Abstract
Background
Low-level, partial resistance is pre-eminent in natural populations, however, the mechanisms underlying this form of resistance are still poorly understood.
Methodology/Principal Findings
In the present study, we used the model pathosystem Pseudomonas syringae pv. tomato DC3000 (Pst) - Arabidopsis thaliana to study the genetic basis of this form of resistance. Phenotypic analysis of a set of Arabidopsis accessions, based on evaluation of in planta pathogen growth revealed extensive quantitative variation for partial resistance to Pst. It allowed choosing a recombinant inbred line (RIL) population derived from a cross between the accessions Bayreuth and Shahdara for quantitative genetic analysis. Experiments performed under two different environmental conditions led to the detection of two major and two minor quantitative trait loci (QTLs) governing partial resistance to Pst and called PRP-Ps1 to PRP-Ps4. The two major QTLs, PRP-Ps1 and PRP-Ps2, were confirmed in near isogenic lines (NILs), following the heterogeneous inbred families (HIFs) strategy. Analysis of marker gene expression using these HIFs indicated a negative correlation between the induced amount of transcripts of SA-dependent genes PR1, ICS and PR5, and the in planta bacterial growth in the HIF segregating at PRP-Ps2 locus, suggesting an implication of PRP-Ps2 in the activation of SA dependent responses.
Conclusions/Significance
These results show that variation in partial resistance to Pst in Arabidopsis is governed by relatively few loci, and the validation of two major loci opens the way for their fine mapping and their cloning, which will improve our understanding of the molecular mechanisms underlying partial resistance.
Introduction
Plants are exposed to a wide variety of pathogens with different invasion strategies. Successful infections are, however, relatively rare because plants have evolved powerful preformed and inducible defence mechanisms to restrict pathogen growth. Nonhost resistance relies on multiple mechanisms, which are beginning to be uncovered [1]–[3]. This defense system shows some similarity with the mammalian innate immunity [4], [5] and is associated with multiple signal transduction events like an oxidative burst, ion fluxes, activation of MAP kinase cascades, with the transcriptional induction of pathogen-responsive genes and with localized callose deposition at the cell wall [6]–[9]. If a pathogen can overcome nonhost resistance, it can spread in its host plant; however, different defense mechanisms in plants can still be activated, leading to complete or partial resistance. Complete resistance, developed in the case of an incompatible interaction, is usually governed by the gene-for-gene system, and also called race-specific resistance. Much research has focused on this form of resistance which is generally inherited as a monogenic trait and is determined by the concomitant presence of a resistance (R) gene in the plant and the corresponding avirulence (avr) gene in the pathogen. Mechanistically, specific resistance relies on the recognition of avr pathogen factors by plant R gene products and the elicitation of local defense responses, often associated with a rapid programmed cell death, called the hypersensitive response (HR). A variety of R genes have been cloned from model and crop plants, and many avr genes have been characterized from bacteria, fungi and oomycetes[10]. Interestingly, although R genes confer resistance to diverse pathogens, their products share structural similarities suggesting the conservation of some signalling events in plant defense [10].
In contrast, the so-called partial resistance is quantitative, presumably non race-specific, and polygenic [11]–[13]. It limits the extent of disease caused by virulent pathogens and constitutes an additional layer of resistance in the absence of R function, during compatible interactions. The genetics of partial resistance has been characterized in many crop plants, such as rice and barley [14], [15] but remains poorly understood in Arabidopsis. One way to increase our knowledge in this field is a genetic study of the quantitative variation in resistance to virulent pathogens.
Although QTL analyses are increasingly used to study complex traits in Arabidopsis, such as developmental and yield traits [16], [17], only a few studies have investigated the genetic bases of quantitative variation in resistance and susceptibility to pathogens [18]–[22]. In most of these studies one or two major QTLs and a few minor loci were identified. In one study investigating plant susceptibility to the fungus Botrytis cinerea, multiple small-to-medium effect QTLs were identified [20], suggesting that multiple mechanisms are involved in susceptibility, or that a large number of polymorphic loci exert an effect on a particular mechanism. In some cases, genes underlying the QTL have been identified. For example, the major QTL for resistance to Plectosphaerella cucumerina was demonstrated to correspond to the ERECTA gene [22] which is implicated in plant development and also contributes to resistance to Ralstonia solanacearum [19].
The Arabidopsis-Pseudomonas interaction is a model pathosystem [23], [24] and has largely contributed to a better understanding of pathogen recognition in plants, pathogen virulence and avirulence determinants, host susceptibility and signal transduction pathways controlling plant defense responses [25]. In a previous study, analysis of natural variation in tolerance indicated that it behaves as a quantitative trait [26]. Tolerance, which can be defined as the ability of the host to endure the presence of the pathogen and to express less severe disease symptoms or less damage [27], differs from resistance in that symptom formation is uncoupled from pathogen growth [28]. When the genetic basis of variation of this trait was analysed in the Arabidopsis accessions Col-0 and No-0, only two minor loci for symptom severity and no QTL for bacterial colonization could be identified [21], although bacterial growth is a key quantitative component of the compatible interaction between Arabidopsis and the endophytic bacterial pathogen Pst.
In this paper, we use QTL analysis to investigate the genetic basis of partial resistance to Pst using the virulent strain DC3000. Natural variation in Arabidopsis for partial resistance to Pst allowed us to identify parental lines exhibiting significant differences in this trait, and to choose a RIL population derived from crosses between the accessions Bay-0 and Shahdara for detailed genetic analysis. Quantitative evaluation of this RIL population after infection with Pst DC3000 showed that partial resistance to Pst is controlled by two major and two minor QTLs. Using the heterogeneous inbred family strategy (HIF) [29], the two major QTLs were validated. To investigate whether these two major loci could influence known signalling pathways, the expression of marker defense genes was analyzed in HIFs, revealing an influence of the PRP-Ps2 locus on the signalling pathway involving salicylic acid.
Results
Natural variation for Pst resistance in Arabidopsis
In order to investigate natural variation of partial resistance to Pst in Arabidopsis thaliana, 27 Arabidopsis accessions were tested for their response to the virulent Pst strain DC3000. The accessions correspond to a core-collection composed of 16 accessions which are estimated to represent most of the variation present within the species Arabidopsis thaliana [30], plus other parental accessions of RIL populations that are publicly-available or under construction (http://www.inra.fr/vast/RILs.htm).
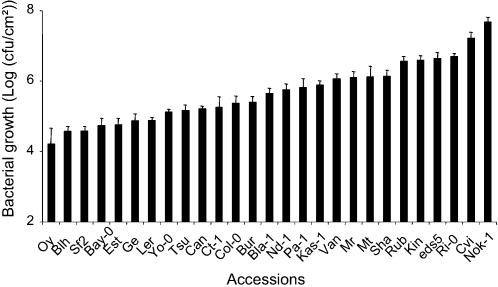
Plant resistance was evaluated by the measurement of bacterial in planta growth 3 days after leaf-infiltration of a bacterial suspension. The chosen inoculation procedure circumvents some layers of resistance operating in a natural infection process, but it is proven to be highly reproducible and allows the quantitative evaluation of resistance and susceptibility. As shown in Figure 1, bacterial growth varied continuously over a range of four orders of magnitude in the different accessions. This result indicates that partial resistance to Pst is a quantitative trait, suggesting that it may be under polygenic control.
Figure 1.
