Abstract
Background
Aeromonas sp. can now be considered relatively common enteropathogens due to the increase of diseases in humans. Aeromonas culicicola is a gram negative rod-shaped bacterium isolated for the first time from the mosquito mid-gut, but subsequently detected in other insects and waters also. Our previous study discovered that A. culicicola harbors three plasmids, which we designated as pAc3249A, pAc3249B and pAc3249C. We investigated and report here the existence and genetic organization of a Conjugal Type IV Secretion System (TFSS) in pAc3249A.
Methodology/Principle Finding
The complete operon is 11,061 bp in length and has G+C content of 47.20% code for 12 ORFs. The gene order and orientation were similar to those found in other bacteria with some differences. We have designated this system as AcTra for Aeromonas culicicola transfer system. BLAST results of ORFs and phylogenetic analysis showed significant similarity towards the respective proteins of the IncI2 plasmid R721 of E. coli. Other bioinformatics studies have been performed to predict conserved motifs/domains, signal peptides, transmembrane helices, etc. of the ORFs.
Conclusions/Significance
BLAST results of ORFs and phylogenetic analysis showed significant similarity towards the respective proteins of the IncI2 plasmid R721 of E. coli.
Introduction
Aeromonas are Gram-negative rod shaped facultative anaerobic bacteria of the family Aeromonadaceae. They are widely distributed in variety of habitats ranging from fresh water to salt water and found in virtually all foods. The past decade has witnessed an explosion of scientific interest in members of the genus Aeromonas and this interest has gone beyond fish pathogenicity. They are now considered as emerging human pathogens suspected to cause gastroenteritis ranging from mild enteritis to cholera like diarrhea [1], [2]. A. hydrophila, A. caviae and A. veronii represents more than 85% of clinical isolates [3], [4].
Several secretion machineries present in Gram-negative bacteria mediate the transport and injection of toxic molecules into target cells. These secretion systems are classified into five types I to V depending on similarities, differences and substrate specificities. These machineries share a common requirement for proteins that utilize ATP as an energy source to drive transport of macromolecules. TFSSs have long been recognized as the systems ancestrally related to bacterial conjugation machines and responsible for the exchange of genetic material [5]. By facilitating conjugative transfer, type IV secretion machineries play crucial roles in the spread of antibiotic resistance genes among bacteria. The bacterial TFSS mediate the transport of macromolecules across the cell envelope of Gram-negative and Gram-positive bacteria [6], [7]. Several important human and plant pathogens have evolved type IV secretion machineries involved in delivering virulence factors (proteins or protein-DNA complexes) to host target cells. The TFSS of Agrobacterium tumefaciens is prototype and is proved to be involved in crown gall disease [8]. Other pathogens are Bordetella pertussis, the agent responsible for whooping cough in children; Helicobacter pylori, responsible for gastric ulcers and stomach cancer; Brucella suis, the causative agent of brucellois and Legionella pneumoniae, the causative agent of Legionnaires' disease were shown to adopt TFSS for virulence [9]–[13]. The phylogenetic and functional relationships evident between type IV and certain conjugal transfer systems has led to the suggestion that these groups form a type IV superfamily of proteins involved in both the conjugal transfer between bacteria and the transit of virulence factors between bacteria and their eukaryotic host [14]. There was only one report of conjugal transfer system in Aeromonas before our study and that was found in a plasmid pFBAOT6, originally isolated from a strain of A. caviae from hospital effluent [15].
In this study, we characterized the sequence of conjugal TFSS from the plasmid pAc3249A of A. culicicola MTCC 3249. This strain harbors three plasmids of different sizes which we designated pAc3249A, pAc3249B and pAc3249C. pAc3249A is circular and the largest, approximately 30 kb in size, found to code for this conjugal transfer system. pAc3249B and pAc3249C are circular, approximately 8.5 kb and 3 kb in size respectively.
Materials and Methods
Bacterial strains and plasmids
Aeromonas species were maintained on Luria Bertani (LB) medium at 30°C. E. coli strains were maintained on LB at 37°C. When required, media were supplemented with ampicillin (100 µg/ml), and tetracyclin (25 µg/ml).
