Abstract
Variation between inbred mice of susceptibility to experimental Trypanosoma cruzi infection has frequently been described, but the immunogenetic background is poorly understood. The outcross of the susceptible parental mouse strains C57BL/6 (B6) and DBA/2 (D2), B6D2F1 (F1) mice, is highly resistant to this parasite. In the present study we show by quantitative PCR that the increase of tissue parasitism during the early phase of infection is comparable up to day 11 between susceptible B6 and resistant F1 mice. A reduction of splenic parasite burdens occurs thereafter in both strains but is comparatively retarded in susceptible mice. Splenic microarchitecture is progressively disrupted with loss of follicles and B lymphocytes in B6 mice, but not in F1 mice. By genotyping of additional backcross offspring we corroborate our earlier findings that susceptibility maps to three loci on Chromosomes 5, 13 and 17. Analysis of gene expression of spleen cells from infected B6 and F1 mice with microarrays identifies about 0.3% of transcripts that are differentially expressed. Assuming that differential susceptibility is mediated by altered gene expression, we propose that the following differentially expressed transcripts from these loci are strong candidates for the observed phenotypic variation: H2-Eα, H2-D1, Ng23, Msh5 and Tubb5 from Chromosome 17; and Cxcl11, Bmp2k and Spp1 from Chromosome 5. Our results indicate that innate mechanisms are not of primary relevance to resistance of F1 mice to T. cruzi infection, and that differential susceptibility to experimental infection with this protozoan pathogen is not paralleled by extensive variation of the transcriptome.
Introduction
Chagas' disease severely affects a considerable number of persons on the American continent, but in the majority of infected, it takes an ‘indeterminate’ course over a long period of time [1]. Genetic factors determining the course and outcome of the infection are thought to be of major influence on the severity of the disease [2]–[6], but the precise background has not been elucidated. Variablity of parasite strains contributes to the complex host-pathogen interaction [3]. As in human disease, the experimental infection has an early parasitaemic phase, which is followed by chronic infection that may or may not lead to the symptoms characteristic of the disease. Some controversy has prevailed over the question whether the severity of the acute phase of the infection and the degree of parasitaemia and/or tissue parasitism correlated with the severity of the chronic complications of Chagas' disease. Recently, it has increasingly been appreciated that the persistence of parasites, rather than the occurence of autoreactive antibodies or cells, determines the degree of tissue destruction [7]–[11]. It was shown that an early phase with high parasitic loads resulted in a late phase with more prominent repercussions on the integrity of affected tissue, with more intense inflammatory infiltrates, more tissue destruction and greater loss of physiological function [12], [13].
The course of experimental T. cruzi infection in inbred strains of mice varies considerably depending on the mouse strain, the route of infection, the parasite strain, and the clone of a given parasite strain [14]–[16]. Other than with Leishmania, no consistent picture has evolved that would relate a certain type of immunologic reactivity with protection from severe disease. It has been noted that certain H2 haplotypes confer a degree of resistance [17]–[19]. The requirement for pro-inflammatory cytokines such as IL-12, IFN-γ, and TNF-α, as well as for MHC-class I and II molecules, CD4+ and CD8+ T lymphocytes and the synthesis of antigen-specific antibodies, for protective immunity has repeatedly been demonstrated [20]–[25]. Generally, a higher degree of expression of anti-inflammatory cytokines such as IL-4, IL-10 and TGF-β was correlated with increased severity of infection, but some conflicting results have been published (e.g., [26], [27]).
In contrast to the parental strains C57BL/6 (B6) and DBA/2 (D2), B6D2F1 (F1) hybrid mice display a considerable degree of resistance to experimental infection in terms of parasitaemia levels and rates of mortality, but precise mechanisms that explained the unusual phenotype of this strain have not been identified. By comparison with susceptible B6 mice, resistance in F1 mice was related to decreased expression of IL-10 and TGF-β in the early phase [21], [28]. However, the isolated analysis of cytokine responses, and the correlation of cytokine expression or regulatory molecules with outcome, bear the danger of focussing on secondary effects or on counter-regulative reactivity, rather than identifying the initial cause for differential outcomes. In the present work, we therefore investigated at which stage of experimental infection tissue parasite loads dissociated between susceptible B6 and resistant F1 mice in order to identify the time point at which the immune responses diverge. We then analysed genomewide expression differences at this time point in the spleen, identified transcriptional correlates for differential outcomes and matched the genomic localisation of these genes with mapped susceptibility loci.
Results
Experimental T. cruzi infection in susceptible B6 and in resistant F1 mice
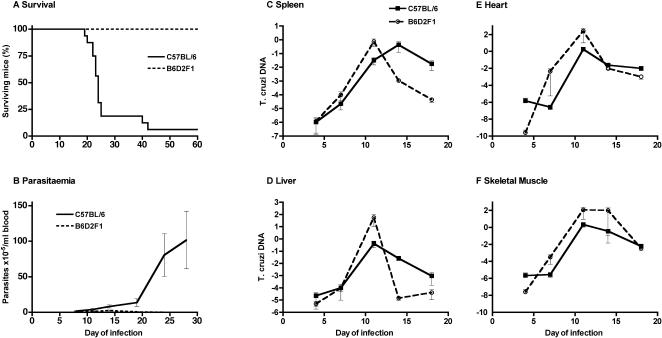
Infection of B6 mice with 104 trypomastigotes of the T. cruzi tulahuen strain caused an acute illness that was usually lethal. Mice succumbed between days 20 and 30 after infection (fig. 1 A). Shortly before death, parasitaemia increased to well over 106/ml (fig. 1 B). In F1 mice, there was a transient para–sitaemic phase around day 13, but parasitaemia did not exceed 106/ml, and all mice invariably survived experimental infection under these conditions (fig 1 A, B). Tissue parasite loads were quantified by real time PCR in various organs during the early phase of the infection (until day 17, fig. 1 C–F). Interestingly, the degree of tissue parasitism was not significantly different in the first 11 days of infection between mouse strains. Resistant F1 mice actually displayed slightly higher parasite loads in all tested tissues up to this stage. After day 11, tissue parasite load declined in resistant mice, while it further increased in spleens of susceptible B6 mice. Thereafter, parasitism decreased in susceptible mice, too, in all tested organs. The rate of parasite clearance in the spleen was lower than in F1 mice. These results indicate that innate resistance is not compromised, if not more effective, in B6 mice. They also indicate that acquired immune responses develop in B6 mice with a delay and appear to be less efficient in containing tissue parasitism, especially in the spleen and liver.
Figure 1. Experimental T. cruzi infection in susceptible B6 and in resistant F1 mice.
Mice were infected with 104 T. cruzi trypomastigotes into the peritoneum.
Survival was monitored up to day 60.
Parasitaemia was determined in a Neubauer chamber.
Tissue parasite loads in spleen, liver, heart and skeletal muscle were determined by real time PCR and are expressed as arbitrary log units of T. cruzi DNA relative to quantity of murine β-actin DNA.
Splenic architecture was severely disrupted, and cellular composition was altered, in susceptible mice infected with T. cruzi
After day 10 of infection, there was a striking difference of parasite loads in the spleen between the mouse strains. Splenomegaly was present in all infected mice to the same extent (total spleen cell numbers increased to about 1–2×108 in either strain) and pointed to a strong involvement of the spleen in the immune response to T. cruzi. We thus assessed by histologic analysis whether splenic architecture was modified, and by flow cytometry whether there were quantitative differences in splenic cellular composition between B6 and F1 mice.
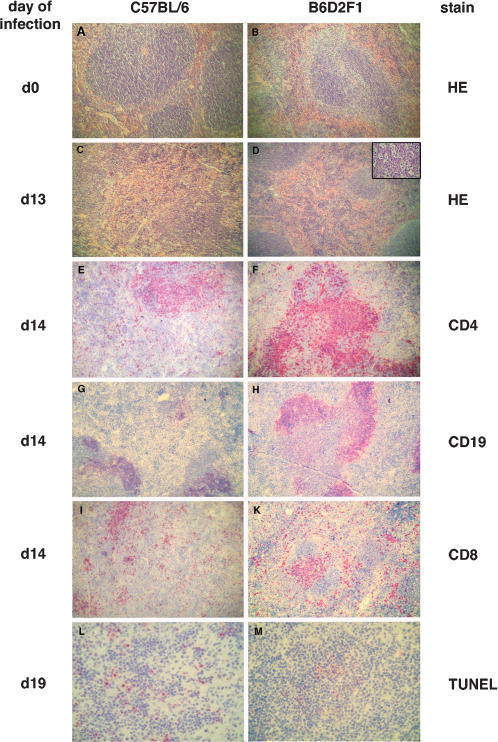
Histologically, the appearance of red and white pulp, the numbers and size of primary follicles, and the distribution of periarteriolar lymphatic sheaths (PALS) was not obviously different in naive mice, and the cellular localisation of cells expressing CD4, CD8, CD19 and CD11b was indistinguishable (fig. 2 A, B and data not shown). By day 13 of an experimental infection with T. cruzi, splenic microarchitecture was observed to be disrupted in B6 mice but not in F1 mice (fig. 2 C, D). The disruption was even more severe on day 23 (not shown). In F1 mice, abundant cells with intracellular apoptotic bodies, resembling tingible body macrophages, were seen in secondary follicles (fig. 2 D, inset), but these were not observed in B6 mice. Periarteriolar sheaths (PALS) were reduced by numbers and smaller in size in B6 mice, but CD4+ cells were found throughout the spleen (fig. 2 E). In F1 mice, some prominent PALS consisting of CD4+ T cells were detectable (fig. 2 F). Follicles with CD19+ B lymphocytes were also reduced in size and numbers in the spleen of infected B6 mice (fig. 2 G). Contrarily, in resistant F1 mice, prominent B cell follicles were present adjacent to PALS (fig. 2 H). CD8 T cells did generally not co-localise to PALS in susceptible mice, but were located in the remains of the red and white pulp (fig. 2 I). In F1 mice, CD8+ cells formed small clusters in the red pulp and co-localised with PALS (fig. 2 K). In summary, the main difference in splenic microarchitecture between mouse strains was the obvious loss of compartmentalised organisation of lyphocytes in susceptible but not in resistant mice.