Natural variation of partial resistance to Pseudomonas syringae pv. tomato DC3000 among Arabidopsis accessions. In planta bacterial growth was assessed three days post inoculation with a bacterial suspension adjusted to 105 cfu/mL. Means and standard errors were calculated from bacterial densities in at least 4 plants.
Among the parental accessions showing very contrasting phenotypes, Ler and Cvi seemed to be good candidates for further QTL analysis, since a well characterised Ler×Cvi RIL population exists [31], [32]. However, the analysis of selected RILs from this population revealed, in accordance with published data [33] that 25% of the RILs are very early-flowering even under short day conditions. This was a major obstacle for the use of this population, because the evaluation of resistance is performed on fully expanded rosette leaves of non flowering plants. As the accessions Bayreuth (Bay-0) and Shahdara (Sha) also exhibited contrasting phenotypes, a F6 RIL population of 420 lines derived from these lines was chosen for QTL analysis [34]. Evaluation of resistance of reciprocal F1 hybrids between Bay-0 and Shahdara revealed that bacterial densities in F1 plants were similar to that evaluated in the parental line Shahdara, and higher than that in Bay-0 (Supplementary Figure 1).
QTL analysis identifies two major and two minor loci for partial resistance
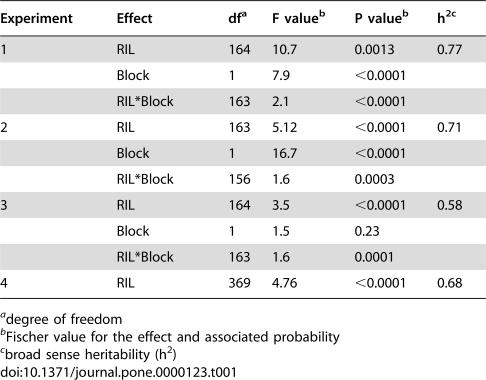
To identify loci responsible for the genetic differences in partial resistance to Pst between Bay-0 and Shahdara, three independent experiments, with two blocks in a complete randomized design, were performed on 165 RILs in greenhouse conditions, and one experiment was performed using 370 RILs under growth chamber conditions (Table 1). A total number of 1730 plants were evaluated for their response to Pst, using generally 14 plants per RIL.
Table 1. The influence of genotype on partial resistance to Pseudomonas syringae pv. tomato DC3000.
| Experiment | Effect | dfa | F valueb | P valueb | h2 c |
| 1 | RIL | 164 | 10.7 | 0.0013 | 0.77 |
| Block | 1 | 7.9 | <0.0001 | ||
| RIL*Block | 163 | 2.1 | <0.0001 | ||
| 2 | RIL | 163 | 5.12 | <0.0001 | 0.71 |
| Block | 1 | 16.7 | <0.0001 | ||
| RIL*Block | 156 | 1.6 | 0.0003 | ||
| 3 | RIL | 164 | 3.5 | <0.0001 | 0.58 |
| Block | 1 | 1.5 | 0.23 | ||
| RIL*Block | 163 | 1.6 | 0.0001 | ||
| 4 | RIL | 369 | 4.76 | <0.0001 | 0.68 |
degree of freedom
Fischer value for the effect and associated probability
broad sense heritability (h2)
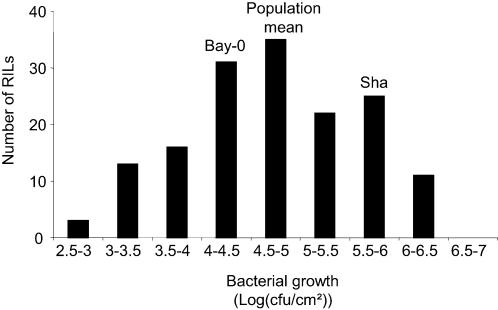
Distributions of the RILs, according to bacterial populations measured in planta (log(cfu/cm2)), showed a continuous variation, suggesting a quantitative and polygenic control of the response (Figure 2). The parental accessions Bay-0 and Shahdara showed, as expected, different levels of bacterial growth. Moreover, transgressive segregation was observed in the RILs. Data obtained from the four experiments performed under different environmental conditions showed significant correlations (Table S1), and within each experiment, the randomized blocks were significantly correlated with R2 values varying from 0.57 to 0.76 (data not shown). Variance analysis of the phenotypic data showed that differences among RILs were highly significant (Table 1). The block effect, except for one of the experiments (experiment 3), and the RIL/block interaction were also significant for all the experiments performed in the greenhouse. Two plants for each RIL were evaluated for their response to Pst in each block, but no significant plant effects could be detected (data not shown). Broad-sense heritabilities calculated from the different experiments, ranged from 0.58 to 0.77, indicating that most of the phenotypic variation appeared to be genetically determined (Table 1).
Figure 2.
Distribution of bacterial growth values in the Bay-0×Shahdara recombinant inbred line (RIL) population. The frequency histogram shows the range of in planta bacterial populations observed in RILs three days post inoculation with Pseudomonas syringae pv. tomato DC3000 in one of the greenhouse experiments. The values obtained for the parental accessions, Bay-0 and Shahdara (Sha), and the genetic mean of the population are indicated.
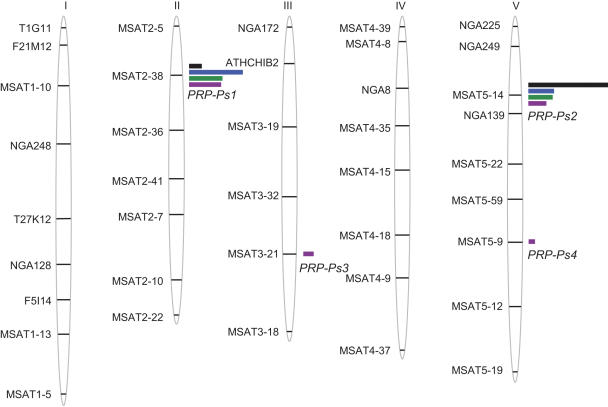
For QTL detection, only composite interval mapping (CIM) results are presented for each experiment, because as compared to other methods, they provide a more accurate estimation of R2 values (phenotypic variances explained by the QTL) and additive effects [35]. Besides, the results of QTL detection obtained for each block of the experiments were the same as those obtained with the adjusted means on blocks (data not shown). Two major QTLs for partial resistance to Pst, PRP-Ps1 (Partial Resistance to Pathogen - Pseudomonas 1) and PRP-Ps2, were detected in all experiments (Figure 3). PRP-Ps1 was localized on chromosome 2 and explained more than 1/4 of the phenotypic variance (up to 42%) except in experiment 1 where it explained only 9%. PRP-Ps2 usually explained less than 20% of the phenotypic variance, whereas its effect was much stronger in experiment 1 where it explained 62% (Table 2). The allelic additive effects of these QTLs are in the same direction, the Bay-0 allele increasing partial resistance compared to the Shahdara allele. In experiment 4, performed under growth chamber conditions, additional minor QTLs were detected on chromosomes III (R2 = 8%) and V (R2 = 5%). In contrast to the major loci, the Shahdara allele at these minor QTLs improves partial resistance to Pst. These observations might explain at least in part, transgressions above the Shahdara phenotypic value and below the Bay-0 phenotypic value in the RIL population (Figure 2).
Figure 3.
Arabidopsis QTLs controlling partial resistance to Pseudomonas syringae pv. tomato DC3000 in the Bay-0×Shahdara recombinant inbred line population. The detected QTLs are represented by bars located at the closest marker position (black, experiment 1; blue, experiment 2; green, experiment 3; purple, experiment 4) on the Bay-0×Shahdara genetic map [34]. The length of the bar is proportional to the QTL effect (R2).