DNA Manipulations
Plasmids were isolated using a Qiagen midi-prep plasmid isolation kit (Qiagen), followed by gel extraction after electrophoresis of the entire plasmid isolation eluate. Restriction endonuclease digestions, ligation, agarose gel electrophoresis were carried out as described in Maniatis et al. [16].The plasmid pAc3249A was digested with BamHI, Sau3AI, HindIII, AluI (New England Biolabs) and ligated into pLitmus29 (New England Biolabs) prepared by respective restriction enzymes digestion and dephosphorylation with Shrimp Alkaline Phosphatase (Roche). Ligated product was transformed into JM 109 competent cells (Invitrogen).
DNA Sequencing and Sequence Assembly
DNA sequencing was carried out on an Applied Biosystems 3730 DNA Analyzer with an ABI PRISM BigDye Terminator cycle sequencing kit (Apllied Biosystems). The clones were initially sequenced with M13 forward and reverse vector specific primers. The sequences were analyzed and assembled using in-house assembly pipeline, which is integration of phred, cross match and phrap. Gaps between contigs were filled by primer walking.
Web Servers and Homology Predictions
Putative coding sequences (CDSs) were identified using glimmer [17]. Functional annotation was done by searching putative protein coding sequences against non-redundant protein database obtained from NCBI using BLASTP [18], against Pfam [19] using hmmer and ProDom [20] database. To identify the functional motifs, each CDS was searched against Prosite [21] database. Secondary structure, disulfide bridges, globularity, and non-standard secondary structure about each putative protein was obtained by using PredictProtein [22] software. The sequence homologs were obtained by PSI-BLAST. Multiple sequence alignment of homologous sequences was carried out using ClustalW [23]. Bootstrap analysis was carried out using SEQBOOT to generate 100 random combinations of the alignments in Phylip. Phylogenetic trees (cladogram) were constructed using parsimony method of Phylip and trees were visualized using TREEVIEW 16. TMHMM server version 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0/) and DAS servers (http://www.sbc.su.se/~miklos/DAS/) were used to perform TMS prediction of all the genes, whereas signal peptide prediction was carried out using SignalP3 (http://www.cbs.dtu.dk/services/SignalP/) and LipoP1 (http://www.cbs.dtu.dk/services/LipoP/). Cello version 2.5 (http://cello.life.nctu.edu.tw/) and PSORT (http://www.psort.org/psortb/) were used to predict cellular localization. The DNA sequence of AcTra is available under the GenBank accession number DQ890522.
Results
Comparison of the AcTra system with other type IV secretion systems
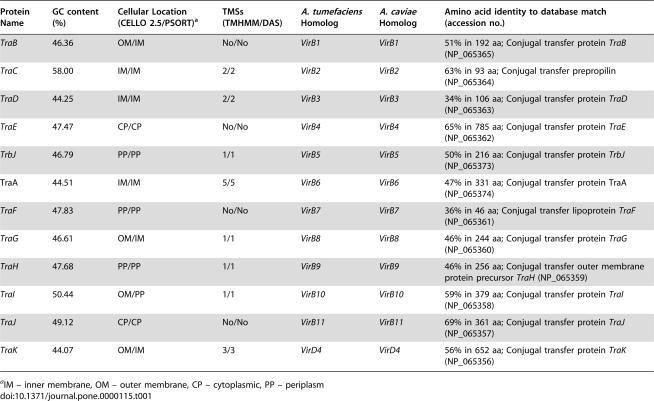
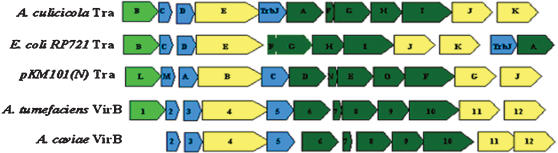
The sequence analysis showed that A. culicicola TFSS operon was 11,061 bp in length, with average G+C content of 47.26% and code for 12 ORFs. We named these ORFs as TraB, TraC, TraD, TraE, TrbJ, TraA, TraF, TraG, TraH, TraI, TraJ, and TraK respectively. G+C content of TraC is exceptionally high and it is 58%. The AcTra gene cluster contains 12 open reading frames (ORFs), homologous to the conjugal transfer system of IncI2 plasmid R721 (Table 1). The arrangement of the genes in A. culicicola is different in some respect from those observed in E. coli. In case of A. culicicola, all 12 proteins were present in continuous stretch whereas in E. coli, TraK through TraB are in sequential manner followed by ~4.5 kb loci coding for some other proteins not associated with TFSS and again followed by TrbJ and TraA. Arrangement of TFSS genes in A. culicicola is very compact with seven overlapping genes. Maximum intergene distance was 39 bases between gene TraC and TraD. Gene arrangement of A. culicicola TFSS and its comparison with other homolog is shown in Fig. 1 whereas the protein analysis details are given in Table 1. The AcTra system resembles other type IV secretion systems including the Tra system of plasmids RP721 and pKM101 of E. coli, VirB-D4 systems of A. tumefaciens, B. henselae, A. caviae and the Ptl system of B. pertussis.