Figure 2. Splenic architecture and cellular composition in susceptible B6 and in resistant F1 mice.
Spleens from B6 mice (left panel) and from F1 mice (right panel) were either fixed in paraformaldehyde, embedded in paraffin, and subsequently stained with hematoxylin and eosin (A–D), or they were shock frozen in liquid nitrogen and analysed by immunohistochemistry (C–M).
Sections are from naive mice (A, B) or from T. cruzi- infected mice (C–M) at the indicated day of infection.
Cryosections were stained for CD4 (E, F), CD19 (G, H), CD8 (I, K), or by the TUNEL reaction (L, M).
Micrographs A–K at original magnification 100×, micrographs L, M and insert in D at 400×; inset in D shows centre of a secondary follice.
The proliferation of T. cruzi within macrophages has been shown to be enhanced in the presence of apoptotic T cells due to a downregulatory effect of apoptotic bodies on trypanocidal effector functions of infected macrophages [29]. We therefore determined the quantitiy and distribution of apoptotic cells in the spleens of infected mice. In spleens from B6 mice, abundant apoptotic cells (identified by the TUNEL reaction) were found throughout the spleen, and they were frequently located sub-capsularly and in the remaining T cell areas, (fig. 2 L). By contrast, apoptotic cell numbers were lower, and they were mainly found in the centre of follicles, in the spleen of F1 mice (fig. 2 M).
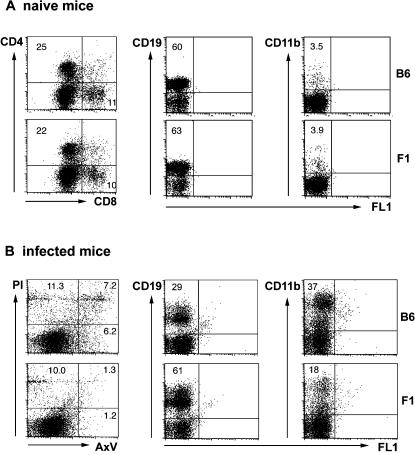
The splenic cellular composition in infected mice was analysed cytometrically with explanted spleen cells to verify the histologic alterations. In naive mice, relative numbers of splenic CD4+ T cells, CD8+ T cells, CD19+ B cells and CD11b+ macrophages did not differ between mice (fig. 3 A). However, cell fractions were notably different later during infection. On day 24 of infection, prior to the detrimental outcome in B6 mice, CD19+ B lymphocytes were decreased at around 30% of cells (fig 3 B) in susceptible B6 mice. Interestingly, in infected F1 mice, the fraction of B cells was similar to that of control mice (about 60%, fig. 3 B). The fraction of CD4+ lymphocytes was 10–15% in both strains (not shown). Splenic CD8+ T cells increased to around 20–25% of cells in B6 mice, and to about 15% in F1 mice (not shown). Strikingly, in B6 mice, relative numbers of CD11b+ macrophages drastically increased more than tenfold, from about 3% in naive mice to 30–50% in infected mice, but in F1 mice, the increase was less pronounced to about 10–20% of spleen cells (fig. 3 B). In summary, a strong bias towards macrophages, and a slight bias towards CD8+ T cells, at the expense of B lymphocytes, was found in the spleen of infected B6 mice, but not in resistant F1 mice.
Figure 3. Cellular composition of the spleen from naive and T. cruzi- infected mice.
Single spleen cell suspensions from naive mice (A) and from mice infected on day 24 with T. cruzi (B) were analysed by flow cytometry.
Cells were stained for both CD4 and CD8, for CD19, for CD11b, or with propidium iodide (PI) and Annexin V (AxV).
Numbers indicate percentage of cells expressing the respective marker.
B6, C57BL/6 mice; F1, B6D2F1 mice.
In naive mice, numbers of apoptotic cells were low at around 1% (determined cytometrically by staining with Annexin V) and did not differ between the two strains (not shown). In infected mice, however, there was a strong increase of Annexin V (AxV) positive cells in B6 mice (fig 3 B). The relative number of cells expressing AxV was 10–20%, with half of these also staining with propidium iodide (PI), indicating that these cells had lost integrity of the cell membrane. In infected F1 mice, AxV positive cells increased to about 2–5%. The proportion of cells stained by PI only was about 10% in either strain, indicating that preparation of samples had equal effects on staining of cells in causing some degree of cell death. In summary, susceptible mice displayed a considerably higher rate of apoptosis in the spleen, possibly accounting for the reduced numbers of B cells.
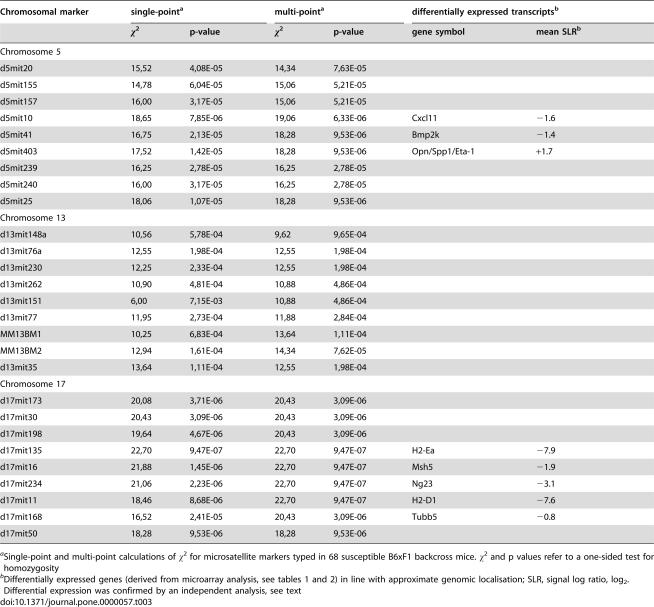
Significant linkage of susceptibility to loci on Chromosomes 5 and 17, and suggestive linkage on Chromosome 13
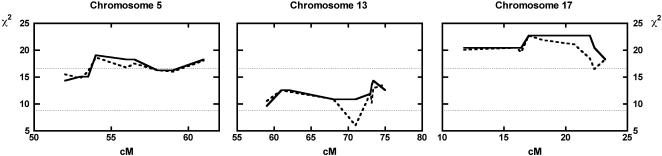
We previously mapped genomic loci as being either suggestively (Chromosome 5) or significantly (Chromosome 17) linked to susceptibility to lethal experimental T. cruzi infection, respectively [30]. The score of a further genomic region on Chromosome 13 did not attain the threshold level for genome-wide linkage but appeared to be involved, too. In an extension study, designed to confirm linkage of a locus previously reported as suggestively linked [31], we phenotyped further male B6xF1 backcross mice, and another 22 mice died during the early phase (up to day 50) of infection. The candidate regions were genotyped with microsatellite markers in these mice, and the resulting data replicated that susceptibility mapped to the reported locus on Chromosome 17, and further substantiated the linkage evidence for the loci on Chromosomes 5 and 13 by raising it to the level of significant and suggestive linkage, respectively (fig. 4).
Figure 4. Murine T. cruzi susceptibility loci map to Chromosomes 5, 13, and 17.
Nine microsatellite markers from the three candidate genomic regions previously identified as suceptibility loci [30] were typed in an additional 22 T. cruzi- susceptible B6xF1 backcross mice (see table 3 for respective markers).
Single-point (dashed line) and multi-point (solid line) calculations of χ2 are shown for all 68 susceptible backcross mice including the 46 from the original study.
Dotted lines at χ2 = 16.56 and χ2 = 8.74 indicate thresholds for suggestive and significant linkage, respectively.
Even though the genes conferring susceptibility are thus mapped, the respective genetic regions are large and the number of candidate genes is substantial, so that safe predictions about the genes themselves are not straightforward. However, it is generally accepted that quantitative differences of transcription determine the outcome of an infection with a complex immunogenetic background. The kinetics of tissue parasite loads during early experimental T. cruzi infection in the two differentially susceptible mouse strains indicate that after about day 10, the implementation of effector immune responses was impaired but not abrogated in susceptible B6 mice. The described differences in parasite proliferation, splenic architecture and splenic cellular composition progressively developed after this time point. Since the spleen appeared to be fundamentally involved in the differential immunity to the parasite, we therefore analysed genome wide expression in explanted spleen cells from either mouse strain in order to identify genes that were differentially transcribed and that mapped to the susceptibility loci. We chose to analyse expression on day 11 as the starting point from which the infection took a divergent course. At this stage, the cellular composition of the spleen was marginally different between mouse strains in that there were more CD8+ cells (7% vs. 4%) in B6 mice, but fewer CD19+ cells, (10% vs. 18%) in B6 mice, while the proportion of CD4+ and of CD11b+ cells was comparable (12% and 17–20%, respectively).
