Abstract
Haploid yeast invades solid agar in response to nutrient limitation. To decipher the cues that underlie invasion, we have developed a single cell invasive growth assay. Using this assay, as well as the traditional plate-washing assay, we show that invasive growth occurs in response to glucose depletion. In the absence of glucose (or other fermentable sugar), individual cells adopted a nonaxial budding pattern and elongated morphology within the first cell divisions, and invasion into the agar was observed in microcolonies containing as few as 10 cells. In support of this observation, we found that glucose suppressed the hyperinvasive growth morphology of STE11-4, pbs2, hsl7, and RAS2V19 mutations. In addition, removal of glucose from YPD medium caused constitutive invasion in wild-type cells. We tested glucose control proteins for a role in invasion and found that Snf1, a protein required for derepression of glucose-repressed genes, was required for invasive growth. The transcription factor Sip4, which interacts with Snf1 and is induced during the diauxic shift, had an inhibitory role on invasive growth, suggesting that multiple mechanisms are required for glucose depletion-dependent invasion.
Several yeast/fungal species have the ability to adopt two growth forms, a vegetative (yeast) form and a pseudohyphal or filamentous form. In pathogenic organisms such as Candida albicans, this morphological transition is important for invasion of host tissue and therefore pathogenesis (1). In nonpathogenic organisms, the transition can occur upon nutrient limitation and has therefore been suggested to be a mechanism to permit foraging for nutrients. Insight into the signal transduction mechanisms that are necessary for the morphological change will contribute to an understanding of basic biological phenomena and of pathogenesis.
The budding yeast Saccharomyces cerevisiae provides a genetic system to investigate the morphological transition from vegetative to filamentous development. In diploid yeast, loss of environmental fixed nitrogen causes the transition from vegetative growth, in which cells are round and bud in a bipolar fashion, to pseudohyphal growth, in which cells are elongated and bud in a unipolar budding pattern (2). In haploid yeast, nutrient limitation causes a similar developmental switch that allows cells to penetrate the surface of an agar medium in a process called invasive growth (see refs. 3–6 for reviews). For simplicity, we will use the term filamentous form to refer to both pseudohyphal and invasive growth, although there are differences between these two growth responses.
At least two signal transduction pathways are required for filamentous growth. One pathway uses a number of components from the pheromone response pathway, notably the p21-activated kinase (PAK) kinase Ste20, the kinases Ste11 and Ste7 from the first two tiers of the mitogen-activated protein (MAP) kinase cascade, and the transcription factor Ste12 (7, 8), but has unique components as well—the MAP kinase Kss1 (9–11) and a second transcription factor Tec1 (12, 13). The second pathway includes the recently identified Gpr1, a presumed receptor of the G protein-coupled receptor (GPCR) class, and its associated Gα subunit, Gpa2 (14, 15). Gpr1 is believed to sense fermentable sugars and is coupled to cAMP-dependent protein kinase A (16). Ras2 also influences signaling through this pathway (17): loss of RAS2 prevents invasion, and hyperactive RAS2V19 causes hyperfilamentation. Few targets of these signal transduction pathways are known, but one is the FLO11 gene, which is required for filamentous growth in haploids and diploids (14, 18, 19). In contrast to the components just enumerated, which are required for filamentous growth, several proteins that appear to function as negative regulators of filamentation have been described. Loss of Hsl1 or Hsl7 leads to hyperfilamentation (20). These proteins interact with the septin ring and inhibit the cell cycle regulator Swe1 (21, 22). In addition, Hsl7 may interact directly with Ste20 to prevent filamentous growth (23).
The signals that trigger filamentous growth are ill-defined. Here, we show by using the standard plate-washing assay that removal of glucose from YPD medium caused constitutive invasive growth in haploid yeast. To determine more precisely the role of glucose in the invasive growth response, we developed a single cell invasive growth assay. Direct observation of cells in the presence or absence of glucose revealed a striking transition from vegetative growth to the filamentous morphology, which was apparent in the first cell divisions. We used this assay to show that invasive growth is caused specifically by loss of fermentable sugars such as glucose. We also found that the filamentous morphology of a number of hyperinvasive growth mutants was completely suppressed by glucose. Investigation of glucose control proteins in invasion revealed a requirement for the global regulatory protein Snf1 and an inhibitory function of the transcription factor Sip4 in invasion.
Materials and Methods
Strains, Plasmids, and Microbiological Techniques.
