Characterization of CD4-Induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res Hum Retroviruses. 2002;18(16):1207–1217. doi: 10.1089/08892220260387959. [DOI] [PubMed] [Google Scholar]
- Yang Z-Y, Chakrabarti BK, Xu L, Welcher B, Kong W-P, Leung K, Panet A, Mascola JR, Nabel GJ. Selective modification of variable loops alters tropism and enhances immunogenicity of human immunodeficiency virus type 1 envelope. J Virol. 2004;78(8):4029–4036. doi: 10.1128/JVI.78.8.4029-4036.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolla-Pazner S, Zhong P, Revesz K, Volsky B, Williams C, Nyambi P, MK G. The cross-clade neutralizing activity of a human monoclonal antibody is determined by the GPGR V3 motif of HIV type 1. AIDS Res Hum Retroviruses. 2004;20(11):1254–1258. doi: 10.1089/aid.2004.20.1254. [DOI] [PubMed] [Google Scholar]