Differential gene expression in the spleens of susceptible B6 and from resistant F1 mice on day 11 of T. cruzi infection
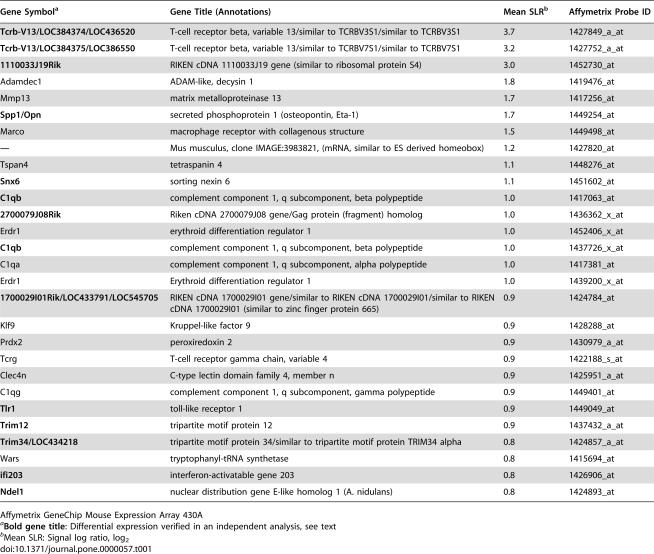
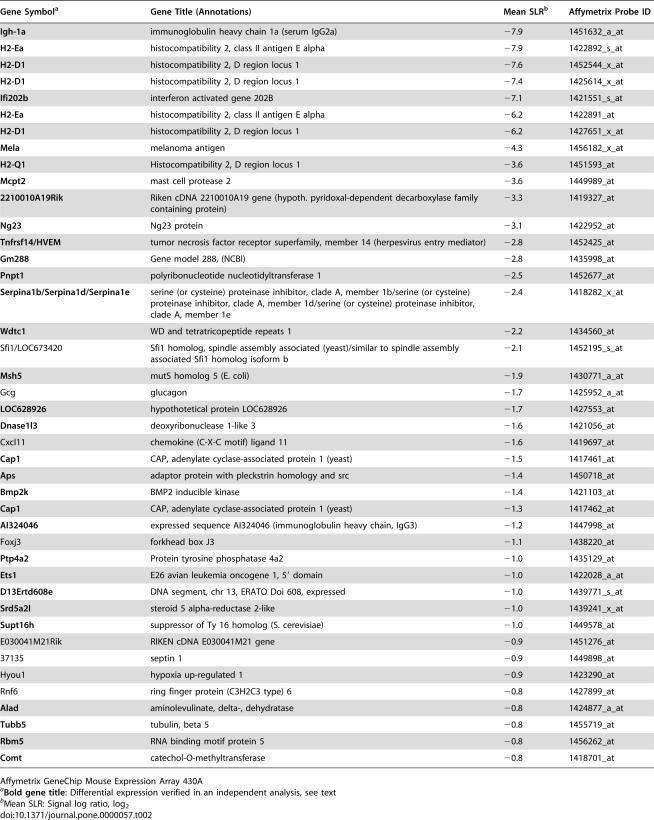
We used mouse expression arrays containing 22690 transcripts and obtained three biologically independent specimen pairs. Data were normalised using target value scaling (target value: 100). Expression was considered significantly different when it showed a concordant regulation in at least 6 of 9 crosswise comparisons and a signal log ratio of 0.8 (fold change of 1.6) or more. Applying these criteria, we found that 28 transcripts were upregulated and 42 transcripts were downregulated in B6 mice, compared to F1 mice (tables 1 and 2). A second, biologically independent experiment was performed in which we analysed the transcription profile of pooled spleen cells from three infected animals of each strain to confirm transcriptional differences. Applying the same statistical algorithms and criteria, we could verify the differential expression of 14 transcripts with increased expression and of 35 transcripts with decreased expression (highlighted by bold gene titles, tables 1 and 2).
Table 1. Transcripts with increased expression in spleens of T. cruzi- infected susceptible B6 mice with respect to resistant F1 mice.
| Gene Symbola | Gene Title (Annotations) | Mean SLRb | Affymetrix Probe ID |
| Tcrb-V13/LOC384374/LOC436520 | T-cell receptor beta, variable 13/similar to TCRBV3S1/similar to TCRBV3S1 | 3.7 | 1427849_a_at |
| Tcrb-V13/LOC384375/LOC386550 | T-cell receptor beta, variable 13/similar to TCRBV7S1/similar to TCRBV7S1 | 3.2 | 1427752_a_at |
| 1110033J19Rik | RIKEN cDNA 1110033J19 gene (similar to ribosomal protein S4) | 3.0 | 1452730_at |
| Adamdec1 | ADAM-like, decysin 1 | 1.8 | 1419476_at |
| Mmp13 | matrix metalloproteinase 13 | 1.7 | 1417256_at |
| Spp1/Opn | secreted phosphoprotein 1 (osteopontin, Eta-1) | 1.7 | 1449254_at |
| Marco | macrophage receptor with collagenous structure | 1.5 | 1449498_at |
| — | Mus musculus, clone IMAGE:3983821, (mRNA, similar to ES derived homeobox) | 1.2 | 1427820_at |
| Tspan4 | tetraspanin 4 | 1.1 | 1448276_at |
| Snx6 | sorting nexin 6 | 1.1 | 1451602_at |
| C1qb | complement component 1, q subcomponent, beta polypeptide | 1.0 | 1417063_at |
| 2700079J08Rik | Riken cDNA 2700079J08 gene/Gag protein (fragment) homolog | 1.0 | 1436362_x_at |
| Erdr1 | erythroid differentiation regulator 1 | 1.0 | 1452406_x_at |
| C1qb | complement component 1, q subcomponent, beta polypeptide | 1.0 | 1437726_x_at |
| C1qa | complement component 1, q subcomponent, alpha polypeptide | 1.0 | 1417381_at |
| Erdr1 | Erythroid differentiation regulator 1 | 1.0 | 1439200_x_at |
| 1700029I01Rik/LOC433791/LOC545705 | RIKEN cDNA 1700029I01 gene/similar to RIKEN cDNA 1700029I01/similar to RIKEN cDNA 1700029I01 (similar to zinc finger protein 665) | 0.9 | 1424784_at |
| Klf9 | Kruppel-like factor 9 | 0.9 | 1428288_at |
| Prdx2 | peroxiredoxin 2 | 0.9 | 1430979_a_at |
| Tcrg | T-cell receptor gamma chain, variable 4 | 0.9 | 1422188_s_at |
| Clec4n | C-type lectin domain family 4, member n | 0.9 | 1425951_a_at |
| C1qg | complement component 1, q subcomponent, gamma polypeptide | 0.9 | 1449401_at |
| Tlr1 | toll-like receptor 1 | 0.9 | 1449049_at |
| Trim12 | tripartite motif protein 12 | 0.9 | 1437432_a_at |
| Trim34/LOC434218 | tripartite motif protein 34/similar to tripartite motif protein TRIM34 alpha | 0.8 | 1424857_a_at |
| Wars | tryptophanyl-tRNA synthetase | 0.8 | 1415694_at |
| ifi203 | interferon-activatable gene 203 | 0.8 | 1426906_at |
| Ndel1 | nuclear distribution gene E-like homolog 1 (A. nidulans) | 0.8 | 1424893_at |
Affymetrix GeneChip Mouse Expression Array 430A
Bold gene title: Differential expression verified in an independent analysis, see text
Mean SLR: Signal log ratio, log2
Table 2. Transcripts with decreased expression in spleens of T. cruzi- infected susceptible B6 mice with respect to resistant F1 mice.