Table 1. Properties of A. culicicola AcTra operon ORFs and their deduced products.
| Protein Name | GC content (%) | Cellular Location (CELLO 2.5/PSORT)a | TMSs (TMHMM/DAS) | A. tumefaciens Homolog | A. caviae Homolog | Amino acid identity to database match (accession no.) |
| TraB | 46.36 | OM/IM | No/No | VirB1 | VirB1 | 51% in 192 aa; Conjugal transfer protein TraB (NP_065365) |
| TraC | 58.00 | IM/IM | 2/2 | VirB2 | VirB2 | 63% in 93 aa; Conjugal transfer prepropilin (NP_065364) |
| TraD | 44.25 | IM/IM | 2/2 | VirB3 | VirB3 | 34% in 106 aa; Conjugal transfer protein TraD (NP_065363) |
| TraE | 47.47 | CP/CP | No/No | VirB4 | VirB4 | 65% in 785 aa; Conjugal transfer protein TraE (NP_065362) |
| TrbJ | 46.79 | PP/PP | 1/1 | VirB5 | VirB5 | 50% in 216 aa: Conjugal transfer protein TrbJ (NP_065373) |
| TraA | 44.51 | IM/IM | 5/5 | VirB6 | VirB6 | 47% in 331 aa; Conjugal transfer protein TraA (NP_065374) |
| TraF | 47.83 | PP/PP | No/No | VirB7 | VirB7 | 36% in 46 aa; Conjugal transfer lipoprotein TraF (NP_065361) |
| TraG | 46.61 | OM/IM | 1/1 | VirB8 | VirB8 | 46% in 244 aa; Conjugal transfer protein TraG (NP_065360) |
| TraH | 47.68 | PP/PP | 1/1 | VirB9 | VirB9 | 46% in 256 aa; Conjugal transfer outer membrane protein precursor TraH (NP_065359) |
| TraI | 50.44 | OM/PP | 1/1 | VirB10 | VirB10 | 59% in 379 aa; Conjugal transfer protein TraI (NP_065358) |
| TraJ | 49.12 | CP/CP | No/No | VirB11 | VirB11 | 69% in 361 aa; Conjugal transfer protein TraJ (NP_065357) |
| TraK | 44.07 | OM/IM | 3/3 | VirD4 | VirD4 | 56% in 652 aa; Conjugal transfer protein TraK (NP_065356) |
IM – inner membrane, OM – outer membrane, CP – cytoplasmic, PP – periplasm
Figure 1.
Comparison of AcuTra operon with conjugal and other type IV secretion system homolog. Different colors are indicative of putative role or location of the protein. Light Green – acetyl transglycosylase; Blue – components of pilus assembly; Yellow – NTPases; Dark Green – proteins forming core components.
Gene Assembly in A. culicicola
The proteins of the type IV secretion machinery can be grouped according to their function and/or their cellular location [5], [9].
Pilus Assembly Components (TraC, TraD and TrbJ)
The TraC is considered as major component of the pilus [14], [24] whereas TrbJ protein is known as a minor component of the pilus structure [25] and essential for TFSS virulence. We observed fully conserved L (position 109), Y (position 113), and Q (position 123) in TrbJ residues might essential for structural and/or functional aspect of TrbJ. Periplasmic form of TrbJ is required for translocation of a DNA substrate to the cell surface [26]. The TraD has not firmly assigned but its cellular localization suggests that it is a minor component of pilus assembly [27]. TraD is a short polypeptide and the multiple alignments for TraD homolog revealed a well conserved (NS)-R-P-A-(LM)-X2-(GN)-(IV)-P motif, which is slightly different from previous report [14]. It also possesses fully conserved D and L at positions 73 and 77 respectively.