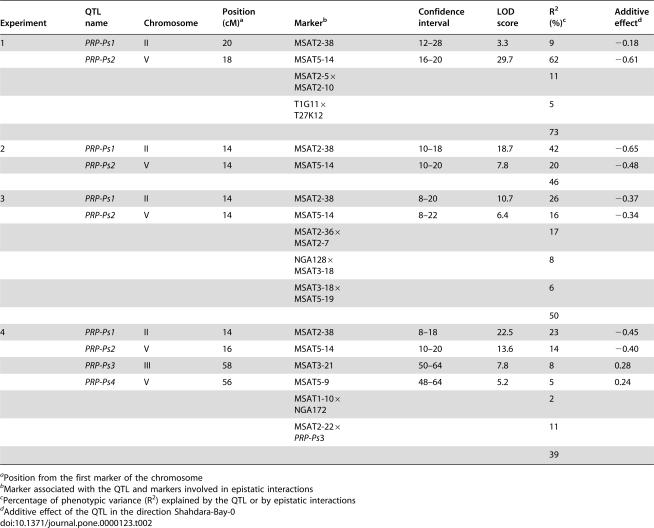
Table 2. Arabidopsis QTLs controlling partial resistance to Pseudomonas syringae pv. tomato DC3000 in the Bay-0×Shahdara recombinant inbred line population.
| Experiment | QTL name | Chromosome | Position (cM)a | Markerb | Confidence interval | LOD score | R2 (%)c | Additive effectd |
| 1 | PRP-Ps1 | II | 20 | MSAT2-38 | 12–28 | 3.3 | 9 | −0.18 |
| PRP-Ps2 | V | 18 | MSAT5-14 | 16–20 | 29.7 | 62 | −0.61 | |
| MSAT2-5×MSAT2-10 | 11 | |||||||
| T1G11×T27K12 | 5 | |||||||
| 73 | ||||||||
| 2 | PRP-Ps1 | II | 14 | MSAT2-38 | 10–18 | 18.7 | 42 | −0.65 |
| PRP-Ps2 | V | 14 | MSAT5-14 | 10–20 | 7.8 | 20 | −0.48 | |
| 46 | ||||||||
| 3 | PRP-Ps1 | II | 14 | MSAT2-38 | 8–20 | 10.7 | 26 | −0.37 |
| PRP-Ps2 | V | 14 | MSAT5-14 | 8–22 | 6.4 | 16 | −0.34 | |
| MSAT2-36×MSAT2-7 | 17 | |||||||
| NGA128×MSAT3-18 | 8 | |||||||
| MSAT3-18×MSAT5-19 | 6 | |||||||
| 50 | ||||||||
| 4 | PRP-Ps1 | II | 14 | MSAT2-38 | 8–18 | 22.5 | 23 | −0.45 |
| PRP-Ps2 | V | 16 | MSAT5-14 | 10–20 | 13.6 | 14 | −0.40 | |
| PRP-Ps3 | III | 58 | MSAT3-21 | 50–64 | 7.8 | 8 | 0.28 | |
| PRP-Ps4 | V | 56 | MSAT5-9 | 48–64 | 5.2 | 5 | 0.24 | |
| MSAT1-10×NGA172 | 2 | |||||||
| MSAT2-22×PRP-Ps3 | 11 | |||||||
| 39 |
Position from the first marker of the chromosome
Marker associated with the QTL and markers involved in epistatic interactions
Percentage of phenotypic variance (R2) explained by the QTL or by epistatic interactions
Additive effect of the QTL in the direction Shahdara-Bay-0
Testing of the 703 possible pairwise interactions revealed highly significant digenic epistatic interactions (Table 2). Most of them occurred between markers with no additive effects and only a few of them were detected between a QTL (or close to the QTL) and a marker of the genetic background.
The total phenotypic variance explained by QTLs with additive effects for partial resistance to Pst ranged from 37% to 67% (data not shown), and increased to 39% to 73% when epistatic interactions were included (Table 2).
Confirmation of the major QTLs PRP-Ps1 and PRP-Ps2 in near isogenic lines
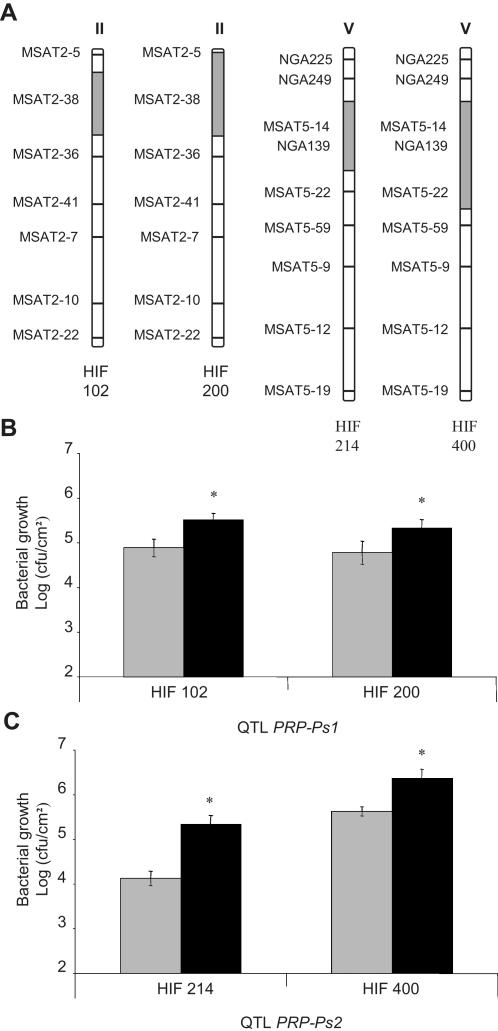
To confirm the effects of the major QTLs PRP-Ps1 and PRP-Ps2 in near isogenic lines (NILs), RILs still segregating for the region of interest (so-called heterogeneous inbred families (HIFs)) were identified. Two sets of HIFs, segregating for the PRP-Ps1 locus surrounding marker MSAT2-38, and two sets segregating for the PRP-Ps2 locus surrounding marker MSAT5-14, were evaluated for partial resistance to Pst (Figure 4A). Each of the chosen RILs displays a different genetic background, a mix of both parental genomes Bay-0 and Shahdara.
Figure 4.
Validation of the major QTLs for partial resistance to Pseudomonas syringae pv. tomato DC3000 with heterogeneous inbred families (HIFs). A HIF 102 segregates around MSAT2-38 and HIF 200 segregates for a region of chromosome II around markers MSAT2-5 and MSAT2-38. HIF 214 segregates for a region of chromosome V around MSAT5-14 and NGA139 and HIF 400 segregates around MSAT5-14, NGA139 and MSAT5-22. Regions for which the HIFs segregate are indicated in hatched boxes and the white regions of the chromosomes represent a mix of both parental genomes Bay-0 and Shahdara. B and C In planta bacterial growth in HIFs fixed for the Bay (grey bar) or Sha (black bar) allele of PRP-Ps1 (B) or PRP-Ps2 (C). Each value is the average of measurement of in planta bacterial growth in at least 8 plants (means and standard errors) from one experiment. Three independent experiments were performed and showed similar results. Asterisks show significant difference in partial resistance (P<0.05).