| Gene Symbola | Gene Title (Annotations) | Mean SLRb | Affymetrix Probe ID |
| Igh-1a | immunoglobulin heavy chain 1a (serum IgG2a) | −7.9 | 1451632_a_at |
| H2-Ea | histocompatibility 2, class II antigen E alpha | −7.9 | 1422892_s_at |
| H2-D1 | histocompatibility 2, D region locus 1 | −7.6 | 1452544_x_at |
| H2-D1 | histocompatibility 2, D region locus 1 | −7.4 | 1425614_x_at |
| Ifi202b | interferon activated gene 202B | −7.1 | 1421551_s_at |
| H2-Ea | histocompatibility 2, class II antigen E alpha | −6.2 | 1422891_at |
| H2-D1 | histocompatibility 2, D region locus 1 | −6.2 | 1427651_x_at |
| Mela | melanoma antigen | −4.3 | 1456182_x_at |
| H2-Q1 | Histocompatibility 2, D region locus 1 | −3.6 | 1451593_at |
| Mcpt2 | mast cell protease 2 | −3.6 | 1449989_at |
| 2210010A19Rik | Riken cDNA 2210010A19 gene (hypoth. pyridoxal-dependent decarboxylase family containing protein) | −3.3 | 1419327_at |
| Ng23 | Ng23 protein | −3.1 | 1422952_at |
| Tnfrsf14/HVEM | tumor necrosis factor receptor superfamily, member 14 (herpesvirus entry mediator) | −2.8 | 1452425_at |
| Gm288 | Gene model 288, (NCBI) | −2.8 | 1435998_at |
| Pnpt1 | polyribonucleotide nucleotidyltransferase 1 | −2.5 | 1452677_at |
| Serpina1b/Serpina1d/Serpina1e | serine (or cysteine) proteinase inhibitor, clade A, member 1b/serine (or cysteine) proteinase inhibitor, clade A, member 1d/serine (or cysteine) proteinase inhibitor, clade A, member 1e | −2.4 | 1418282_x_at |
| Wdtc1 | WD and tetratricopeptide repeats 1 | −2.2 | 1434560_at |
| Sfi1/LOC673420 | Sfi1 homolog, spindle assembly associated (yeast)/similar to spindle assembly associated Sfi1 homolog isoform b | −2.1 | 1452195_s_at |
| Msh5 | mutS homolog 5 (E. coli) | −1.9 | 1430771_a_at |
| Gcg | glucagon | −1.7 | 1425952_a_at |
| LOC628926 | hypothotetical protein LOC628926 | −1.7 | 1427553_at |
| Dnase1l3 | deoxyribonuclease 1-like 3 | −1.6 | 1421056_at |
| Cxcl11 | chemokine (C-X-C motif) ligand 11 | −1.6 | 1419697_at |
| Cap1 | CAP, adenylate cyclase-associated protein 1 (yeast) | −1.5 | 1417461_at |
| Aps | adaptor protein with pleckstrin homology and src | −1.4 | 1450718_at |
| Bmp2k | BMP2 inducible kinase | −1.4 | 1421103_at |
| Cap1 | CAP, adenylate cyclase-associated protein 1 (yeast) | −1.3 | 1417462_at |
| AI324046 | expressed sequence AI324046 (immunoglobulin heavy chain, IgG3) | −1.2 | 1447998_at |
| Foxj3 | forkhead box J3 | −1.1 | 1438220_at |
| Ptp4a2 | Protein tyrosine phosphatase 4a2 | −1.0 | 1435129_at |
| Ets1 | E26 avian leukemia oncogene 1, 5′ domain | −1.0 | 1422028_a_at |
| D13Ertd608e | DNA segment, chr 13, ERATO Doi 608, expressed | −1.0 | 1439771_s_at |
| Srd5a2l | steroid 5 alpha-reductase 2-like | −1.0 | 1439241_x_at |
| Supt16h | suppressor of Ty 16 homolog (S. cerevisiae) | −1.0 | 1449578_at |
| E030041M21Rik | RIKEN cDNA E030041M21 gene | −0.9 | 1451276_at |
| 37135 | septin 1 | −0.9 | 1449898_at |
| Hyou1 | hypoxia up-regulated 1 | −0.9 | 1423290_at |
| Rnf6 | ring finger protein (C3H2C3 type) 6 | −0.8 | 1427899_at |
| Alad | aminolevulinate, delta-, dehydratase | −0.8 | 1424877_a_at |
| Tubb5 | tubulin, beta 5 | −0.8 | 1455719_at |
| Rbm5 | RNA binding motif protein 5 | −0.8 | 1456262_at |
| Comt | catechol-O-methyltransferase | −0.8 | 1418701_at |
Affymetrix GeneChip Mouse Expression Array 430A
Bold gene title: Differential expression verified in an independent analysis, see text
Mean SLR: Signal log ratio, log2
Over all, only about 0.3% of transcripts present on the microarray were found to be differentially expressed between the two mouse strains. Among these were a number of genes that are involved in immune responses or inflammatory processes, such as: the T cell receptor (TCR) Vβ 13; the TCR gamma chain; osteopontin (Opn, or Spp1, secreted phosphoprotein 1); complement component 1 subcomponents; the herpes virus entry mediator (Hvem), a member of the TNF receptor superfamily and the ligand of the B and T lymphocyte attenuator (BTLA) [32]; Cxcl11, a T cell recruiting CXC chemokine (also known as I-TAC); Toll-like receptor 1 (Tlr1); Clec4n, a C type lectin-like receptor (also termed dectin-2). Among the transcripts with decreased expression, the strongest differences were expectedly found for genes not encoded in the genome of B6 mice, such as Igh-1a (an allotype of the γ2a gene encoding the heavy chain of Ig2a immunoglobulins, [33]), H2-Ea (encoding H2-Eα, an MHC class II protein) [34], [35], and H2-D1 (encoding an MHC class Ia protein not expressed in mice with the H2b haplotype [36]).
Only a few differentially transcribed genes map to the susceptibility loci on Chromosomes 5 and 17 (table 3). Three transcripts map to the locus on Chromosome 5, with two (the IFN-inducible chemokine Cxcl11, and Bmp2k, bone morphogenetic protein 2-inducible kinase) being down-regulated, and one (Spp1) being upregulated, in susceptible mice. Expression of Bmp2k and Spp1 transcripts were concordantly regulated in the second microarray experiment. Cxcl11 mRNA was not detectable in either pool of RNA in this experiment, but it was found to be about four-fold decreased in infected B6 mice in an RT-PCR assay (O. Steinmetz, personal communication), corroborating the results of the gene array in which a signal log ratio of −1.6 was determined (corresponding to a three-fold decrease, table 3). By contrast, expression levels of two other CXC chemokines (Cxcl9 and Cxcl10) did not vary between mice in either analysis.
Table 3. susceptibility loci in B6 mice, and transcripts of genes encoded in these genomic regions that are differentially expressed between T. cruzi- infected B6 and F1 mice.
| Chromosomal marker | single-pointa | multi-pointa | differentially expressed transcriptsb | |||
| χ2 | p-value | χ2 | p-value | gene symbol | mean SLRb | |
| Chromosome 5 | ||||||
| d5mit20 | 15,52 | 4,08E-05 | 14,34 | 7,63E-05 | ||
| d5mit155 | 14,78 | 6,04E-05 | 15,06 | 5,21E-05 | ||
| d5mit157 | 16,00 | 3,17E-05 | 15,06 | 5,21E-05 | ||
| d5mit10 | 18,65 | 7,85E-06 | 19,06 | 6,33E-06 | Cxcl11 | −1.6 |
| d5mit41 | 16,75 | 2,13E-05 | 18,28 | 9,53E-06 | Bmp2k | −1.4 |
| d5mit403 | 17,52 | 1,42E-05 | 18,28 | 9,53E-06 | Opn/Spp1/Eta-1 | +1.7 |
| d5mit239 | 16,25 | 2,78E-05 | 16,25 | 2,78E-05 | ||
| d5mit240 | 16,00 | 3,17E-05 | 16,25 | 2,78E-05 | ||
| d5mit25 | 18,06 | 1,07E-05 | 18,28 | 9,53E-06 | ||
| Chromosome 13 | ||||||
| d13mit148a | 10,56 | 5,78E-04 | 9,62 | 9,65E-04 | ||
| d13mit76a | 12,55 | 1,98E-04 | 12,55 | 1,98E-04 | ||
| d13mit230 | 12,25 | 2,33E-04 | 12,55 | 1,98E-04 | ||
| d13mit262 | 10,90 | 4,81E-04 | 10,88 | 4,86E-04 | ||
| d13mit151 | 6,00 | 7,15E-03 | 10,88 | 4,86E-04 | ||
| d13mit77 | 11,95 | 2,73E-04 | 11,88 | 2,84E-04 | ||
| MM13BM1 | 10,25 | 6,83E-04 | 13,64 | 1,11E-04 | ||
| MM13BM2 | 12,94 | 1,61E-04 | 14,34 | 7,62E-05 | ||
| d13mit35 | 13,64 | 1,11E-04 | 12,55 | 1,98E-04 | ||
| Chromosome 17 | ||||||
| d17mit173 | 20,08 | 3,71E-06 | 20,43 | 3,09E-06 | ||
| d17mit30 | 20,43 | 3,09E-06 | 20,43 | 3,09E-06 | ||
| d17mit198 | 19,64 | 4,67E-06 | 20,43 | 3,09E-06 | ||
| d17mit135 | 22,70 | 9,47E-07 | 22,70 | 9,47E-07 | H2-Ea | −7.9 |
| d17mit16 | 21,88 | 1,45E-06 | 22,70 | 9,47E-07 | Msh5 | −1.9 |
| d17mit234 | 21,06 | 2,23E-06 | 22,70 | 9,47E-07 | Ng23 | −3.1 |
| d17mit11 | 18,46 | 8,68E-06 | 22,70 | 9,47E-07 | H2-D1 | −7.6 |
| d17mit168 | 16,52 | 2,41E-05 | 20,43 | 3,09E-06 | Tubb5 | −0.8 |
| d17mit50 | 18,28 | 9,53E-06 | 18,28 | 9,53E-06 | ||
Single-point and multi-point calculations of χ2 for microsatellite markers typed in 68 susceptible B6xF1 backcross mice. χ2 and p values refer to a one-sided test for homozygosity
Five transcripts that were significantly reduced in B6 mice map to the locus on Chromosome 17, which covers the murine MHC complex (table 3). All differences were confirmed in the second microarray experiment with pooled RNA. One additional gene (Glo1, glyoxalase 1) close to the locus on Chromosome 17 was insignificantly (SLR−0.6, equivalent to a 1.5 fold decrease) but consistently regulated in that it was decreased in all 9 comparisons between the 3 individual mice of either strain, and the regulation was observed for several transcripts of the gene. It maps centromerically to the suceptibility locus at about 16 cM.