Yeast strains are listed in Table 1. All strains were isogenic with HYL333 and 334 of the Σ1278b background (provided by G. Fink, Whitehead Institute for Biomedical Research, Cambridge, MA). Gene disruptions were performed by digestion of one-step replacement plasmids containing elm1∷URA3, kss1∷URA3, hsl1∷URA3, hsl7∷URA3, snf1∷URA3, ste11∷URA3, ste12∷URA3, or ste20∷URA3, or by PCR-derived disruption fragments. The ste12∷URA3, ste11∷URA3, ste20∷URA3, kss1∷URA3, and STE11-4 integration constructs were reported previously (24) as were pRS315 and 316 plasmids (25). Gene disruptions were confirmed by PCR Southern analysis and by phenotype. Plasmids conferring hyperinvasive growth were analyzed by sequence analysis (26). Yeast and bacterial strains were propagated by using standard methods (27). Yeast extract/peptone/dextrose (YPD) and synthetic complete dextrose (SCD) media have been described (28), and were made at the same time as other media for consistency. Low nitrogen synthetic low ammonia histidine dextrose (SLAHD) medium was made as described (2). Yeast transformations were performed as described (29). Bacterial transformations, bacterial DNA preparations, and plasmid constructions were performed by standard methods (30). HYL333 and 334 are parents of congenic strains used for other studies of invasive growth (G. Fink, personal communication). The congenic strains had the same characteristics as the parent strain (e.g., Snf1-dependent invasion, and invasion in response to glucose depletion) but had less elongated cell morphology. We have not explored this difference. Fermentable sugar-dependent changes in bud pattern and cell elongation were also observed in Sc252JHa, W303, and 246-1-1 strain backgrounds (not shown).
Table 1.
Yeast strains
| Strain | Relevant genotype | Reference |
|---|---|---|
| HYL333 | MATa ura3-52 | G. Fink |
| HYL334 | MATα ura3-52 | G. Fink |
| 1 | HYL333 leu2 | A. Goehring |
| 2 | HYL333/HYL334 | This study |
| 3 | HYL333 ste12 | This study |
| 4 | HYL333 ste20 | This study |
| 5 | HYL333 ste4 | This study |
| 6 | HYL333 ste5 | This study |
| 7 | HYL333 STE11-4 | This study |
| 8 | HYL333 RAS2V19 | This study |
| 9 | HYL333 hsl1 | This study |
| 10 | HYL333 hsl7 | This study |
| 11 | HYL333 grr1 | This study |
| 12 | HYL333 snf1 | This study |
| 13 | HYL333 snf1 hsl7 | This study |
| 14 | HYL333 snf1 hsl1 | This study |
| 15 | HYL333/334 snf1/snf1 | This study |
| 16 | HYL333 snf1 STE11-4 | This study |
| 17 | HYL333 snf1 pbs2 | This study |
| 18 | HYL333 snf1 grr1 | This study |
| 19 | HYL333 snf1 sip4 | This study |
| 20 | HYL333 snf4 | This study |
| 21 | HYL333 mig1 | This study |
| 22 | HYL333 hap4 | This study |
| 23 | HYL333 sip1 | This study |
| 24 | HYL333 sip4 | This study |
| 25 | HYL333 sok2 | This study |
Single Cell Invasive Growth Assay.
Synthetic complete medium lacking glucose (SC medium) was used to examine growth habit. Cells were capable of more than 20 cell divisions on this medium, presumably because of poor carbon sources that contaminate the agar. Cells were grown to stationary phase in SCD liquid medium, washed twice with water, and spread onto freshly poured SCD or SC medium at a concentration of 104 cells/plate. Plates were incubated at 25°C for 24 h and the occurrence of yeast form (yf) and filamentous form (ff) microcolonies was assessed by microscopic examination. A microcolony was designated yf if every cell of that microcolony was round, and if no unipolar buds were observed, and ff if more than 50% of the cells were elongated and if more than 50% of the cells were budding in a unipolar manner. Microcolonies that did not fit into either of the above categories were rare (less than 10%) and were not considered part of the total percentage. The fraction of cells exhibiting unipolar budding and elongated morphology was determined by examining at least 200 cells from microcolonies at the 20-cell (or less) stage. Plasmids were maintained on SC medium by selection for the appropriate auxotrophic marker, which had no effect on filament formation.
Plate-Washing Assay.
The plate-washing assay was performed as described (8). Care was taken not to break the plane of the agar. Invasion was allowed to proceed for 3 days at 30°C, and plates were washed with water and rubbed vigorously with a wet finger to remove cells that did not invade the agar. Invasion into SC medium was monitored by plate washing, except that S plates were incubated for 24 h at 25°C, washed as above, and examined by microscopy.
Microscopy.
Standard microbiological techniques were used (31). For experiments that required observation by the ×40 or ×100 objectives, cells were scraped using 4 ml of distilled water, concentrated by centrifugation, resuspended in 20 μl of water, and visualized by microscopy. For bud scar staining, cells were collected as above, resuspended in 1 μg/ml Calcofluor (Sigma) for 10 min, washed twice in water, and visualized by fluorescence microscopy.
Results
Glucose Depletion Causes Constitutive Invasion.