As shown in Figure 4B and 4C, all the HIF lines carrying the Bay allele at PRP-Ps1 or PRP-Ps2 showed significantly lower bacterial colonization than the corresponding NILs carrying the Sha alleles. In planta bacterial growth was between 5 to 10 fold lower when the Bay-0 alleles were present. These results confirmed the quantitative contribution of both loci to partial Pst resistance and validated the major QTLs, PRP-Ps1 and PRP-Ps2.
PRP-Ps2 influences SA-dependent gene expression
Genetic analyses revealed that the balance between signalling components such as ethylene (ET), jasmonates (JA) and salicylic acid (SA) is crucial to modulate and adapt the various defense mechanisms to a given pathogen [36]. While JA and ET are important positive regulators in the resistance to necrotrophic pathogens, SA has been demonstrated to play a central role in resistance to biotrophs [36], [37]. Partial resistance to Pst relies to a large part on SA dependent defense responses and is negatively regulated by the JA pathway.
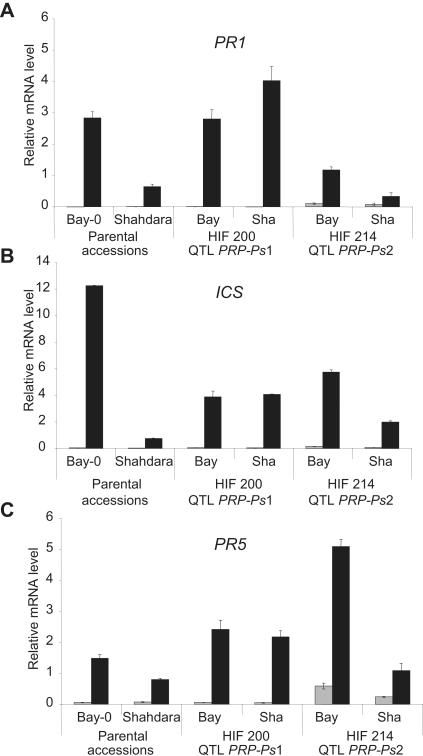
We therefore analysed to what extent PRP-Ps1 and PRP-Ps2 influenced signalling through these pathways. The expression of marker genes of SA and JA/ET signalling was investigated in the lines HIF200 and HIF214 segregating at the PRP-Ps1 or PRP-Ps2 locus, respectively. For SA-dependent signalling, the expression of the defense genes PR1 (Pathogenesis Related 1), PR5 (Pathogenesis-Related 5) and ICS (IsoChorismate Synthase) was analysed in parental ecotypes and in HIFs before and 24 h after infection with virulent Pst (Figure 5) [38], [39]. As expected, all three marker genes showed stronger pathogen-responsive expression in the more resistant Bay-0 accession than in Shadhara (Figure 5). Interestingly, they were also expressed at significantly higher levels in HIF214 plants that carry the Bay allele of PRP-Ps2, than in HIF214 plants carrying the Sha allele. Thus, the differences observed between the HIFs for in planta bacterial growth at the PRP-Ps2 locus can be correlated with those observed for expression of SA-dependent genes, suggesting an implication of PRP-Ps2 in the activation of SA dependent responses. In contrast, there was no significant difference in expression levels of SA marker genes when HIF200 plants were analysed, suggesting that the PRP-Ps1 locus does not influence SA-dependent gene expression.
Figure 5.
Expression analysis of the defense marker genes PR1 (A), ICS (B) and PR5 (C) in HIFs and parental accessions. The transcript levels were determined by Q-RT-PCR with cDNA generated from leaves before (grey) and after (black) inoculation with Pst DC3000 at 5.105 cfu/mL. The expression value of the individual genes was normalized by using the expression level of β-Tubulin4 as an internal standard. Mean mRNA levels of three plants are shown with corresponding standard errors. Similar results were obtained from two biological experiments.
For JA- and ET-dependent signal transduction pathways, the expression of PR3 (Pathogenesis-Related 3) and VSP (Vegetative Storage Protein) was analysed [38], [40]. Although these genes were induced upon inoculation, no correlation between resistance level and gene induction level could be observed when parental accessions or the HIFs were compared (data not shown).
Discussion
In natural populations there is a prevalence of low level, partial resistance that often prevents pathogens from reaching seriously damaging levels. Several studies have identified major genes controlling partial resistance in various crops [41]–[43]. However, the molecular mechanisms underlying these loci were not identified.
In the model plant Arabidopsis, while the characterization of simply inherited R genes mediating strain-specific recognition of pathogens and complete resistance has been intensively investigated, the mechanisms that control partial resistance are poorly understood. Evidence has however been obtained that the different types of resistance already described (nonhost, complete, partial) have several features in common. For example, genes necessary for R-gene mediated resistance are also involved in non-host and partial resistances in Arabidopsis. For example, eds1, pad4 and sag101 mutants, which are impaired in R-gene dependent resistance, show enhanced susceptibility to the virulent bacterium Pst and allow invasive growth of non-host powdery mildew isolates [2], [44]–[46]. In addition, whole genome gene expression analysis [47] and the study of the expression of individual genes [48], indicate similar transcriptional changes in compatible and incompatible interactions with Pst, with most differences in the defense transcriptome being due to the kinetics and the amplitude of the response. In addition, defense mechanisms involved in partial, complete and nonhost resistances, are modulated by many type III effector proteins (TTEs) delivered by bacteria during infection [49], [50]. Interestingly, the outcome of the interaction between effectors and defense mechanisms depends not only on the nature of the effector but also on the kinetics of its delivery by the bacteria [51]. In conclusion, these findings point to a complex network of regulators and regulatory mechanisms operating for the different forms of resistance in Arabidopsis. Although genetic analyses have clearly identified several R genes and some EDS genes acting in complete resistance pathways and affecting other types of resistance, to our knowledge and with the exception of the work of Kover already mentioned [21], no such a direct genetic approach has been conducted for partial resistance to the model pathogen Pst in Arabidopsis, which should reveal major actors of this resistance.
The most important finding of this study was the discovery in Arabidopsis of two major loci governing variation in partial resistance to Pst, in the Bay-0×Shahdara RIL population. In addition, the validation of these loci opens the way for their fine mapping and their cloning, which will improve our understanding of the molecular mechanisms underlying partial resistance.
PRP-Ps1 and PRP-Ps2, two major traits controlling partial resistance to Pseudomonas
Two major QTLs and two minor loci controlling partial resistance to Pst have been identified through our quantitative analysis. For the two major QTLs, the Bay-0 allele enhances the resistance to Pst. On the contrary, for the two minor QTLs, PRP-Ps3 and PRP-Ps4, the Shahdara allele enhances the resistance to Pst. Alleles from the susceptible parent that enhance disease resistance were already reported for several host-pathogen interactions [52]–[55]. This may also explain transgression towards susceptibility in the RIL population.
Highly significant correlations between experiments validate reproducibility of the phenotypic evaluations. The homogeneous behaviour of RILs also contributed to confer high heritabilities, indicating that the variations observed are mostly genetically controlled and that the estimation of partial resistance by phenotypic evaluations is reliable. However, the detected QTLs explained from 39% to 73% of the phenotypic variance, and comparison with the narrow sense heritabilities (from 0.58 to 0.77) suggests that not all the genetic variance is explained by these QTLs. This may result either i) from the choice of the significance threshold (2.7) which could have prevented the detection of minor QTLs, or ii) from the underestimation of the QTL contribution. Another explanation could be the involvement of the epistatic interactions detected in our study and which can potentially have an important contribution in genetic variation in complex traits [56].