No significant expression differences were found to map to the putative locus on Chromosome 13 (cytogenetic band 13D2.3), but three transcripts of an expressed gene (4833420G17Rik) were reproducibly regulated below the threshold (SLR+0.4, eqivalent to a 1.3 fold increase). It encodes a protein with a gonadotropin beta chain-like domain. The slightly increased expression in B6 mice was confirmed in the pooled RNA microarray analysis. Another transcript from this locus, Paip1 (polyadelynate binding protein-interacting protein 1, a translation factor), was downregulated in 5 of 9 comparisons only (SLR−1.9), but this could not be substantiated in the second experiment.
Numerous gene transcripts have been shown to be involved in immunity of Chagas' disease and its experimental models. A few of these displayed differential expression below the threshold level, and there was concordant regulation in the microarray experiment with pooled RNA samples. Several transcripts of inducible TGF-β were increased in B6 mice (SLR+0.6). This is noteworthy as it supports results of a previous investigation on the differences of the immune response of B6 and F1 mice to T. cruzi in which increased TGF-β expression was shown to enhance susceptibilty [28]. Transcription signals of CD44 (coding for an adhesion factor on activated T and B cells; SLR+0.4), and of granzyme B (SLR+0.4) were slightly upregulated in B6 mice. A downregulated transcription signal in B6 mice was found for the beta chain of the IL-2 receptor (SLR−0.5).
Discussion
Disease following infection with a pathogen is the result of a complex interaction between the host and the infectious agent [37]. Both host and pathogen genomes are involved in determining the severity of a sickness, and enviromental factors also contribute to the course of an ailment. Genetic variation within human cohorts complicates matters with respect to the identification of genetic sources for disease severity, and in general, several gene loci, splice variants and variation of gene copy numbers are involved [38]. It is thus conceivable that the identification of suceptibility genes to infectious diseases in man has not been fruitful thus far, despite of the progress that genome sequencing has achieved. Therefore, it may be a more straightforward approach to pinpoint genes that affect the severity of infectious disease in animal models, and then to assess whether orthologous mechanisms are at work in the human situation [39]. Since a hosts genotype manifests via transcription of genes, candidate genes may be identified by a combination of mapping data and transcriptional profiling. Determining the gene expression differences from susceptibility loci is thus a hypothesis-driven experiment with the advantage that the multiple testing problem of microarray analyses disappears and that the verification of the gene expression data by independent methods is not strictly required [40].
Morphological and cytometrical analysis of the organ with the greatest differences in parasite loads, the spleen, revealed a disruption of normal anatomical structure and modulation of cellular composition in susceptible B6 mice. In addition, a substantial increase of apoptotic cells was observed, possibly accounting further for the increased parasite burden, since the presence of apoptotic cells were connected with increased parasite proliferation, and with decreased trypanocidal activity of infected macrophages [29]. B cell apoptosis has also been shown to correlate with reduced numbers of B cells and with a suppressed immune response to T. cruzi [41]. The loss of B cells and of splenic follicles make a decreased humoral immune response to the parasite a candidate pathogenetic pathway. Indeed, on day 17 of infection, serologic reactivity to parasite antigens was decreased in B6 mice for antibodies of all IgG subclasses, except for IgG2b, when compared with F1 mice (unpublished observation). The requirement of an effective humoral response for the resolution of experimental T. cruzi infection has been demonstrated [42]–[44]. The bias towards macrophages at the expense of B cells in the spleen of susceptible mice reproduces results of a previous study with parasites of differential virulence. In this study, virulent parasites, but not avirulent ones, caused a reduction of CD19+ B cells, and an increase of Mac3+ macrophages, in the spleen of infected mice [45].
We analysed genome wide expression in the spleen, prior to an insufficient immune response to the parasite, and prior to the profound alterations in cellular composition, in order to identify transcriptional correlates of susceptibility that mapped to the genomic susceptibility loci. We identified three candidate genes from the susceptibility locus on Chromosome 5, and five from the locus on Chromosome 17, with significantly different expression between differentially resistant strains of mice. The organisation of secondary lymphoid organs, such as the spleen, is regulated by compartmentalised expression of lymphoid tissue chemokines, enabling effective communication between lymphocytes and antigen-presenting cells to mount protective immune responses to pathogens. It was therefore interesting to find that the expression of T cell attractant CXC chemokine ligand Cxcl11 mRNA was decreased in suscptible mice. CXCL11 is the most potent agonist of the chemokine receptor CXCR3 that is predominantly found on activated T cells, Th1 type T cells, peripheral T cells and a subset of B cells [46], [47]. It is produced by monocytoid cells, epithelial cells, neutrophils and endothelial cells, and its expression is induced by IFN-γ. It is involved in generating Th1 type immune responses. CXCL11 was also reported to be a natural antagonist of CCR3 [48], [49] and of CCR5 [47], indicating a possible role in counteracting the effects of other inflammatory chemokines such as RANTES, MIP-1α and MIP-1β. More recently, CCR5 expression was shown to augment the inflammatory reaction and to participate in host protection in experimental Chagas' disease [50], [51]. It was now published that CXCL11 also binds to a newly identified chemokine receptor, CXCR7, that is expressed on activated endothelial and other cells, providing a growth and survival advantage and adhesion properties [52]. The role of CXCL11 in T. cruzi infection has not been investigated, but the two other CXCR3- binding chemokines, CXCL9 and CXCL10, were found to contribute to a protective immune response in mice [53], [54]. Decreased expression of Cxcl11 mRNA in susceptible mice may be involved in the insufficient immune response observed in B6 mice by preventing adequate association between T cells and antigen-presenting cells and/or B cells, or by reduced antagonism of CCR3- and/or CCR5- mediated inflammatory reactions.
A further gene from the locus on Chromosome 5 with decreased expression in B6 mice was Bmp2k. Bone morphogenetic proteins play a role in skeletal development. Signalling involves transcription of Bmp2k which encodes a serin/threonin kinase. Activation of BMP2K attenuates osteocalcin expression and reduces osteoblast differentiation [55]. Its role in infections in general, and its possible role in determing the outcome of experimental T. cruzi infection, remain to be determined.
Osteopontin (OPN; also termed secreted phosphoprotein 1, SPP1, or early T lymphocyte activation 1, ETA-1) was primarily described as an early adhesion receptor expressed after T cell activation and mediating macrophage migration [56], [57]. It is regarded as an essential factor in inducing Th1 type immune responses and has been described as a regulator of resistance to microbial infection [58]–[61]. Recently, its role in Th1 type viral immunity was disputed [62], and it was shown that SPP1 mediated IFN-α production in plasmacytoid dendritic cells [63]. Several cells express Spp1 following viral or bacterial infection, including dendritic cells, T cells and macrophages, and it functions as a cytokine and anti-apoptotic growth factor, involved in adhesion, chemotaxis, cell differentiation and inflammation. Osteopontin was also found to participate in collagen fibrillogenesis and matrix re-organisation in wound healing [64]. Transcription of Spp1 was increased in T. cruzi susceptible B6 mice, indicating its role in determining the strong Th1 type immune response. Possibly, its expression is increased secondary to the severe disruption of splenic architecture and due to an increased re-modelling activity.
Several genes within the susceptibility locus on Chromosome 17 were expressed at reduced levels in susceptible mice. The MHC H2-D1 and H2-Eα genes are not encoded in B6 mice and signals were thus drastically decreased. MHC encoded proteins are essential for the presentation of antigens to T lymphocytes. MHC polymorphism is believed to be driven by pathogen-based mechanisms [65], and it has been associated with increased resistance to a model infection [66]. The heterozygosity of the intercross F1 at the MHC locus (H2b/d) implies that a greater number of antigens can potentially be presented to antigen-specific T cells, resulting in a larger pool of lymphoytes that are able to respond to the pathogen. Since a protective response to T. cruzi infection requires both cellular and humoral acquired immune effector mechanisms, it follows that both MHC class I and class II genes are strong candidates for determining susceptibility. However, the presence of a heterozygous state as such did not necessarily confer resistance [14]. Certain H2 haplotypes were associated with a protective effect under given experimental conditions (reviewed by Campbell [3]). In the present study, though, both parental strains succumbed to infection [15], [30], and neither H2 haplotype (neither H2b nor H2d) was thus protective in itself. In man, the influence of HLA haplotypes on disease progression has been demonstrated [6], [67], [68].
The other genes with decreased expression within the locus on Chromosome 17 have not been assigned a role in immunity so far. Tubb5 codes for the β5 isotype of the β-tubulin family and is expressed at low levels ubiquitously [69]. Msh5 codes for a member of the mismatch repair family of proteins, that is expressed mainly in gonads and has a role in chromosome pairing during meiosis; deficiency was associated with apoptosis of testicular and ovarian cells and sterility [70], [71]. The expressed sequence Ng23 has not yet been classified regarding function or process [72].