Proteins forming core components (TraA, TraF-TraI)
The TraA protein is highly hydrophobic with five predicted TMSs and a large central predicted periplasmic loop whose secondary structure is important for DNA substrate translocation [28]. The TraA homologs are relatively poorly conserved with no fully conserved residues. TraF is a small lipoprotein, has a signal peptide and no predicted TMS. TraF interacts with TraH and stabilizes several Tra subunits [29], [30]. TraG is an inner membrane protein with an N-terminal TMS. The TraH subunit is hydrophilic and possesses three functional domains also reported in other species [31]. The N-terminal periplasmic domain of TraH is highly conserved which is required for channel activity and pilus biogenesis [31] whereas; C-terminal plays an important role in interaction with TraF [30]. The TraI is situated in periplasm and possess one TMS. Hydrophobic C-terminal region of TraI is conserved and also possesses coiled-coil structure, which is important for interaction of TraI with bitopic TraG [32] and TraF-TraH dimer [33]. N-terminal region of TraI is hydrophobic and is poorly conserved whereas C-termini exhibits short sequences of alternating hydrophilic and hydrophobic residues forming putative β-sheet structure. Within this C-terminal region are four fully conserved and additional nine well-conserved glycyl residues as reported before [14]. These observations would have some structural and functional implications.
NTPases (TraE, TraJ and TraK)
These protein exhibits highest sequence conservation among TFSS components. The TraE subunit of A. culicicola does not have any predicted TMS but possesses Walker A and Walker B nucleotide binding domains (P-loop) and is localized in cytoplasm. Sequence analysis of TraJ protein reveals the presence of highly conserved hydrophobic domains and typical nucleotide binding domains important for ATPase activity. ATPase activity lies in the hexameric form of TraJ, which is stimulated by lipids [34]. The TraK also possesses conserved motifs and nucleotide binding domains. TraK is situated in the inner membrane and there are three N-terminal TMSs. The ATPase activity of TraE, TraJ and TraK energize TFSS function [9], [24].
The TraB is dispensable for TFSS function [35], [36] and not required for transfer of TFSS substrates. TraB protein analysis predicted conserved transglycosylase domain [36], [37]. It is basically situated in inner membrane or sometimes on outer membrane also and contributes to channel assembly. Three conserved motifs- a) (VI)-X7-(VIL)-E-S, b) (LIF)-X2-C-X-(SN)-(LI) and c) S-X-Y, have been observed, previous one at N-terminal region and latter two at relatively central portion of the protein.
Phylogenetic Relationship
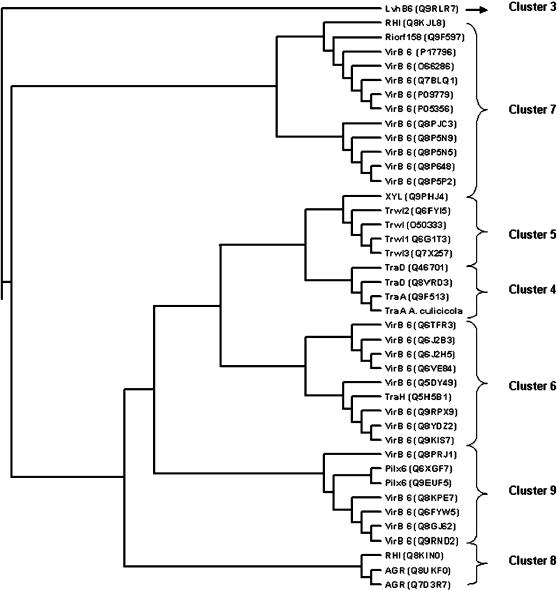
Phylogenetic analysis suggests that the TFSSs have evolved from a common ancestral system with virtually no shuffling of constituents even between sequence-divergent systems. We carried out phylogenetic analysis of A. culicicola TFSS proteins with homologs identified by PSI-BLAST. Trees were generated using phylip with hundred iterations. Topology of all the trees (data not shown) is more or less same and it has been observed that A. culicicola grouped with E. coli and Haemophilus influenzae in cluster 4 proposed by Cao and Saier (Fig. 2) [14].
Figure 2.
Phylogenetic tree for the TraA, drawn using parsimony method of Phylip. Text in parenthesis indicates Accession number of respective gene.