Strong interaction with the environment is one of the hallmarks of quantitative traits and it is not uncommon that the magnitude of QTLs varies with changing environmental conditions or even, that QTLs are only expressed in a given environment and not in others [57], [58]. Environmental effects on epistatic loci are even more pronounced [59], [60]. Therefore plant resistance and susceptibility are strongly influenced by environmental conditions, and must be studied in controlled experimental setups. The fact that, in another study investigating natural variation in tolerance of Arabidopsis to Pst, different resistance levels were found for several accessions [26], may be due to such environmental effects. Although environment-genotype interactions were not evaluated in our study, we detected significant “experiment” and “block” effects (Table 1) suggesting that environmental conditions influence partial resistance to Pst. In addition, the relative contributions of the major QTLs, PRP-Ps1 and PRP-Ps2, to the overall phenotypic values showed some variation between experiment 1 and experiments 2, 3 and 4, and the detected epistatic interactions were not consistent for all experiments. However, despite the apparent effects of the environment on partial resistance, the two major QTLs, PRP-Ps1 and PRP-Ps2, were detected in four independent experiments whatever the environmental conditions, and could be confirmed in NILs using the powerful HIF strategy [29]. The fact that the effects of PRP-Ps1 and PRP-Ps2 can be detected in segregating populations opens the way for their fine-mapping and their molecular cloning. Knowing the molecular identity of PRP-Ps1 and PRP-Ps2 will not only give new, fundamental insights into partial resistance, but also allow to study on a molecular level, the plasticity of partial resistance.
Candidate loci
Two major QTLs, and two minor loci accounting for about one half of the phenotypic variation have been identified through our quantitative analysis, suggesting that a few major mechanisms are controlling partial resistance to Pst. This is surprisingly similar to complete resistance in which the genetics of the plant-pathogen interaction is under the control of one or a few major genes. Consequently, it would be interesting to investigate whether putative disease resistance proteins, or other candidate genes related to different types of resistance (as described earlier), are found at the genomic location of the QTLs identified in this study. However, since no pathogen resistance QTL has been cloned as such and only a few genes acting as resistance QTL in defined interactions have been identified, the molecular nature of such genes remains elusive.
Partial resistance QTLs frequently co-localize with known R gene loci and it is assumed that many QTLs correspond to weak or defeated R-genes with, in most cases, the canonical NB-LRR domains[61]. Since resistance genes are highly polymorphic within species and populations [62], such QTLs may correspond to allelic variants of qualitative resistance genes with intermediate phenotypes [63]. For example, the rice Xa21D gene confers partial resistance to bacterial blight, while other Xa21 alleles confer complete resistance [64]. In barley, clustering of major resistance gene and resistance QTL has also been reported [65]. PRP-Ps2, PRP-Ps3 and PRP-Ps4 co-localize with regions containing hypothetical NB-LRR resistance genes (http://niblrrs.ucdavis.edu/At-RGenes) [66], making them potential candidate genes. Thus, within the confidence interval defined for the QTL PRP-Ps4 on chromosome V, is located a cluster of resistance genes, and among them the R gene RPS4. In addition, PRP-Ps2 and PRP-Ps3 co-localize with a resistance QTL to powdery mildew, RPW11 [18], and with a QTL detected for tolerance to Pst associated with symptom severity [21], respectively.
In addition to a role in pathogen perception, partial resistance QTLs may also operate in signal transduction. Multiple signalling elements of resistance have been identified by genetic means and a central role of SA production and signal transduction is well established [67]. However, none of the identified genes nor eds, co-localize with the PRP-Ps QTLs.
Partial resistance QTLs might also correspond to genes involved in constitutive defense responses. The cell wall is generally believed to be an efficient physical barrier against microbial attack. Alterations in cell wall structure could therefore result in altered partial resistance. QTL analysis employing the Bay-Sha population has detected multiple loci influencing cell wall structure [68] and some of them co-localize to PRP-Ps QTLs. PRP-Ps1 co-localizes with the ARH-1 QTL for the Ara-Rha ratio reflecting difference in RGI (rhamnogalacturonan I) structure and PRP-Ps3 and PRP-Ps4 co-localize with two minor QTLs, HLD-3 and HLD-4 (HLD for dark-grown hypocotyl length) for hypocotyl length reflecting cell elongation.
In summary, a number of genes putatively associated with the plant defense system, co-localize with some of the PRP-Ps loci. However, the confidence intervals found for these QTLs involve rather large genomic regions, which need to be reduced using HIFs to define candidate genes more precisely.
Gene expression/pathways controlled by PRP-Ps1 and PRP-Ps2
The central role of SA-dependent defense responses in resistance to Pst is well established [36]. Mutants affected in SA production or signal transduction show reduced resistance to Pst [39]. The finding that the resistance level in HIFs differing in the PRP-Ps2 locus, correlates with the expression of marker genes of the SA response is therefore particularly interesting. It indicates that the PRP-Ps2 locus might be involved in the activation of inducible defence responses associated with partial resistance, but it raises the question at what level PRP-Ps2 is acting; in SA production, in SA signalling or up-stream of SA? Quantification of SA levels and responsiveness of HIFs to SA application should provide answers to this question. PRP-Ps1, in contrast, seems to act independently of SA. At least SA pathway marker genes show no differential expression in an HIF segregating for this locus. Therefore, PRP-Ps1 may act in constitutively expressed defense responses or in SA-independent pathways necessary for complete resistance. Such pathways have been revealed by mutant and transcriptome analysis [38], [69] and their elements are beginning to be identified [70].
In this context, an interesting recent study aimed at identifying loci controlling transcriptome variation by global eQTL analysis [71]. Categorizing eQTLs has the potential to enable reverse genetics for the identification of genes controlling quantitative traits and may also help to enhance the rate of QTL cloning. Interestingly in this analysis, several eQTLs detected on chromosomes II, III and V are related to networks involved in plant-pathogen responses, suggesting that they may influence variation in plant-pathogen interactions in the Bay-0×Sha population. By mapping QTLs for partial Pst resistance in the same RIL population that has been used for global gene expression analysis, the relationship between partial resistance to Pst and genetic architecture of eQTLs could be evaluated. More generally, combining QTL mapping with whole genome expression analysis to identify transcripts corresponding to QTLs will give a more complete picture of the complex genetic architecture of quantitative traits [57].
In the future, molecular cloning of the QTLs identified in our study will certainly help to understand the molecular basis of quantitative variation of partial resistance to Pseudomonas syringae in Arabidopsis, a model pathosystem in the plant pathology field.
Methods
Plant material and growth conditions
The Bay-0×Shahdara RIL population and the heterogeneous inbred families (HIFs) derived from the RIL population were developed by Loudet et al. [34] and unpublished. A complete description of the RIL population is available at http://dbsgap.versailles.inra.fr/vnat/Fichier_collection/Rech_rils_pop.php. HIFs were obtained as previously described [72]. The reciprocal F1 hybrids derived from the cross between Bay-0 and Shahdara (F1 BS) and between Shahdara and Bay-0 (F1 SB) were generated in our laboratory. Plants were grown in a growth chamber at 22°C, with a 9h light period and a light intensity of 190 µmol/m2/s. All experiments were performed with 4 to 5 week-old plants.