The immunogenetic background of susceptibility of inbred mice to experimental infections in general, and to experimental infection with T. cruzi in particular, has not been delineated despite progress in analysing numerous immunologic players and pathways. The present work aimed at identifying candidates that direct the many secondary variations of the immune response that have been described. Further work is needed to substantiate these proposals and to clarify the role of susceptibility genes in the evolution of an ineffective immune response. It is important to recognise that the results are strictly related to the common model that has been used in this study. It is certainly probable that alternate mechanisms apply in other inbred mice and with other stocks and strains of T. cruzi.
Materials and Methods
Parasites and mice
The T. cruzi tulahuen strain was used in all experiments. Mouse experiments were registered at and approved by the Federal Health Authorities of the State of Hamburg. Parasites were maintained in Balb/c mice by fortnightly passage. Trypomastigotes were counted in a Neubauer chamber following lysis in a solution of NH4Cl (0.89% w/v) [73], and blood was appropriately diluted with phosphate buffered saline (PBS) for an experimental infection with 104 parasites. C57BL/6 (B6) and C57BL/6xDBA/2 (B6D2F1, F1) mice were obtained from Charles River (Sulzbach, Germany) and infected at the age of 6–8 weeks by inoculation of parasites into the peritoneum in a volume of 200 µl. Parasitaemia was determined in 2 µl of tail vein blood after lysis in 18 µl of NH4Cl.
Quantitative PCR
Relative tissue parasite burdens were determined by real time PCR in specimens of about 20 mg of tissue. DNA extraction was performed with a Gentra Puregene™ tissue kit (Biozym, Hessisch Oldendorf, Germany) according to the manufacturer's instructions. DNA was dissolved in 50 µl of H2O, and a 100 fold dilution was used for amplification. T. cruzi PCR was performed on an Abi Prism 7700 SDS Instrument (Applied Biosystems, Weiterstadt, Germany) as previously described [30]. The quantity of host DNA was determined by real time PCR of β-actin from the same samples as described [30], with the exception that thermal cycles were 20 s at 95°C and 40 s at 58°C. The quantity of parasite DNA in a given sample is expressed in relation to its content of β-actin DNA.
Histology
Specimens from the spleen were obtained from naive and from experimentally infected mice, fixed in buffered 10% formalin and embedded in paraffin. Sections were stained with hematoxylin-eosin (HE) and examined by light microscopy.
Immunohistochemistry
Spleen specimens were shock frozen in liquid nitrogen and stored at −80°C until further processing. Sections of 5–8 µm were stained with one of the following antibodies (obtained from Beckton Dickinson, Heidelberg, Germany): anti-CD4; anti-CD8; or anti-CD19; antibodies were diluted 1∶50 in PBS. Sections were then fixed in paraformaldehyde, and a secondary biotinylated anti-rat Ig antibody was applied for 30 min. Detection was achieved with streptavidin/horseradish peroxidase conjugate and developed with the peroxidase substrate diaminobenzidine (Dako, Hamburg, Germany). The TUNEL reaction on some sections was performed with the ‘In Situ Cell Death Detection Kit, AP’ according to the manufacturers instructions (Roche, Mannheim Germany) Sections were counterstained with hematoxylin and analysed by optical microscopy.
Flow cytometry
Spleen cell surface expression was analysed by staining of cell suspensions with antibodies against either CD4 (Cy5-conjugated) and CD8 (FITC-conjugated), against CD19 (PE-conjugated), or against CD11b (PE-conjugated). Some samples were stained with Annexin V and with propidium iodide to determine fractions of apoptotic cells. All reagents were obtained from Becton Dickinson (Heidelberg, Germany). Cells were fixed in 1% paraformaldehyde and analysed with a FACSCalibur flow cytometer (Beckton Dickinson, Mountain View, CA, USA) using the CellQuest software. Results are expressed as the percentage of cells analysed that were positive for the surface marker of interest.
Microsatellite genotyping
As an extension of our previous study [30], a further 22 male backcross mice (B6xF1) were experimentally infected and identified as being susceptible to a lethal outcome. Genomic DNA of these animals was subjected to microsatellite genotyping to confirm and fine map the genomic regions on Chromosomes 17, 5, and 13, previously identified as putatively linked to susceptibility. Nine polymorphic markers for each region were analysed, as described elsewhere [30]. The markers MM13BM1 and MM13BM2 were identified on mouse Chromosome 13 and amplified with the follwing primer pairs: MM13BM1F (5′atagccaccaacaggcattg) and MM13BM1R (5′FAM-tcggctttccctaaactgtg) as well as MM13BM2F (5′gcttaggcaatgggtctgatag) and MM13BM2R (5′HEX-tttgggtttgtatatgtctgactg). For statistical analysis, genotyping data of the newly typed 22 animals were combined with the original data set obtained from 46 susceptible mice in the previous study. Chi-square statistics were computed comparing the numbers of homozygous versus heterozygous mice. The construction of the genetic maps was performed with MAPMAKER/EXP v3.0b.
Microarray Analysis
Procedures for cDNA synthesis, labelling and hybridization were carried out according to the manufacturer's protocol (Affymetrix). All experiments were performed using Affymetrix mouse genome GeneChip 430A. Three mice of either strain (B6 and F1, respectively) from three independent experiments were sacrificed on day 11, the spleen was removed, and single cell suspensions were prepared. All procedures were performed at 4°C. RNA was prepared with a Qiagen RNeasy kit (Qiagen, Hilden, Germany) according to the manufacturers instructions, and the quality of the preparation was checked by agarose gel electrophoresis. In brief, 5 µg of total RNA were used for first strand cDNA synthesis with an HPLC-purified T7-(dT)24 primer (One-Cycle cDNA Synthesis Kit, Affymetrix). Synthesis of biotin-labeled cRNA was carried out using the IVT Labeling Kit (Affymetrix) and then purified (Sample Cleanup Module, Affymetrix). For hybridization, 15 µg of fragmented cRNA was incubated with the chip in 200 µl of hybridization solution in Hybridization Oven 640 (Affymetrix) at 45°C for 16 hours. GeneChips were then washed and stained using the Affymetrix Fluidics Station according to the GeneChip Expression Analysis Technical Manual. Microarrays were scanned with the Agilent GeneArray Scanner, and the signals were processed using the GeneChip expression analysis algorithm (v.2; Affymetrix). To compare samples and experiments, the trimmed mean signal of each array was scaled to a target intensity of 100. Absolute and comparison analyses were performed with Affymetrix MAS (version 5.0, Affymetrix) software using default parameters. To assist in the identification of genes that were positively or negatively regulated in the experiment, we selected genes that showed a signal log ratio of at least 0.8 (increase) or −0.8 (decrease) compared to the baseline (B6 compared to F1) [74]. Annotations were further analyzed with interactive query analysis at www.affymetrix.com. Pathways and other functional groupings of genes were evaluated for differential regulation using the visualisation tool GenMAPP (UCSF) as described elsewhere [75], [76].
Acknowledgments
We thank Klara Tenner-Racz and Paul Racz for help with histological and immunohistochemical analysis of spleen sections. The technical assistance of Kristin Reeck, Ulricke Klauenberg and Petra Eggers is gratefully acknowledged.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the German Federal Ministry of Science and Education through the National Genome Research Network.