Discussion
A. culicicola harbors three uncharacterized plasmids. Their role and importance in A. culicicola has not determined yet. We investigated conjugal transfer system in one among those plasmids namely pAc3249A. This is the first report of complete analysis of conjugal type IV secretion system in A. culicicola. It would be interesting to find the genes present on this plasmid as it has conjugation machinery for self transmission. Our system has shown highest homology to E. coli rather than A. caviae which indicated that A. culicicola might have acquired this plasmid through lateral or horizontal transfer from E. coli in mosquito mid-gut, its site of isolation. Complete sequencing and characterization of pAc3249A would reveal the role of conjugal transfer system and eventually the role of pAc3249A in A. culicicola.
Acknowledgments
We are kindly thankful to CSIR (Council of Scientific and Industrial Research), India for providing research fellowship to Dayananda KM.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors received no financial support.
References
- 1.Figueras MJ, Guarro J, Martinez-Murcia A. Clinically relevant Aeromonas species. Clin Infect Dis. 2000;30:988–989. doi: 10.1086/313839. [DOI] [PubMed] [Google Scholar]
- 2.Janda JM. Aeromonas and Plesiomonas. In: Sussman M, editor. Molecular medical microbiology. San Diego, Calif: Academic Press; 2001. pp. 1237–1270. [Google Scholar]
- 3.Janda JM, Abbott SL. Evolving concepts regarding the genus Aeromonas: an expanding panorama of species, disease presentations, and unanswered questions. Clin Infect Dis. 1998;27:332–344. doi: 10.1086/514652. [DOI] [PubMed] [Google Scholar]
- 4.Joseph SW, Carnahan AM. Update on the genus Aeromonas. ASM News. 2000;66:218–233. [Google Scholar]
- 5.Christie PJ. Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol Microbiol. 2001;40:294–305. doi: 10.1046/j.1365-2958.2001.02302.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cascales E, Christie PJ. The versatile bacterial type IV secretion systems. Nat Rev Microbiol. 2003;1:137–49. doi: 10.1038/nrmicro753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grohmann E, Muth G, Espinosa M. Conjugative plasmid transfer in Gram-positive bacteria. Microb Mol Biol Rev. 2003;67:277–301. doi: 10.1128/MMBR.67.2.277-301.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zupan J, Muth TR, Draper O, Zambryski PC. The transfer of DNA from Agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J. 2000;23:11–28. doi: 10.1046/j.1365-313x.2000.00808.x. [DOI] [PubMed] [Google Scholar]
- 9.Baron C, O'Callaghan D, Lanka E. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol Microbiol. 2002;43:1359–1366. doi: 10.1046/j.1365-2958.2002.02816.x. [DOI] [PubMed] [Google Scholar]
- 10.Burns DL. Type IV transporters of pathogenic bacteria. Curr Opin Microbiol. 2003;6:29–34. doi: 10.1016/s1369-5274(02)00006-1. [DOI] [PubMed] [Google Scholar]
- 11.Covacci A, Telford JL, Giudice GD, Parsonnet J, Rappuoli R. Helicobacter pylori virulence and genetic geography. Science. 1999;284:1328–1333. doi: 10.1126/science.284.5418.1328. [DOI] [PubMed] [Google Scholar]
- 12.Boschiroli ML, Ouahrani-Bettache S, Foulongne V, Michaux-Charachon S, Bourg G, et al. Type IV secretion and Brucella virulence. Vet Microbiol. 2002;90:341–348. doi: 10.1016/s0378-1135(02)00219-5. [DOI] [PubMed] [Google Scholar]
- 13.Roy CR, Tilney LG. The road less traveled: transport of Legionella to the endoplasmic reticulum. J Cell Biol. 2002;158:415–419. doi: 10.1083/jcb.200205011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao TB, Saier MH. Conjugal type IV macromolecular transfer systems of Gram-negative bacteria: organismal distribution, structural constraints and evolutionary conclusions. Microbiology. 2001;147:3201–3214. doi: 10.1099/00221287-147-12-3201. [DOI] [PubMed] [Google Scholar]
- 15.Rhodes G, Parkhill J, Bird C, Ambrose K, Jones MC, et al. Complete Nucleotide Sequence of the Conjugative Tetracycline Resistance Plasmid pFBAOT6, a Member of a Group of IncU Plasmids with Global Ubiquity. Appl Environ Microbiol. 2004;70:7497–7510. doi: 10.1128/AEM.70.12.7497-7510.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritz EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring. Harbor, NY: Cold Spring Harbor laboratory; 1984. [Google Scholar]