Bacterial strains, plant inoculation procedure and bacterial growth measurement
Pseudomonas syringae pv. tomato (Pst) DC3000 strain was grown at 29°C on KingB's medium supplemented with 50 µg/mL of rifampicin [73]. For the determination of in planta bacterial growth, we used an inoculum of 4.104 cfu/mL. For gene expression analysis, we used an inoculum of 5.105 cfu/mL. Plant inoculations and in planta bacterial growth analysis were performed essentially as described previously with two to four plants per RIL and per experiment [74]. In order to analyze in planta bacterial densities in hundreds of plants in parallel, leaf discs were harvested in 96 deep-well microtiter plates. Bacteria were extracted by addition of 0.2% Silwet L-77 and shaking for 30 minutes. Subsequent serial dilutions and depositions on culture medium were performed by using microtiter plates and multichannel pipettes.
Experimental design for Bay-0xShahdara RIL population and HIFs
The RIL population was evaluated by performing 4 experiments under different conditions. Three experiments were conducted with 165 RILs of the population under greenhouse conditions and another one with 370 RILs under growth chamber conditions. For the three experiments performed in greenhouse, each RIL was evaluated in a complete randomized block design. Two plants of each RIL per block with two blocks in each experiment were evaluated for partial resistance to Pst. For the other experiment in growth chamber, complete randomized design of RILs with two plants for each RIL was used.
HIFs were also evaluated for partial resistance to Pst by in planta bacterial growth analysis. At least, 32 plants of each HIF were tested in two to three independent experiments.
In the case of the evaluations of the F1 hybrids, 16 plants were tested for each cross.
Statistical analyses
Data was analyzed for each block and each experiment. Adjusted means of disease scores (lsmeans) of RILs in blocks were estimated from variance analysis (ANOVA). Phenotypic correlations among the blocks, the experiments, and the variables were calculated. When replicates were available for an experiment, broad sense heritabilities (h2) were estimated from the mean square (MS) of ANOVA using the formula adapted from Gallais [75]:
where σ2 g is the genetic variance (MSg-MSgr)/rn, σ2 gr the genotype*block interaction (MSgr-MSe)/n, σ2 e the environmental variance (MSe), n the number of plants and r the number of replicates.
Data analyses were performed with Statistical Analysis System (SAS) software (SAS Institute Inc., North Carolina, USA). Variance analysis of in planta bacterial growth data was performed using PROC GLM of SAS with randomized effects.
QTL detection
The QTL was detected on the lsmeans of in planta bacterial growth data for each experiment, and on the means of the in planta bacterial growth data of each block (when the block effect was highly significant, P<0.01). Variance analysis (LR), Interval mapping (IM) and composite interval mapping (CIM) were performed with the QTL Cartographer software Version 1.17 (Basten et al, North Carolina State University, USA) for each trait. Interval mapping (IM) and composite interval mapping (CIM) were also performed with PLABQTL Version 1.2 (Utz and Melchinger, University of Hohenheim, Germany). After performing 1000 permutations with ANOVA, a LOD threshold of 2.7 was used to declare a putative QTL significant. For CIM, 2 to 4 of the most informative markers per trait were chosen as cofactors. For each trait, a multiway ANOVA was performed with molecular markers near the QTL peaks to estimate the total percentage of phenotypic variation (R2) explained by the significant QTLs. QTLs were named PRP-Ps for Partial Resistance to Pathogen - Pst. Confidence intervals of the detected QTLs were estimated from the PLABQTL software.
In addition to additive effects, digenic epistasis was tested with a two-factor ANOVA model with an interaction between pairs of markers. With the PROC GLM of SAS software, 703 interaction tests were performed and a significance level of P<0.001 (0.7 false positive) was chosen for detecting digenic epistasis. Global R2 was estimated with full ANOVA including all additive effects and digenic epistatic effects.
RNA extraction and Q-RT-PCR analysis
Material for RNA analysis was ground in liquid nitrogen and total RNA was isolated using the Macherey-Nagel Nucleospin RNA plant kit (Macherey-Nagel, Hoerdt, France) according to the manufacturer's recommendations. Reverse transcription was performed with 1 µg of total RNA using the superscript reverse transcriptase II (Invitrogen, Carlsbad, CA, USA). Quantitative PCR was run on a Lightcycler system (Roche Diagnostics, Meylan, France) according to the manufacturer's recommendations with the following conditions: 1 cycle: 9 min at 95°C for; 45 cycles: 5 sec at 95°C, 10 sec at 65°C and 20 sec at 72°C. β-Tubulin4 was used as an internal standard. The primer sets used are for PR1 (locus At2g14610, forward primer: GGAGCTACGCAGAACAACTAAGA, reverse primer: CCCACGAGGATCATAGTTGCAACTGA), for PR5 (locus At1g75040, forward primer: CGGTACAAGTGAAGGTGCTCGTT, reverse primer: GCCTCGTAGATG GTTACAATGTCA), and for ICS (At1g74710, forward primer: GCCGTCTCTGAAC TCAAATCTCAA, reverse primer: GTTACGAGCAAGAACAACCTTGTT). Specificity of the amplifications was verified by melting curve analysis. Efficiency of the amplification was verified by the analysis of standard curves.
Supporting Information
Analysis of the F1 progeny of reciprocal crosses between Bay-0 and Shahdara for partial resistance to Pseudomonas syringae pv. tomato DC3000. In planta bacterial growth was assessed three days post inoculation in the F1 generation of reciprocal crosses between Bay-0 and Shahdara. Each value is the average of in planta bacterial growth of at least 4 plants (means and standard errors).
(0.62 MB EPS)
Phenotypic correlations among experiments
(0.04 MB DOC)
Acknowledgments
We would like to thank Vera Saliba-Colombani and Fabien Nogué for providing some of the HIF lines. We are also grateful to Olivier Bouchez, Noëllie Journot and Raimana Louette for technical help, to Yves Marco and Clare Gough for critical reviewing of the manuscript, and to Claudine Balagué for helpful discussions.
Footnotes
Funding: This work has been financially supported by a GABI-Genoplante-MCyt contract on Plant Genome Research.