References
- 1.Miles MA. New World Trypanosomiasis. In: Cox FEG, Wakelin D, Gillespie SH, Despommier DD, editors. Topley & Wilson's Microbiology and Microbial Infections. London: Hodder Arnold; 2005. pp. 376–398. [Google Scholar]
- 2.Williams-Blangero S, Vandeberg JL, Blangero J, Teixeira AR. Genetic epidemiology of seropositivity for Trypanosoma cruzi infection in rural Goias, Brazil. Am J Trop Med Hyg. 1997;57:538–543. doi: 10.4269/ajtmh.1997.57.538. [DOI] [PubMed] [Google Scholar]
- 3.Campbell DA, Westenberger SJ, Sturm NR. The determinants of Chagas disease: connecting parasite and host genetics. Curr Mol Med. 2004;4:549–562. doi: 10.2174/1566524043360249. [DOI] [PubMed] [Google Scholar]
- 4.Fae KC, Drigo SA, Cunha-Neto E, Ianni B, Mady C, et al. HLA and beta-myosin heavy chain do not influence susceptibility to Chagas disease cardiomyopathy. Microbes Infect. 2000;2:745–751. doi: 10.1016/s1286-4579(00)00501-3. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Mestre MT, Layrisse Z, Montagnani S, Acquatella H, Catalioti F, et al. Influence of the HLA class II polymorphism in chronic Chagas' disease. Parasite Immunol. 1998;20:197–203. [PubMed] [Google Scholar]
- 6.Deghaide NH, Dantas RO, Donadi EA. HLA class I and II profiles of patients presenting with Chagas' disease. Dig Dis Sci. 1998;43:246–252. doi: 10.1023/a:1018829600200. [DOI] [PubMed] [Google Scholar]
- 7.Zhang L, Tarleton RL. Parasite persistence correlates with disease severity and localization in chronic Chagas' disease. J Infect Dis. 1999;180:480–486. doi: 10.1086/314889. [DOI] [PubMed] [Google Scholar]
- 8.Vago AR, Macedo AM, Adad SJ, Reis DD, Correa-Oliveira R. PCR detection of Trypanosoma cruzi DNA in oesophageal tissues of patients with chronic digestive Chagas' disease. Lancet. 1996;348:891–892. doi: 10.1016/S0140-6736(05)64761-7. [DOI] [PubMed] [Google Scholar]
- 9.Tarleton RL, Zhang L, Downs MO. “Autoimmune rejection” of neonatal heart transplants in experimental Chagas disease is a parasite-specific response to infected host tissue. Proc Natl Acad Sci U S A. 1997;94:3932–3937. doi: 10.1073/pnas.94.8.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marinho CR, Bucci DZ, Dagli ML, Bastos KR, Grisotto MG, et al. Pathology affects different organs in two mouse strains chronically infected by a Trypanosoma cruzi clone: a model for genetic studies of Chagas' disease. Infect Immun. 2004;72:2350–2357. doi: 10.1128/IAI.72.4.2350-2357.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buckner FS, Wilson AJ, Van Voorhis WC. Detection of live Trypanosoma cruzi in tissues of infected mice by using histochemical stain for beta-galactosidase. Infect Immun. 1999;67:403–409. doi: 10.1128/iai.67.1.403-409.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guarner J, Bartlett J, Zaki SR, Colley DG, Grijalva MJ, et al. Mouse model for Chagas disease: immunohistochemical distribution of different stages of Trypanosoma cruzi in tissues throughout infection. Am J Trop Med Hyg. 2001;65:152–158. doi: 10.4269/ajtmh.2001.65.152. [DOI] [PubMed] [Google Scholar]
- 13.Marinho CR, D'Imperio Lima MR, Grisotto MG, Alvarez JM. Influence of acute-phase parasite load on pathology, parasitism, and activation of the immune system at the late chronic phase of Chagas' disease. Infect Immun. 1999;67:308–318. doi: 10.1128/iai.67.1.308-318.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wrightsman R, Krassner S, Watson J. Genetic control of responses to Trypanosoma cruzi in mice: multiple genes influencing parasitemia and survival. Infect Immun. 1982;36:637–644. doi: 10.1128/iai.36.2.637-644.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trischmann TM, Bloom BR. Genetics of murine resistance to Trypanosoma cruzi. Infect Immun. 1982;35:546–551. doi: 10.1128/iai.35.2.546-551.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powell MR, Wassom DL. Host genetics and resistance to acute Trypanosoma cruzi infection in mice. I. Antibody isotype profiles. Parasite Immunol. 1993;15:215–221. doi: 10.1111/j.1365-3024.1993.tb00603.x. [DOI] [PubMed] [Google Scholar]
- 17.Wrightsman RA, Krassner SM, Watson JD, Manning JE. Role of the H-2s haplotype in survival of mice after infection with Trypanosoma cruzi. Infect Immun. 1984;44:351–354. doi: 10.1128/iai.44.2.351-354.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andrade V, Barral-Netto M, Andrade SG. Patterns of resistance of inbred mice to Trypanosoma cruzi are determined by parasite strain. Braz J Med Biol Res. 1985;18:499–506. [PubMed] [Google Scholar]
- 19.Andrade LO, Machado CR, Chiari E, Pena SD, Macedo AM. Trypanosoma cruzi: role of host genetic background in the differential tissue distribution of parasite clonal populations. Exp Parasitol. 2002;100:269–275. doi: 10.1016/s0014-4894(02)00024-3. [DOI] [PubMed] [Google Scholar]
- 20.Tarleton RL, Koller BH, Latour A, Postan M. Susceptibility of beta 2-microglobulin-deficient mice to Trypanosoma cruzi infection. Nature. 1992;356:338–340. doi: 10.1038/356338a0. [DOI] [PubMed] [Google Scholar]
- 21.Silva JS, Morrissey PJ, Grabstein KH, Mohler KM, Anderson D, et al. Interleukin 10 and interferon gamma regulation of experimental Trypanosoma cruzi infection. J Exp Med. 1992;175:169–174. doi: 10.1084/jem.175.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rottenberg ME, Bakhiet M, Olsson T, Kristensson K, Mak T, et al. Differential susceptibilities of mice genomically deleted of CD4 and CD8 to infections with Trypanosoma cruzi or Trypanosoma brucei. Infect Immun. 1993;61:5129–5133. doi: 10.1128/iai.61.12.5129-5133.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michailowsky V, Silva NM, Rocha CD, Vieira LQ, Lannes-Vieira J, et al. Pivotal role of interleukin-12 and interferon-gamma axis in controlling tissue parasitism and inflammation in the heart and central nervous system during Trypanosoma cruzi infection. Am J Pathol. 2001;159:1723–1733. doi: 10.1016/s0002-9440(10)63019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holscher C, Kohler G, Muller U, Mossmann H, Schaub GA, et al. Defective nitric oxide effector functions lead to extreme susceptibility of Trypanosoma cruzi-infected mice deficient in gamma interferon receptor or inducible nitric oxide synthase. Infect Immun. 1998;66:1208–1215. doi: 10.1128/iai.66.3.1208-1215.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graefe SE, Jacobs T, Gaworski I, Klauenberg U, Steeg C, et al. Interleukin-12 but not interleukin-18 is required for immunity to Trypanosoma cruzi in mice. Microbes Infect. 2003;5:833–839. doi: 10.1016/s1286-4579(03)00176-x. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Tarleton RL. Characterization of cytokine production in murine Trypanosoma cruzi infection by in situ immunocytochemistry: lack of association between susceptibility and type 2 cytokine production. Eur J Immunol. 1996;26:102–109. doi: 10.1002/eji.1830260116. [DOI] [PubMed] [Google Scholar]
- 27.Soares MB, Silva-Mota KN, Lima RS, Bellintani MC, Pontes-de-Carvalho L, et al. Modulation of chagasic cardiomyopathy by interleukin-4: dissociation between inflammation and tissue parasitism. Am J Pathol. 2001;159:703–709. doi: 10.1016/S0002-9440(10)61741-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silva JS, Twardzik DR, Reed SG. Regulation of Trypanosoma cruzi infections in vitro and in vivo by transforming growth factor beta (TGF-beta). J Exp Med. 1991;174:539–545. doi: 10.1084/jem.174.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freire-de-Lima CG, Nascimento DO, Soares MB, Bozza PT, Castro-Faria-Neto HC, et al. Uptake of apoptotic cells drives the growth of a pathogenic trypanosome in macrophages. Nature. 2000;403:199–203. doi: 10.1038/35003208. [DOI] [PubMed] [Google Scholar]
- 30.Graefe SE, Meyer BS, Muller-Myhsok B, Ruschendorf F, Drosten C, et al. Murine susceptibility to Chagas' disease maps to chromosomes 5 and 17. Genes Immun. 2003;4:321–325. doi: 10.1038/sj.gene.6363972. [DOI] [PubMed] [Google Scholar]
- 31.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 32.Sedy JR, Gavrieli M, Potter KG, Hurchla MA, Lindsley RC, et al. B and T lymphocyte attenuator regulates T cell activation through interaction with herpesvirus entry mediator. Nat Immunol. 2005;6:90–98. doi: 10.1038/ni1144. [DOI] [PubMed] [Google Scholar]
- 33.Morgado MG, Cam P, Gris-Liebe C, Cazenave PA, Jouvin-Marche E. Further evidence that BALB/c and C57BL/6 gamma 2a genes originate from two distinct isotypes. Embo J. 1989;8:3245–3251. doi: 10.1002/j.1460-2075.1989.tb08484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones PP, Murphy DB, McDevitt HO. Variable synthesis and expression of E alpha and Ae (E beta) Ia polypeptide chains in mice of different H-2 haplotypes. Immunogenetics. 1981;12:321–337. doi: 10.1007/BF01561674. [DOI] [PubMed] [Google Scholar]
- 35.Liao G, Wang J, Guo J, Allard J, Cheng J, et al. In silico genetics: identification of a functional element regulating H2-Ealpha gene expression. Science. 2004;306:690–695. doi: 10.1126/science.1100636. [DOI] [PubMed] [Google Scholar]
- 36.Allcock RJ, Martin AM, Price P. The mouse as a model for the effects of MHC genes on human disease. Immunol Today. 2000;21:328–332. doi: 10.1016/s0167-5699(00)01654-6. [DOI] [PubMed] [Google Scholar]
- 37.Casadevall A, Pirofski L. Host-pathogen interactions: the attributes of virulence. J Infect Dis. 2001;184:337–344. doi: 10.1086/322044. [DOI] [PubMed] [Google Scholar]