Previous reports showed that nutrient limitation causes haploid invasive growth in yeast (8), so we investigated various nutrients and found that removal of glucose from YPD medium (YP medium) caused constitutive invasion of haploid yeast by the plate-washing assay (Fig. 1). Invasion was observed within the first 16 h of growth on YP, compared with more than 48 h for YPD. Microscopic examination revealed filamentous structures on YP within the first cell divisions (not shown). Addition of glucose or other fermentable sugars (fructose or mannose) to YP medium prevented the hyperinvasive growth (not shown). Removal of other components from rich medium, such as yeast extract (PD) or peptone (YD) did not cause hyperinvasive growth of yeast in the presence of glucose (not shown).
Figure 1.
Removal of glucose causes constitutive invasion. Equal concentrations of cells were spotted onto YPD (plus glucose) or YP medium (minus glucose), incubated for 16 h and 48 h, and photographed. Growth of cells on YP presumably occurred using poor carbon sources present in the yeast extract or agar.
A Single Cell Invasive Growth Assay.
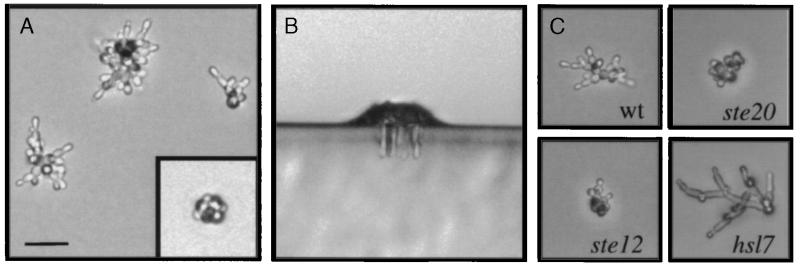
We observed microcolonies on synthetic medium lacking glucose (SC medium, Fig. 2A). These microcolonies were composed of elongated cells that had a unipolar budding pattern and had invaded the agar, as revealed by plate washing and bisection of the agar followed by microscopic examination (Fig. 2B). Invasion into the agar was detected in microcolonies containing as few as 10 cells. The ff morphology was identical to that previously reported (8), as well as the morphology of cells that invaded YPD medium (not shown). In contrast, microcolonies on synthetic medium containing glucose (SCD) contained yf cells, round cells that budded axially, none of which invaded the agar (Fig. 2A Inset). Glucose spotted at the center of an SC plate allowed for observation of both cell types on the same plate: yf microcolonies in the region of diffused glucose, and ff microcolonies outside the region of diffused glucose. A distinct transition marked the boundary between yf and ff microcolonies; a single microcolony displaying both characteristics was rarely observed.
Figure 2.
Single cell invasive growth assay. (A) Cells of the Σ1278b background were spread onto SC medium and incubated for 24 h. Microcolonies were visualized by light microscopy at ×200. (Inset) Typical example of a microcolony formed in the presence of glucose at the same magnification. (B) Agar invasion on medium lacking glucose. Wild-type cells were spread onto SC medium and incubated for 24 h at 25°C. Plates were cut with a razor blade, and sections were set perpendicular to the plane of invasion for direct visualization by light microscopy. (C) Invasive growth mutants have phenotypes on medium lacking glucose. Equal concentrations of cells were spread onto medium lacking glucose. Mutant genotypes are as follows: Upper Left, wild type; Upper Right, ste20; Lower Left, ste12; and Lower Right, hsl7. A, B, and C are at the same scale, with the scale bar in A representing 40 μm.
Mutants defective for functions known to be required for invasive growth were examined by the single cell assay to confirm the legitimacy of the assay. Both ste20 and ste12 mutants were defective in the formation of elongated cells and in unipolar budding (Fig. 2C). The defect in unipolar budding was more severe for ste20 mutants than for ste12 mutants by the single cell assay (Fig. 2C), consistent with results obtained by the plate-washing assay (not shown and 7, 8). Loss of STE genes, such as STE4 and STE5, which are not required for invasion, also did not influence the morphological transition seen by the single cell assay (not shown). We examined hyperinvasive growth mutants and found that disruption of HSL7 resulted in microcolonies containing hyperelongated cells by the single cell assay (Fig. 2C). Thus, mutations that diminish and mutations that enhance invasive growth had parallel consequences on cellular phenotypes as revealed by the single cell assay.
We examined the effect of different stimuli on yf and ff growth. Spotting lower concentrations of glucose resulted in proportionately smaller halos of yf microcolonies, demonstrating a direct relationship between glucose concentration and yeast form growth (Table 2). Fermentable sugars (sucrose, mannose, and fructose) caused yf growth, although the halo of yf rescue was smaller than for glucose. Nonfermentable carbon sources (such as glycogen and glycerol) were incapable of conferring yf growth (Table 2). Medium lacking fixed nitrogen, amino acids, or nucleotides did not cause ff growth, and addition of SDS or salt did not induce ff growth in the presence of glucose (Table 2). Finally, spotting of either yeast extract or peptone did not prevent the ff morphology, whereas YPD medium restored yf growth to the same degree as glucose alone (Table 2).