References
- 1.Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 2.Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 3.Stein M, Dittgen J, Sanchez-Rodriguez C, Hou BH, Molina A, et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurnberger T, Brunner F, Kemmerling B, Piater L. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol Rev. 2004;198:249–266. doi: 10.1111/j.0105-2896.2004.0119.x. [DOI] [PubMed] [Google Scholar]
- 5.Nurnberger T, Brunner F. Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr Opin Plant Biol. 2002;5:318–324. doi: 10.1016/s1369-5266(02)00265-0. [DOI] [PubMed] [Google Scholar]
- 6.Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 1999;18:265–276. doi: 10.1046/j.1365-313x.1999.00265.x. [DOI] [PubMed] [Google Scholar]
- 7.Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415:977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 8.Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, et al. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- 9.Li X, Lin H, Zhang W, Zou Y, Zhang J, et al. Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc Natl Acad Sci U S A. 2005;102:12990–12995. doi: 10.1073/pnas.0502425102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 11.Keen NT. Gene-for-gene complementarity in plant-pathogen interactions. Annual Review of Genetic. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 12.Kamoun S, Huitema E, Vleeshouwers VGAA. Resistance to oomycetes: a general role for the hypersensitive response? Trends in Plant Science. 1999;4:196–200. doi: 10.1016/s1360-1385(99)01404-1. [DOI] [PubMed] [Google Scholar]
- 13.Hulbert SH, Webb CA, Smith SM, Sun Q. RESISTANCE GENE COMPLEXES: Evolution and Utilization. Annual Review of Phytopathology. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- 14.Inukai T, Vales MI, Hori K, Sato K, Hayes PM. RMo1 confers blast resistance in barley and is located within the complex of resistance genes containing Mla, a powdery mildew resistance gene. Mol Plant Microbe Interact. 2006;19:1034–1041. doi: 10.1094/MPMI-19-1034. [DOI] [PubMed] [Google Scholar]
- 15.Li ZK, Arif M, Zhong DB, Fu BY, Xu JL, et al. Complex genetic networks underlying the defensive system of rice (Oryza sativa L.) to Xanthomonas oryzae pv. oryzae. PNAS. 2006;103:7994–7999. doi: 10.1073/pnas.0507492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alonso-Blanco C, Koornneef M. Naturally occuring variation in Arabidopsis: an underexploited resource for plant genetics. Trends in Plant Science. 2000;5:22–29. doi: 10.1016/s1360-1385(99)01510-1. [DOI] [PubMed] [Google Scholar]
- 17.Koornneef M, Alonso-Blanco C, Vreugdenhil D. Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Biol. 2004;55:141–172. doi: 10.1146/annurev.arplant.55.031903.141605. [DOI] [PubMed] [Google Scholar]
- 18.Wilson IW, Schiff CL, Hughes DE, Somerville SC. Quantitative Trait Loci Analysis of Powdery Mildew Disease Resistance in the Arabidopsis thaliana Accession Kashmir-1. Genetics. 2001;158:1301–1309. doi: 10.1093/genetics/158.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godiard L, Sauviac L, Torii KU, Grenon O, Mangin B, et al. ERECTA, an LRR receptor-like kinase protein controlling development pleiotropically affects resistance to bacterial wilt. Plant J. 2003;36:353–365. doi: 10.1046/j.1365-313x.2003.01877.x. [DOI] [PubMed] [Google Scholar]
- 20.Denby KJ, Kumar P, Kliebenstein DJ. Identification of Botrytis cinerea susceptibility loci in Arabidopsis thaliana. Plant J. 2004;38:473–486. doi: 10.1111/j.0960-7412.2004.02059.x. [DOI] [PubMed] [Google Scholar]
- 21.Kover PX, Wolf JB, Kunkel BN, Cheverud JM. Genetic architecture of Arabidopsis thaliana response to infection by Pseudomonas syringae. Heredity. 2005;94:507–517. doi: 10.1038/sj.hdy.6800651. [DOI] [PubMed] [Google Scholar]
- 22.Llorente F, Alonso-Blanco C, Sanchez-Rodriguez C, Jorda L, Molina A. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 2005;43:165–180. doi: 10.1111/j.1365-313X.2005.02440.x. [DOI] [PubMed] [Google Scholar]
- 23.Quirino BF, Bent AF. Deciphering host resistance and pathogen virulence: the Arabidopsis/Pseudomonas interaction as a model. Molecular Plant Pathology. 2003;4:517–530. doi: 10.1046/j.1364-3703.2003.00198.x. [DOI] [PubMed] [Google Scholar]
- 24.Nomura K, Melotto M, He SY. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr Opin Plant Biol. 2005;8:361–368. doi: 10.1016/j.pbi.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 25.Katagiri F, Thilmony R, He SY. The Arabidopsis thaliana-Pseudomonas syringae interaction. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. pp. 1–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kover PX, Schaal BA. Genetic variation for disease resistance and tolerance among Arabidopsis thaliana accessions. Proc Natl Acad Sci U S A. 2002;99:11270–11274. doi: 10.1073/pnas.102288999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parlevliet JE. Components of resistance that reduce the rate of epidemic development. Annual Review of Phytopathology. 1979;17:203–222. [Google Scholar]
- 28.Buell CR, Sommerville SC. Use of Arabidopsis recombinant inbred lines reveals a monogenic and a novel digenic resistance mechanism to Xanthomonas campestris pv campestris. Plant J. 1997;12:21–29. doi: 10.1046/j.1365-313x.1997.12010021.x. [DOI] [PubMed] [Google Scholar]
- 29.Tuinstra MR, Ejeta G, Goldsbrough PB. Heterogeneous inbred family (HIF) analysis: a method for developping near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet. 1997;95:1005–1011. [Google Scholar]
- 30.McKhann HI, Camilleri C, Berard A, Bataillon T, David JL, et al. Nested core collections maximizing genetic diversity in Arabidopsis thaliana. Plant J. 2004;38:193–202. doi: 10.1111/j.1365-313X.2004.02034.x. [DOI] [PubMed] [Google Scholar]
- 31.Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C, et al. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 1998;14:259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- 32.Alonso-Blanco C, Koornneef M, Stam P. The use of recombinant inbred lines (RILs) for genetic mapping. Methods Mol Biol. 1998;82:137–146. doi: 10.1385/0-89603-391-0:137. [DOI] [PubMed] [Google Scholar]
- 33.Alonso-Blanco C, El-Assal SE, Coupland G, Koornneef M. Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics. 1998;149:749–764. doi: 10.1093/genetics/149.2.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loudet O, Chaillou S, Camilleri C, Bouchez D, Daniel-Vedele F. Bay-0×Shahdara recombinant inbred line population: a powerful tool for the genetic dissection of complex traits in Arabidopsis. Theor Appl Genet. 2002;104:1173–1184. doi: 10.1007/s00122-001-0825-9. [DOI] [PubMed] [Google Scholar]
- 35.Zeng ZB. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kunkel BN, Brooks DM. Cross talk between signaling pathways in pathogen defense. Curr Opin Plant Biol. 2002;5:325–331. doi: 10.1016/s1369-5266(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 37.Thomma BP, Penninckx IA, Broekaert WF, Cammue BP. The complexity of disease signaling in Arabidopsis. Curr Opin Immunol. 2001;13:63–68. doi: 10.1016/s0952-7915(00)00183-7. [DOI] [PubMed] [Google Scholar]
- 38.Glazebrook J, Chen W, Estes B, Chang HS, Nawrath C, et al. Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 2003;34:217–228. doi: 10.1046/j.1365-313x.2003.01717.x. [DOI] [PubMed] [Google Scholar]
- 39.Nawrath C, Métraux J-P. Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. The Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gu Y-Q, Wildermuth MC, Chakravarthy S, Loh Y-T, Yang C, et al. Tomato Transcription Factors Pti4, Pti5, and Pti6 Activate Defense Responses When Expressed in Arabidopsis. Plant Cell. 2002;14:817–831. doi: 10.1105/tpc.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young ND. QTL mapping and quantitative disease resistance in plants. Annual Review of Phytopathology. 1996;34:479–501. doi: 10.1146/annurev.phyto.34.1.479. [DOI] [PubMed] [Google Scholar]