- 17.Salzberg S, Delcher A, Kasif S, White O. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 1998;26:544–548. doi: 10.1093/nar/26.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic Local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, et al. The Pfam Protein Families Database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonnhammer ELL, Kahn D. Modular arrangement of proteins as inferred from analysis of homology. Protein Science. 1994;3:482–492. doi: 10.1002/pro.5560030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hulo N, Bairoch A, Bulliard V, Cerutti L, De Castro E, et al. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rost B, Yachdav G, Liu J. The PredictProtein Server. Nucleic Acids Res 32(Web Server issue) 2003:W321–W326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuo-Bin L. ClustalW-MPI: ClustalW Analysis Using Distributed and Parallel Computing. Bioinformatics. 2003;19:1585–1586. doi: 10.1093/bioinformatics/btg192. [DOI] [PubMed] [Google Scholar]
- 24.Christie PJ, Vogel JP. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt-Eisenlohr H, Domke N, Baron C. TraC of IncN plasmid pKM101 associates with membranes and extracellular high-molecular-weight structures in Escherichia coli. J Bacteriol. 1999;181:5563–71. doi: 10.1128/jb.181.18.5563-5571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cascales E, Christie PJ. Definition of a bacterial type IV secretion pathway for a DNA substrate. Science. 2004;304:1170–73. doi: 10.1126/science.1095211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shamaei-Tousi A, Cahill R, Frankel G. Interaction between Protein subunits of the Type IV Secretion System of Bartonella henselae. J Bacteriol. 2004;186:4796–4801. doi: 10.1128/JB.186.14.4796-4801.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ. Agrobacterium tumefaciens TraA domains direct the ordered export of a DNA substrate through a type IV secretion system. J Mol Biol. 2004;341:961–77. doi: 10.1016/j.jmb.2004.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baron C, Thorstenson YR, Zambryski PC. The lipoprotein VirB7 interacts with VirB9 in the membranes of Agrobacterium tumefaciens. J Bacteriol. 1997;179:1211–18. doi: 10.1128/jb.179.4.1211-1218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spudich GM, Fernandez D, Zhou XR, Christie PJ. Intermolecular disulde bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc Natl Acad Sci USA. 1996;93:7512–17. doi: 10.1073/pnas.93.15.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jakubowski S, Cascales E, Krishnamoorthy V, Christie PJ. Agrobacterium tumefaciens VirB9, an outer-membrane-associated component of a type IV secretion system, regulates substrate selection and T-pilus biogenesis. J Bacteriol. 2005;187:3486–95. doi: 10.1128/JB.187.10.3486-3495.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Das A, Xie YH. The Agrobacterium T-DNA transport pore proteins VirB8, VirB9, and VirB10 interact with one another. J Bacteriol. 2000;182:758–63. doi: 10.1128/jb.182.3.758-763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beaupre CE, Bohne J, Dale EM, Binns AN. Interactions between VirB9 and VirB10 membrane proteins involved in movement of DNA from Agrobacterium tumefaciens into plant cells. J Bacteriol. 1997;179:78–89. doi: 10.1128/jb.179.1.78-89.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krause S, Pansegrau W, Lurz R, de la Cruz F, Lanka E. Enzymology of type IV macromolecule secretion systems: the conjugative transfer regions of plasmids RP4 and R388 and the cag pathogenicity island of Helicobacter pylori encode structurally and functionally related nucleoside triphosphate hydrolases. J Bacteriol. 2000;182:2761–2770. doi: 10.1128/jb.182.10.2761-2770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayer M, Iberer R, Bischof K, Rassi E, Stabentheiner E. Functional and mutational analysis of p19, a DNA transfer protein with muramidase activity. J Bacteriol. 2001;183:3176–83. doi: 10.1128/JB.183.10.3176-3183.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger BR, Christie PJ. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: VirB2 through VirB10 are essential virulence genes. J Bacteriol. 1994;176:3646–60. doi: 10.1128/jb.176.12.3646-3660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koraimann G. Lytic transglycosylases in macromolecular transport systems of Gram-negative bacteria. Cell Mol Life Sci. 2003;60:2371–88. doi: 10.1007/s00018-003-3056-1. [DOI] [PMC free article] [PubMed] [Google Scholar]