Table 2.
Effects of stimuli on the filamentous morphology
| Stimulus added* | % yf† | % ff‡ | yf radius§ |
|---|---|---|---|
| Glu (1 M) | >95 | <5 | 20 |
| Glu (0.25 M) | >95 | <5 | 15 |
| Glu (0.06 M) | >95 | <5 | 4 |
| Sucrose (1 M) | >95 | <5 | 12 |
| Mannose (1 M) | >95 | <5 | 11 |
| Glycogen (100%) | <5 | >95 | 0 |
| Glycerol (100%) | <5 | >95 | 0 |
| Glu (1 M) + 0.1% SDS | >95 | <5 | 20 |
| Glu (1 M) + 1 M NaCl¶ | >95 | <5 | 20 |
| YPD | >95 | <5 | 20 |
| YP | <5 | >95 | 0 |
| Peptone (10%) | <5 | >95 | 0 |
| Yeast extract (10%) | <5 | >95 | 0 |
| Stimulus removed‖ | |||
| Nitrogen | 90 | 10 | NA |
| Amino acids | >95 | <5 | NA |
| Uracil | >95 | <5 | NA |
Stimulus (10 μl) was spotted onto discs and placed on SC medium; Glu, glucose.
% yf denotes round cells and axial budding for >200 microcolonies adjacent to the disc.
% ff denotes elongated cells and unipolar budding for >200 microcolonies adjacent to the disc.
Distance (in mm) from the center of a disc to the point at which only ff colonies were observed.
Some unipolar budding (<20%) was observed.
SCD medium lacking the noted ingredient; for nitrogen, SLAHD medium was prepared as described in Materials and Methods; NA, not applicable.
Suppression of Hyperinvasive Growth Mutant Phenotypes by Glucose.
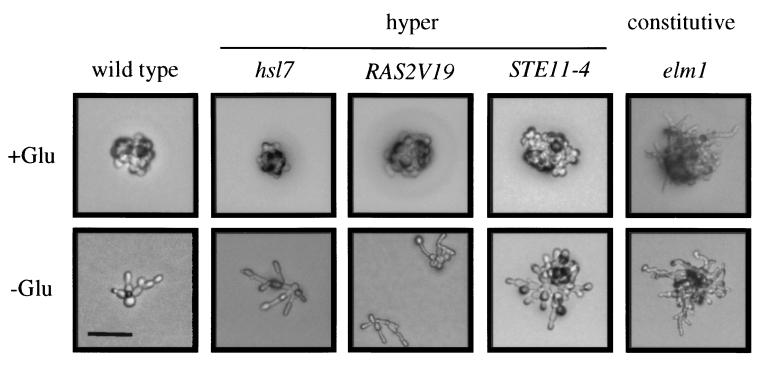
We examined a number of hyperinvasive growth mutants by the single cell assay to determine the effect of glucose on the morphology of these mutants. We found complete suppression of the morphological phenotypes of hsl1 and hsl7 by glucose (Fig. 3). This suppression is consistent with the observation that the elongated cell morphology of hsl1 and hsl7 mutants was observed only in stationary phase but not during exponential growth (21). Hyperactivation of the Ras pathway by integration of the RAS2V19 allele (32) or by deletion of BCY1 (33) caused an abnormal cell morphology, including hyperelongated cells, a phenotype that was completely suppressed by glucose (Fig. 3). The ff morphology of the STE11-4 and sok2 mutants (24, 34) was also suppressed by glucose (Fig. 3). In contrast, elm1 and grr1 mutants (35, 36) had an irregular morphology composed of hyperelongated cells and bent cells in the presence or absence of glucose (Fig. 3). Thus, a distinction can be made between constitutive (grr1 and elm1) and hyperinvasive (hsl1, hsl7, bcy1, RAS2V19, sok2 and STE11-4) mutants based on behavior in glucose. In support of this distinction, invasion of constitutive mutants was observed within the first cell divisions on YPD (not shown).
Figure 3.
Suppression of hyperinvasive growth mutant phenotypes by glucose. Equal concentrations of cells were grown for 16 h on SCD or SC medium. A 5-μl spot of 1 M glucose was placed onto a filter at the center of SC medium to allow observation of both yf and ff cell types. The RAS2V19 allele was compared on YP and YPD medium because the invasive growth phenotype was more apparent on rich medium. All pictures were at the same scale, with the bar in the lower left-hand corner representing 40 μm.