- 38.Cooke GS, Hill AV. Genetics of susceptibility to human infectious disease. Nat Rev Genet. 2001;2:967–977. doi: 10.1038/35103577. [DOI] [PubMed] [Google Scholar]
- 39.Kellam P, Weiss RA. Infectogenomics: insights from the host genome into infectious diseases. Cell. 2006;124:695–697. doi: 10.1016/j.cell.2006.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knudsen S. Guide to analysis of DNA microarray data. Chichester: John Wiley & Sons Inc; 2004. p. 184. [Google Scholar]
- 41.Zuniga E, Motran C, Montes CL, Diaz FL, Bocco JL, et al. Trypanosoma cruzi-induced immunosuppression: B cells undergo spontaneous apoptosis and lipopolysaccharide (LPS) arrests their proliferation during acute infection. Clin Exp Immunol. 2000;119:507–515. doi: 10.1046/j.1365-2249.2000.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar S, Tarleton RL. The relative contribution of antibody production and CD8+ T cell function to immune control of Trypanosoma cruzi. Parasite Immunol. 1998;20:207–216. doi: 10.1046/j.1365-3024.1998.00154.x. [DOI] [PubMed] [Google Scholar]
- 43.Trischmann TM. Non-antibody-mediated control of parasitemia in acute experimental Chagas' disease. J Immunol. 1983;130:1953–1957. [PubMed] [Google Scholar]
- 44.Trischmann TM. Role of cellular immunity in protection against Trypanosoma cruzi in mice. Parasite Immunol. 1984;6:561–570. doi: 10.1111/j.1365-3024.1984.tb00825.x. [DOI] [PubMed] [Google Scholar]
- 45.Alba Soto CD, Mirkin GA, Solana ME, Gonzalez Cappa SM. Trypanosoma cruzi infection modulates in vivo expression of major histocompatibility complex class II molecules on antigen-presenting cells and T-cell stimulatory activity of dendritic cells in a strain-dependent manner. Infect Immun. 2003;71:1194–1199. doi: 10.1128/IAI.71.3.1194-1199.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cole KE, Strick CA, Paradis TJ, Ogborne KT, Loetscher M, et al. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high affinity binding to CXCR3. J Exp Med. 1998;187:2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petkovic V, Moghini C, Paoletti S, Uguccioni M, Gerber B. I-TAC/CXCL11 is a natural antagonist for CCR5. J Leukoc Biol. 2004;76:701–708. doi: 10.1189/jlb.1103570. [DOI] [PubMed] [Google Scholar]
- 48.Xanthou G, Duchesnes CE, Williams TJ, Pease JE. CCR3 functional responses are regulated by both CXCR3 and its ligands CXCL9, CXCL10 and CXCL11. Eur J Immunol. 2003;33:2241–2250. doi: 10.1002/eji.200323787. [DOI] [PubMed] [Google Scholar]
- 49.Loetscher P, Pellegrino A, Gong JH, Mattioli I, Loetscher M, et al. The ligands of CXC chemokine receptor 3, I-TAC, Mig, and IP10, are natural antagonists for CCR3. J Biol Chem. 2001;276:2986–2991. doi: 10.1074/jbc.M005652200. [DOI] [PubMed] [Google Scholar]
- 50.Hardison JL, Wrightsman RA, Carpenter PM, Kuziel WA, Lane TE, et al. The CC chemokine receptor 5 is important in control of parasite replication and acute cardiac inflammation following infection with Trypanosoma cruzi. Infect Immun. 2006;74:135–143. doi: 10.1128/IAI.74.1.135-143.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Machado FS, Koyama NS, Carregaro V, Ferreira BR, Milanezi CM, et al. CCR5 plays a critical role in the development of myocarditis and host protection in mice infected with Trypanosoma cruzi. J Infect Dis. 2005;191:627–636. doi: 10.1086/427515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burns JM, Summers BC, Wang Y, Melikian A, Berahovich R, et al. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hardison JL, Wrightsman RA, Carpenter PM, Lane TE, Manning JE. The chemokines CXCL9 and CXCL10 promote a protective immune response but do not contribute to cardiac inflammation following infection with Trypanosoma cruzi. Infect Immun. 2006;74:125–134. doi: 10.1128/IAI.74.1.125-134.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Talvani A, Ribeiro CS, Aliberti JC, Michailowsky V, Santos PV, et al. Kinetics of cytokine gene expression in experimental chagasic cardiomyopathy: tissue parasitism and endogenous IFN-gamma as important determinants of chemokine mRNA expression during infection with Trypanosoma cruzi. Microbes Infect. 2000;2:851–866. doi: 10.1016/s1286-4579(00)00388-9. [DOI] [PubMed] [Google Scholar]
- 55.Kearns AE, Donohue MM, Sanyal B, Demay MB. Cloning and characterization of a novel protein kinase that impairs osteoblast differentiation in vitro. J Biol Chem. 2001;276:42213–42218. doi: 10.1074/jbc.M106163200. [DOI] [PubMed] [Google Scholar]
- 56.Patarca R, Freeman GJ, Singh RP, Wei FY, Durfee T, et al. Structural and functional studies of the early T lymphocyte activation 1 (Eta-1) gene. Definition of a novel T cell-dependent response associated with genetic resistance to bacterial infection. J Exp Med. 1989;170:145–161. doi: 10.1084/jem.170.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh RP, Patarca R, Schwartz J, Singh P, Cantor H. Definition of a specific interaction between the early T lymphocyte activation 1 (Eta-1) protein and murine macrophages in vitro and its effect upon macrophages in vivo. J Exp Med. 1990;171:1931–1942. doi: 10.1084/jem.171.6.1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nau GJ, Liaw L, Chupp GL, Berman JS, Hogan BL, et al. Attenuated host resistance against Mycobacterium bovis BCG infection in mice lacking osteopontin. Infect Immun. 1999;67:4223–4230. doi: 10.1128/iai.67.8.4223-4230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maeno Y, Nakazawa S, Dao le D, Van Tuan N, Giang ND, et al. Osteopontin is involved in Th1-mediated immunity against Plasmodium falciparum infection in a holoendemic malaria region in Vietnam. Acta Trop. 2006;98:305–310. doi: 10.1016/j.actatropica.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 60.Koguchi Y, Kawakami K, Uezu K, Fukushima K, Kon S, et al. High plasma osteopontin level and its relationship with interleukin-12-mediated type 1 T helper cell response in tuberculosis. Am J Respir Crit Care Med. 2003;167:1355–1359. doi: 10.1164/rccm.200209-1113OC. [DOI] [PubMed] [Google Scholar]
- 61.Ashkar S, Weber GF, Panoutsakopoulou V, Sanchirico ME, Jansson M, et al. Eta-1 (osteopontin): an early component of type-1 (cell-mediated) immunity. Science. 2000;287:860–864. doi: 10.1126/science.287.5454.860. [DOI] [PubMed] [Google Scholar]
- 62.Abel B, Freigang S, Bachmann MF, Boschert U, Kopf M. Osteopontin is not required for the development of Th1 responses and viral immunity. J Immunol. 2005;175:6006–6013. doi: 10.4049/jimmunol.175.9.6006. [DOI] [PubMed] [Google Scholar]
- 63.Shinohara ML, Lu L, Bu J, Werneck MB, Kobayashi KS, et al. Osteopontin expression is essential for interferon-alpha production by plasmacytoid dendritic cells. Nat Immunol. 2006;7:498–506. doi: 10.1038/ni1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liaw L, Birk DE, Ballas CB, Whitsitt JS, Davidson JM, et al. Altered wound healing in mice lacking a functional osteopontin gene (spp1). J Clin Invest. 1998;101:1468–1478. doi: 10.1172/JCI1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Potts WK, Slev PR. Pathogen-based models favoring MHC genetic diversity. Immunol Rev. 1995;143:181–197. doi: 10.1111/j.1600-065x.1995.tb00675.x. [DOI] [PubMed] [Google Scholar]
- 66.Messaoudi I, Guevara Patino JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between mhc polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298:1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 67.Colorado IA, Acquatella H, Catalioti F, Fernandez MT, Layrisse Z. HLA class II DRB1, DQB1, DPB1 polymorphism and cardiomyopathy due to Trypanosoma cruzi chronic infection. Hum Immunol. 2000;61:320–325. doi: 10.1016/s0198-8859(99)00177-9. [DOI] [PubMed] [Google Scholar]
- 68.Nieto A, Beraun Y, Collado MD, Caballero A, Alonso A, et al. HLA haplotypes are associated with differential susceptibility to Trypanosoma cruzi infection. Tissue Antigens. 2000;55:195–198. doi: 10.1034/j.1399-0039.2000.550301.x. [DOI] [PubMed] [Google Scholar]
- 69.Luduena RF. Multiple forms of tubulin: different gene products and covalent modifications. Int Rev Cytol. 1998;178:207–275. doi: 10.1016/s0074-7696(08)62138-5. [DOI] [PubMed] [Google Scholar]
- 70.Edelmann W, Cohen PE, Kneitz B, Winand N, Lia M, et al. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet. 1999;21:123–127. doi: 10.1038/5075. [DOI] [PubMed] [Google Scholar]
- 71.de Vries SS, Baart EB, Dekker M, Siezen A, de Rooij DG, et al. Mouse MutS-like protein Msh5 is required for proper chromosome synapsis in male and female meiosis. Genes Dev. 1999;13:523–531. doi: 10.1101/gad.13.5.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mouse Genome Database (MGD), Mouse Genome Informatics Web Site, The Jackson Laboratory, http://www.informatics.jax.org. Sept. 2006. [Google Scholar]
- 73.Hoff R. A method for counting and concentrating living Trypanosoma cruzi in blood lysed with ammonium chloride. J Parasitol. 1974;60:527–528. [PubMed] [Google Scholar]
- 74.Rajagopalan D. A comparison of statistical methods for analysis of high density oligonucleotide array data. Bioinformatics. 2003;19:1469–1476. doi: 10.1093/bioinformatics/btg202. [DOI] [PubMed] [Google Scholar]
- 75.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, et al. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bonner AE, Lemon WJ, You M. Gene expression signatures identify novel regulatory pathways during murine lung development: implications for lung tumorigenesis. J Med Genet. 2003;40:408–417. doi: 10.1136/jmg.40.6.408. [DOI] [PMC free article] [PubMed] [Google Scholar]