- 42.Bai Y, Huang CC, van der Hulst R, Meijer-Dekens F, Bonnema G, et al. QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorum G1.1601 co-localize with two qualitative powdery mildew resistance genes. Mol Plant Microbe Interact. 2003;16:169–176. doi: 10.1094/MPMI.2003.16.2.169. [DOI] [PubMed] [Google Scholar]
- 43.Nguyen TTT, Koizumi S, La TN, Zenbayashi KS, Ashizawa T, et al. Pi35(t), a new gene conferring partial resistance to leaf blast in the rice cultivar Hokkai 188. Theor Appl Genet. 2006 doi: 10.1007/s00122-006-0337-8. in press. [DOI] [PubMed] [Google Scholar]
- 44.Jirage D, Tootle TL, Reuber TL, Frost LN, Feys BJ, et al. Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salycilic acid signaling. Proceedings of The National Academy of Sciences of USA. 1999;96:13583–13588. doi: 10.1073/pnas.96.23.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Falk A, Feys BJ, Frost LN, Jones JDG, Daniels MJ, et al. EDS1, an essential component of R gene-mediated disease resistance in Arabidopsis has homology to eukaryotic lipases. Proceedings of The National Academy of Sciences of USA. 1999;96:3292–3297. doi: 10.1073/pnas.96.6.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feys BJ, Wiermer M, Bhat RA, Moisan LJ, Medina-Escobar N, et al. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell. 2005;17:2601–2613. doi: 10.1105/tpc.105.033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, et al. Quantitative Nature of Arabidopsis Responses during Compatible and Incompatible Interactions with the Bacterial Pathogen Pseudomonas syringae. Plant Cell. 2003;15:317–330. doi: 10.1105/tpc.007591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamb CJ, Ryals JA, Ward ER, Dixon RA. Emerging strategies for enhancing crop resistance to microbial pathogens. Biotechnology (N Y) 1992;10:1436–1445. doi: 10.1038/nbt1192-1436. [DOI] [PubMed] [Google Scholar]
- 49.Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 51.de Torres M, Mansfield JW, Grabov N, Brown IR, Ammouneh H, et al. Pseudomonas syringae effector AvrPtoB suppresses basal defence in Arabidopsis. The Plant Journal. 2006;47:368–382. doi: 10.1111/j.1365-313X.2006.02798.x. [DOI] [PubMed] [Google Scholar]
- 52.Foulongne M, Pascal T, Pfeiffer F, Kervella J. QTLs for powdery mildew resistance in peach×Prunus davidiana crosses: consistency across generations and environments. Molecular Breeding. 2003;12:33–50. [Google Scholar]
- 53.Arahana VS, Graef GL, Specht JE, Steadman JR, Eskridge KM. Identification of QTLs for resistance to Sclerotinia sclerotiorum in soybean. Crop Science. 2001;41:180–188. [Google Scholar]
- 54.Al-Chaarani GR, Roustaee A, Gentzbittel L, Mokrani L, Barrault G, et al. A QTL analysis of sunflower partial resistance to downy mildew (Plasmopara halstedii) and black stem (Phoma macdonaldii) by the use of recombinant inbred lines (RILs). Theoretical and Applied Genetics. 2002;104:490–496. doi: 10.1007/s001220100742. [DOI] [PubMed] [Google Scholar]
- 55.Perchepied L, Dogimont C, Pitrat M. Strain-specific and recessive QTLs involved in the control of partial resistance to Fusarium oxysporum f. sp. melonis race 1.2 in a recombinant inbred line population of melon. Theor Appl Genet. 2005;111:65–74. doi: 10.1007/s00122-005-1991-y. [DOI] [PubMed] [Google Scholar]
- 56.Kroymann J, Donnerhacke S, Schnabelrauch D, Mitchell-Olds T. Evolutionary dynamics of an Arabidopsis insect resistance quantitative trait locus. Proc Natl Acad Sci U S A. 2003;100(Suppl 2):14587–14592. doi: 10.1073/pnas.1734046100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mackay TF. The genetic architecture of quantitative traits: lessons from Drosophila. Curr Opin Genet Dev. 2004;14:253–257. doi: 10.1016/j.gde.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 58.Mitchell-Olds T, Schmitt J. Genetic mechanisms and evolutionary significance of natural variation in Arabidopsis. Nature. 2006;441:947–952. doi: 10.1038/nature04878. [DOI] [PubMed] [Google Scholar]
- 59.Liao CY, Wu P, Hu B, Yi KK. Effects of genetic background and environment on QTLs and epistasis for rice (Oryza sativa L.) panicle number. Theoretical and Applied Genetics. 2001;103:104–111. [Google Scholar]
- 60.Mauricio R. Ontogenetics of QTL: the genetic architecture of trichome density over time in Arabidopsis thaliana. Genetica. 2005;123:75–85. doi: 10.1007/s10709-002-2714-9. [DOI] [PubMed] [Google Scholar]
- 61.Gebhardt C, Valkonen JPT. Organization of genes controlling disease resistance in the potato genome. Annual Review of Phytopathology. 2001;39:79–102. doi: 10.1146/annurev.phyto.39.1.79. [DOI] [PubMed] [Google Scholar]
- 62.Jones JD. Putting knowledge of plant disease resistance genes to work. Curr Opin Plant Biol. 2001;4:281–287. doi: 10.1016/s1369-5266(00)00174-6. [DOI] [PubMed] [Google Scholar]
- 63.Robertson A. Understanding the relationship between qualitative and quantitative genetics. In: Helentjaris T, Burr B, editors. Development and application of molecular markers to problems in plant genetics. New York, USA: Cold Spring Harbour Lab. Press; 1989. pp. 81–88. [Google Scholar]
- 64.Wang GL, Ruan DL, Song WY, Sideris S, Chen L, et al. Xa21D encodes a receptor-like molecule with a leucine-rich repeat domain that determines race-specific recognition and is subject to adaptive evolution. Plant Cell. 1998;10:765–779. doi: 10.1105/tpc.10.5.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Williams KJ. The molecular genetics of disease resistance in barley. Aust J Agric Res. 2003;54:1065–1079. [Google Scholar]
- 66.Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hammond-Kosack KE, Parker JE. Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotechnol. 2003:177–193. doi: 10.1016/s0958-1669(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 68.Mouille G, Witucka-Wall H, Bruyant MP, Loudet O, Pelletier S, et al. Quantitative trait Loci analysis of primary cell wall composition in Arabidopsis. Plant Physiol. 2006;141:1035–1044. doi: 10.1104/pp.106.079384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Glazebrook J, Rogers EE, Ausubel FM. Use of Arabidopsis for genetic dissection of plant defense responses. Annual Review of Genetic. 1997;31:547–569. doi: 10.1146/annurev.genet.31.1.547. [DOI] [PubMed] [Google Scholar]
- 70.Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kliebenstein DJ, West MA, van Leeuwen H, Loudet O, Doerge RW, et al. Identification of QTLs controlling gene expression networks defined a priori. BMC Bioinformatics. 2006;7:308. doi: 10.1186/1471-2105-7-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Loudet O, Gaudon V, Trubuil A, Daniel-Vedele F. Quantitative trait loci controlling root growth and architecture in Arabidopsis thaliana confirmed by heterogeneous inbred family. Theor Appl Genet. 2005;110:742–753. doi: 10.1007/s00122-004-1900-9. [DOI] [PubMed] [Google Scholar]
- 73.Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. the plant Cell. 1991;3:49–59. doi: 10.1105/tpc.3.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lorrain S, Lin B, Auriac MC, Kroj T, Saindrenan P, et al. VASCULAR ASSOCIATED DEATH1, a novel GRAM domain-containing protein, is a regulator of cell death and defense responses in vascular tissues. The Plant Cell. 2004;16:2217–2232. doi: 10.1105/tpc.104.022038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gallais A. In: Théorie de la sélection en amélioration des plantes. Gallais A, Bannerot H, editors. Paris, France: Masson; 1990. p. 588. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of the F1 progeny of reciprocal crosses between Bay-0 and Shahdara for partial resistance to Pseudomonas syringae pv. tomato DC3000. In planta bacterial growth was assessed three days post inoculation in the F1 generation of reciprocal crosses between Bay-0 and Shahdara. Each value is the average of in planta bacterial growth of at least 4 plants (means and standard errors).
(0.62 MB EPS)
Phenotypic correlations among experiments
(0.04 MB DOC)