Cell Elongation and Unipolar Budding upon Glucose Starvation.
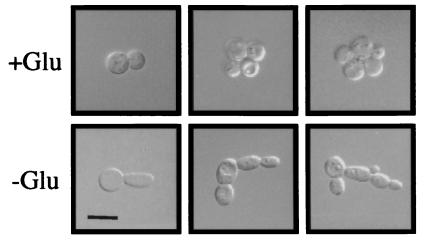
We used the single cell assay to examine the first cell divisions with or without glucose. Individual cells were placed onto SC or SCD medium (by micromanipulation) and examined for characteristics of yf or ff in subsequent cell divisions. The first bud produced on SC medium was 1.5 times as long (and thinner) as on SCD medium (Fig. 4). In fact, more than 90% of all daughters produced on SC medium were elongated. Elongated cells were observed infrequently (<1%) on SCD medium. Budding pattern of cells grown with or without glucose was compared by direct visualization of microcolonies and by bud scar staining and counting. The progenitor cell sometimes lost the axial budding pattern on SC medium (Fig. 4); 25% of new buds formed either at the opposite pole or at a site randomly chosen from the first bud (Table 3). More strikingly, the bud pattern of daughter cells was unipolar on SC medium (greater than 85% of buds counted; Table 3). The second and third buds of the daughter were adjacent to the first distal bud greater than 95% of the time. Both the progenitor cell and its daughter cells budded in an exclusively axial pattern on SCD (≈98%). Spotting glucose onto a plate containing ff microcolonies caused new daughters to exhibit a spherical morphology and axial budding pattern within the first cell division (not shown).
Figure 4.
Cells grown with or without glucose have strikingly different morphologies in the first cell divisions. Cells were grown on SCD or SC medium, scraped off the plates, centrifuged, and photographed. Typical examples of microcolonies were shown. All pictures were at the same scale, with the bar in the Lower Left corner representing 10 μm.
Table 3.
Bud site selection is influenced by glucose
| Cell | Bud | Bud pattern, %*
|
||
|---|---|---|---|---|
| Axial | Random | Opposite | ||
| M† | 2nd, 3rd, or 4th | 75 | 5 | 20 |
| D1 | 1st | 15 | 0 | 85 |
| D1 | 2nd | 98 | 0 | 2 |
| D2 | 1st | 13 | 0 | 87 |
| D2 | 2nd | 98 | 0 | 2 |
Cells were grown for 24 h on SC medium, scraped from the plates, and stained with Calcofluor as described in Materials and Methods. Cells on SCD medium were observed to have >98% axial budding.
For cells chosen, M, mother; D1, 1st daughter; D2, 2nd daughter.
Glucose Control Proteins Are Important for Proper Invasion.
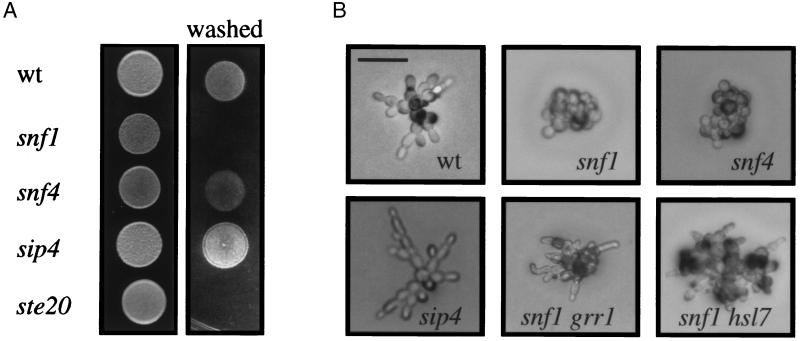
To begin to determine the genetic requirements for invasion as a response to glucose depletion, we disrupted genes known to be involved in glucose control. The Snf1 protein is required for derepression of glucose-repressed genes and for global changes in the cell upon glucose starvation (see ref. 37). We found that snf1 mutants had a severe invasive growth defect, comparable to the invasive growth defect of ste20 (Fig. 5A). In both the plate washing and single cell assays, snf1 mutants did not invade the agar, form elongated cells, or bud in a unipolar pattern. We also found that loss of Grr1 (35), a functional antagonist of Snf1, completely suppressed the invasive growth defect of snf1 mutants (Fig. 5B). Partial suppression of the snf1 invasive growth defect and yf morphology was observed by disruption of HSL7 (Fig. 5B), or by introduction of the hyperactive STE11-4 allele (not shown). The snf1 STE11-4 strain grew more slowly than strains containing either single mutation (not shown).
Figure 5.
Snf1 and Sip4 are required for proper invasion. (A) Equal concentrations of cells were spotted onto YPD medium and grown for 3 days at 30°C. The plates were photographed, washed, and photographed again. (B) Equal concentrations of cells were spread onto SC medium, incubated for 16 h at 25°C, and photographed. For B, all pictures were taken at the same scale, with the bar in the Upper Left corner representing 40 μm.
Snf1 is known to interact with Snf4 to control a subset of Snf1 functions (38), but we found that disruption of SNF4 had a less severe defect in invasion (Fig. 5A). The snf4 mutants showed a defect in ff growth by the single cell assay (Fig. 5B), suggesting that Snf4 is required for a subset of Snf1 invasive growth functions. Snf1 also associates with and activates Sip4 (39), a transcription factor required for activation of genes induced by the diauxic shift from fermentation to respiration (40). We found that loss of SIP4 had a hyperinvasive growth phenotype by the traditional and single cell assays (Fig. 5 A and B). Disruption of SIP4 in a snf1 mutant partially restored invasion; however, the sip4 snf1 cells did not invade to the same extent as wild-type or sip4 cells. Loss of HAP4, MIG1, or SIP1 had no effect on invasive growth by either plate-washing or single cell assays (not shown), suggesting that a specific subset of glucose control proteins are required for invasion.
Discussion
Glucose Depletion Is a Trigger for Haploid Invasive Growth.
We have obtained evidence that depletion of glucose (or other fermentable sugars) causes haploid invasive growth. We initially observed by the plate-washing assay that wild-type cells exhibited constitutive invasion on rich medium lacking glucose. Subsequent experiments demonstrated that, on synthetic medium lacking glucose, cells adopted an elongated morphology, a unipolar budding pattern, and invaded the agar-phenotypes that were completely suppressed by glucose. Strikingly, the elongated cell morphology of most hyperinvasive growth mutants was also suppressed by glucose. In keeping with a role for glucose in the control of invasion, we showed that the glucose control proteins Snf1 and Sip4 had dramatic, albeit opposite effects on invasive growth (see below). In contrast, the absence of fixed nitrogen, amino acids, or nucleotides did not cause invasion. The Ras pathway is required for filamentation (2) and may connect to glucose depletion by sensing the level of cAMP in the cell, which is influenced by glucose concentration (41). Indeed, the FLO11 gene, which is required for invasion and is a known target of both the RAS and STE20-dependent signal transduction pathways (19), is transcriptionally repressed in glucose rich environments (42).
Why does invasion occur better into rich than synthetic medium? We show that invasion does not occur constitutively on rich medium but begins when glucose levels become limiting for vegetative growth. Invasion may be more robust on rich medium because other nutrients (e.g., poor carbon sources, nucleotides, and amino acids) stimulate the growth of cells that are in the filamentous form. In fact, addition of yeast extract (a component of rich medium) to SC medium stimulated filamentation (unpublished results). Our observations suggest that cell density is not a factor in the transition to ff growth, nor does an excreted factor cause invasion, because a single cell can adopt ff growth.
Single Cell Invasive Growth Assay.
Previous studies of haploid invasive growth in yeast have relied exclusively on a plate-washing assay (8) to determine the extent of invasion. We have developed a single cell invasive growth assay that has the same genetic and physiological requirements as the plate-washing assay and that allows direct examination of filament formation in haploids. Several benefits of this assay in the study of filamentation in yeast are worth noting. First, the assay is penetrant in wild-type cells: greater than 95% of microcolonies exhibit ff growth in the absence of glucose, and the morphological changes in bud pattern and cell elongation are visible in virtually every cell. This penetrant phenotype is in contrast to diploid pseudohyphal growth, where only a subset of wild-type cells (5–25%) is observed to be in ff growth in nitrogen-limiting environments (see 17, 43). Second, the assay allows quantitative analysis of unipolar budding and cell elongation in haploid yeast undergoing the transition to filamentous growth. In fact, the precise lineage of budding events can be followed by time course examination of an individual cell. Finally, the ability to study filamentation in haploids will facilitate genetic analysis.
The morphological transition upon glucose depletion is rapid. First, there is a change from axial to a nonaxial budding pattern in the first cell deprived of glucose. This deviation from axial budding can be explained by loss of a transient axial mark on starvation (44). Indeed, we observed a delay in the formation of the first bud on medium lacking glucose (not shown). Second, the daughter cells adopt an exclusively unipolar budding pattern on glucose deprivation. This pattern is not what one would expect from the loss of an axial signal (J. Pringle, personal communication) and suggests that haploid yeast have a mechanism to relate glucose levels to the budding machinery, one that involves Ste20 (and to a lesser extent Ste12). Finally, an increase in the cell length is observed in the first daughter produced in a glucose-deprived environment and is apparent in all subsequent daughters. Previous reports demonstrate that a G2 delay causes cell elongation and that this occurs in ff cells (45). This may be part of the explanation here, but we also provide evidence to implicate Ste20 and Ste12 in the establishment of elongated cells.
Role of Glucose-Control Proteins in Invasion.
We found that Snf1, a protein kinase whose activity is stimulated in glucose-limiting environments, is required for invasive growth. The Snf4 protein, which binds to and regulates Snf1, is only partly required: snf4 mutants undergo invasive growth as assessed by the plate-washing assay but show only a limited response by the single cell assay. Although Snf1 and Snf4 have similar phenotypes and share overlapping functions, there is evidence for distinct functions as well (46). Loss of the glucose control protein Grr1 (35), whose mutant phenotype is known to suppress some Snf1 functions (47), completely suppressed the invasive growth defect of snf1 mutants. One target of Snf1 is Sip4 (39), a transcription factor required at the diauxic shift (40). Sip4 is not required for filamentation; rather, sip4 mutants exhibit a hyperinvasive growth phenotype. A second target of Snf1, which may be more relevant to Snf1's role in filamentous growth, is Msn1, itself required for filamentous growth (48). In support of this possibility, we find that overexpression of Msn1 can partially suppress the snf1 invasive growth defect (unpublished data).
Snf1 may have a role in the morphological transition to ff growth aside from its role in sensing glucose depletion. Snf1 is required for pseudohyphal growth in diploids (not shown). These cells form filaments in response to nitrogen limitation, a nutrient limitation that is not thought to influence Snf1 activity. Mutations that confer hyperinvasive growth, such as STE11-4, pbs2, and hsl7 partially bypass the invasive growth defect of a snf1 mutation. Because these mutations are not known to impinge on glucose sensing or metabolism, we infer that the snf1 invasive growth defect is in part unrelated to the role of Snf1 in glucose sensing. In further support of this point of view, snf1 STE11-4 mutants grow more slowly than either single mutant, implying a genetic relationship between the STE invasive growth pathway and Snf1 function. Thus, Snf1 may influence invasive growth by multiple mechanisms. At least one fungal pathogen had been shown to require Snf1 for virulence (49), giving further credence to the role of glucose control proteins in fungal morphogenesis.
Acknowledgments
We thank Gerald Fink, John Pringle, James Broach, Sarat Chandarlapaty, Mark Johnston, Michael Grunstein, Alan Meyers, Tom Stevens, April Goehring, Ira Herskowitz, and Sean O'Rourke for providing advice, strains, and/or plasmids. Thanks also to Greg Smith, Dave Mitchell, Hilary Kemp, Megan Keniry, David Rivers, Frank Stahl, and Elizabeth Monika for helpful comments and suggestions. This work was supported by research (GM-30027 for G.F.S.) and training (GM19188 for P.J.C.) grants from the U.S. Public Health Service.
Abbreviations
- ff
filamentous form
- yf
yeast form
- YPD
yeast extract/peptone/dextrose
- SCD
synthetic complete dextrose
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
See commentary on page 13461.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240345197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240345197
References
- 1.Silar P, Daboussi M J. Trends Genet. 1999;15:141–145. doi: 10.1016/s0168-9525(99)01698-4. [DOI] [PubMed] [Google Scholar]
- 2.Gimeno C J, Ljungdahl P O, Styles C A, Fink G R. Cell. 1992;68:1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 3.Madhani H D, Fink G R. Trends Cell Biol. 1998;8:348–353. doi: 10.1016/s0962-8924(98)01298-7. [DOI] [PubMed] [Google Scholar]
- 4.Banuett F. Microbiol Mol Biol Rev. 1998;62:249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin D E, Errede B. Curr Opin Cell Biol. 1995;7:197–202. doi: 10.1016/0955-0674(95)80028-x. [DOI] [PubMed] [Google Scholar]
- 6.Kron S J, Gow N A. Curr Opin Cell Biol. 1995;7:845–855. doi: 10.1016/0955-0674(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 7.Liu H, Styles C A, Fink G R. Science. 1993;262:1741–1744. doi: 10.1126/science.8259520. [DOI] [PubMed] [Google Scholar]
- 8.Roberts R L, Fink G R. Genes Dev. 1994;8:2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 9.Cook J G, Bardwell L, Thorner J. Nature (London) 1997;390:85–88. doi: 10.1038/36355. [DOI] [PubMed] [Google Scholar]
- 10.Tedford K, Kim S, Sa D, Stevens K, Tyers M. Curr Biol. 1997;7:228–238. doi: 10.1016/s0960-9822(06)00118-7. [DOI] [PubMed] [Google Scholar]
- 11.Madhani H D, Styles C A, Fink G R. Cell. 1997;91:673–684. doi: 10.1016/s0092-8674(00)80454-7. [DOI] [PubMed] [Google Scholar]
- 12.Gavrias V, Andrianopoulos A, Gimeno C J, Timberlake W E. Mol Microbiol. 1996;19:1255–1263. doi: 10.1111/j.1365-2958.1996.tb02470.x. [DOI] [PubMed] [Google Scholar]
- 13.Madhani H D, Fink G R. Science. 1997;275:1314–1317. doi: 10.1126/science.275.5304.1314. [DOI] [PubMed] [Google Scholar]
- 14.Tamaki H, Miwa T, Shinozaki M, Saito M, Yun C W, Yamamoto K, Kumagai H. Biochem Biophys Res Commun. 2000;267:164–168. doi: 10.1006/bbrc.1999.1914. [DOI] [PubMed] [Google Scholar]
- 15.Ansari K, Martin S, Farkasovsky M, Ehbrecht I M, Kuntzel H. J Biol Chem. 1999;274:30052–30058. doi: 10.1074/jbc.274.42.30052. [DOI] [PubMed] [Google Scholar]
- 16.Lorenz M C, Pan X, Harashima T, Cardenas M E, Xue Y, Hirsch J P, Heitman J. Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mosch H U, Fink G R. Genetics. 1997;145:671–684. doi: 10.1093/genetics/145.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo W S, Dranginis A M. Mol Biol Cell. 1998;9:161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rupp S, Summers E, Lo H J, Madhani H, Fink G F. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma X J, Lu Q, Grunstein M. Genes Dev. 1996;10:1327–1340. doi: 10.1101/gad.10.11.1327. [DOI] [PubMed] [Google Scholar]
- 21.McMillan J N, Longtine M S, Sia R A, Theesfeld C L, Bardes E S, Pringle J R, Lew D J. Mol Cell Biol. 1999;19:6929–6939. doi: 10.1128/mcb.19.10.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shulewitz M J, Inouye C J, Thorner J. Mol Cell Biol. 1999;19:7123–7137. doi: 10.1128/mcb.19.10.7123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fujita T, Tonouchi A, Hiroko T, Inose F, Nagashima T, Satoh R, Tanaka S. Proc Natl Acad Sci USA. 1999;96:8522–8527. doi: 10.1073/pnas.96.15.8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevenson B J, Rhodes N, Errede B, Sprague G F., Jr Genes Dev. 1992;6:1293–1304. doi: 10.1101/gad.6.7.1293. [DOI] [PubMed] [Google Scholar]
- 25.Sikorski R S, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman F, Fink G R, Hicks J B. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 28.Rose M D, Winston F, Hieter P. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. [Google Scholar]
- 29.Gietz R D, Schiestl R H, Willems A R, Woods R A. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 31.Stevenson B J, Ferguson B, De Virgilio C, Bi E, Pringle J R, Ammerer G, Sprague G F., Jr Genes Dev. 1995;9:2949–2963. doi: 10.1101/gad.9.23.2949. [DOI] [PubMed] [Google Scholar]
- 32.Mosch H U, Roberts R L, Fink G R. Proc Natl Acad Sci USA. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan X, Heitman J. Mol Cell Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ward M P, Gimeno C J, Fink G R, Garrett S. Mol Cell Biol. 1995;15:6854–6863. doi: 10.1128/mcb.15.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Blacketer M J, Madaule P, Myers A M. Genetics. 1995;140:1259–1275. doi: 10.1093/genetics/140.4.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koehler C M, Myers A M. FEBS Lett. 1997;408:109–114. doi: 10.1016/s0014-5793(97)00401-8. [DOI] [PubMed] [Google Scholar]
- 37.Carlson M. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 38.Jiang R, Carlson M. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 39.Vincent O, Carlson M. EMBO J. 1999;18:6672–6681. doi: 10.1093/emboj/18.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y, Davis C, Broach J R. EMBO J. 1998;17:6942–6951. doi: 10.1093/emboj/17.23.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gagiano M, vanDyk D, Bauer F F, Lambrechts M G, Pretorius I S. Mol Microbiol. 1999;31:103–116. doi: 10.1046/j.1365-2958.1999.01151.x. [DOI] [PubMed] [Google Scholar]
- 43.Kron S J, Styles C A, Fink G R. Mol Biol Cell. 1994;5:1003–1022. doi: 10.1091/mbc.5.9.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chant J, Pringle J R. J Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loeb J D J, Karentseva T A, Pan T, SepulvedaBecerra M, Liu H P. Genetics. 1999;153:1535–1546. doi: 10.1093/genetics/153.4.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Estruch F, Treitel M A, Yang X, Carlson M. Genetics. 1992;132:639–650. doi: 10.1093/genetics/132.3.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Flick J S, Johnston M. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Estruch F, Carlson M. Nucleic Acids Res. 1990;18:6959–6964. doi: 10.1093/nar/18.23.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tonukari N J, Scott-Craig J S, Walton J D. Plant Cell. 2000;12:237–248. doi: 10.1105/tpc.12.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]