Abstract
IFNs play critical roles in host defense by modulating the expression of various genes via signal transducer and activator of transcription factors. We show that IFNα/β activates another important transcription factor, NF-κB. DNA-binding activity of NF-κB was induced by multiple type 1 IFNs and was promoted by IFN in a diverse group of human, monkey, rat, and murine cells. Human IFN promoted NF-κB activation in murine cells that express the human IFNα/β receptor-1 signal-transducing chain of the type 1 IFN receptor. IFN promotes inhibitor of kappa B α (IκBα) serine phosphorylation and degradation, and stimulates NF-κB DNA-binding and transcriptional activity. Importantly, IFN promotes cell survival by protecting cells against a variety of proapoptotic stimuli, such as virus infection and antibody-mediated crosslinking. Expression of superrepressor forms of IκBα, besides inhibiting IFN-mediated NF-κB activation and IκBα degradation, also enhanced apoptotic cell death in IFN-treated cells. We conclude that NF-κB activation by IFNα/β is integrated into a signaling pathway through the IFNα/β receptor-1 chain of the type 1 IFN receptor that promotes cell survival in apposition to various apoptotic stimuli.
Keywords: apoptosis, viral infection
IFNs are multifunctional cytokines that block viral infection, inhibit cell proliferation, and modulate cell differentiation. IFNs are produced by a variety of cells in response to infections by viruses, mycoplasma, and bacteria as well as in response to noninfectious agents. IFNs are clinically useful in the treatment of diverse diseases, including certain forms of cancer, laryngeal and genital papillomas, chronic viral hepatitis, and multiple sclerosis. In general, IFN is an important model for the study of how genes are regulated by polypeptide ligand binding to cell surface receptors.
Type 1 IFNs (IFN α, β, and ω) bind to a common cell-surface receptor, whereas the receptor for type 2 IFN (IFNγ) is distinct (1). The type 1 IFN receptor is composed of IFNα/β receptor (IFNAR)-1 and IFNAR-2 chains (2–4) that undergo rapid ligand-dependent tyrosine phosphorylation. IFNs transduce signals from the cell surface, resulting in selective gene induction (5–7) through the activation of Janus tyrosine kinases and signal transducer and activator of transcription (STAT) factors (8, 9). Here, we describe another IFN-signaling pathway that also leads to altered gene expression that involves the NF-κB transcription factor.
NF-κB regulates the expression of genes involved in immune and inflammatory responses by binding to cis-acting κB sites in the promoters and enhancers of these genes. Under most circumstances, NF-κB lies dormant in the cytoplasm of unstimulated cells by virtue of its interaction with inhibitor of kappa B (IκB) proteins. Viruses, cytokines, lipopolysaccharides, and other stimulating agents promote dissociation of the inactive NF-κB–IκB complexes via the serine phosphorylation and degradation of IκB. These events lead to the unmasking of the nuclear localization sequence of NF-κB, thereby allowing NF-κB to enter the nucleus and bind DNA. NF-κB-regulated genes play an important role in suppressing apoptosis (10–13). Because several IFN-regulated genes have κB elements in their promoters and enhancers (14), we examined whether NF-κB activation is an important pathway through which IFNs modulate immunity, inflammation, and cell survival.
In the present study, we identify an IFN-signaling pathway involving the NF-κB transcription factor. This pathway protects cells against proapoptotic agents, showing that IFN is a cell-survival factor. IFN receptor signaling induces the serine phosphorylation and degradation of IκBα, leading to the stimulation of NF-κB DNA-binding and transcriptional activity. These data show that signals generated through NF-κB are indispensable for IFN-dependent cell survival (antiapoptotic). Because the use of IFN to treat human diseases is often limited by its inability to efficiently induce cell death, our identification of the mechanism through which IFN-induced death is limited is of great importance.
Materials and Methods
Biological Reagents and Cell Culture.
Recombinant human IFNα (IFNCon1), IFNα2, and IFNβ (Betaseron) were provided by Amgen Biologicals, Hoffmann–La Roche, and Berlex Biosciences, respectively. Human IFNα1 and IFNα8 were obtained from PBL Biomedical Laboratories (New Brunswick, NJ). IFN activities are expressed in international reference units/ml as assayed by protection against the cytopathic effect of vesicular stomatitis virus (VSV) on human fibroblasts by using the National Institutes of Health human IFNα standard for reference. Anti-Rel and IκBα antibodies were generously provided by N. Rice (National Cancer Institute, Frederick, MD). Human Daudi cells were maintained in static suspension cultures at 2–15 × 105 cells per ml in RPMI medium 1640 supplemented with 10% defined calf serum (HyClone).
Transfection Conditions and Constructs.
High-efficiency transient transfection of cells was accomplished by electroporation (capacitance 300 μF; 250 V) with 500 μg of salmon sperm DNA and 20 μg of plasmid DNA for each sample. IκBαM has S32A/S36A and Ser to Ala mutations in the COOH-terminal PEST motif (region rich in P, E, S, and T) of IκBα (12). IκBαΔN is an IκBα deletion mutant with two potential serine phosphorylation sites removed from the NH2 terminus (15). The pUX-CAT 3XHLAκB chloramphenicol acetyltransferase (CAT) reporter construct contains three tandemly repeated copies of the NF-κB site from the HLA-B7 gene (16).
NF-κB Activity Measurements.
Nuclei were extracted with buffer (20 mM Tris⋅HCl, pH 7.85/250 mM sucrose/0.4 M KCl/1.1 mM MgCl2/5 mM β-mercaptoethanol/1 mM NaF/1 mM Na3VO4/1 mM PMSF/5 μg/ml soybean trypsin inhibitor/5 μg/ml leupeptin/1.75 μg/ml benzamidine), and extracts were frozen and stored at −80°C (17). For electrophoretic mobility-shift assay (EMSA), the nuclear extracts were incubated with a 32P-labeled κB probe (5′-AGTTGAGGGGACTTTCCCAGG-3′) derived from an NF-κB binding sequence in the Ig gene promoter (18). To define the presence of specific Rel proteins, nuclear extracts were preincubated with a 1:25 dilution of anti-Rel antibodies at 25°C for 20 min and then subjected to EMSA. Gels were quantitated by PhosphorImager autoradiography. For reporter gene assays, cells were transiently cotransfected by electroporation with the pUX-CAT 3XHLAκB CAT reporter construct (16) and the appropriate expression vector. After 48 h, the cells were treated with IFNCon1 (5,000 units/ml) for 15 min and assayed for CAT activity. After TLC, radioactivity was measured by PhosphorImager autoradiography.
IκBα Degradation.
At various times after IFNα treatment, 1 × 108 cells were lysed directly in Laemmli buffer and equivalent amounts of protein were subjected to SDS/PAGE. Proteins were transferred to poly(vinylidene difluoride) membranes, immunoblotted with specific affinity-purified rabbit anti-IκBα antibody, and visualized by chemiluminescence with the enhanced chemiluminescence reagent (Amersham Pharmacia).
Antiviral, Antiproliferative, and Apoptosis Assays.
For determining antiviral activity, cell cultures (5 × 105 cells per ml) were preincubated overnight with IFNCon1, followed by infection with VSV for 1.5 h at 0.1 plaque-forming units per cell. At 24-h postinfection, the virus yield in the medium was assayed by plaque formation on indicator Vero cells (19). The antiproliferative action of IFN was assayed by treating Daudi cells (5 × 104 cells per well of 24-well plates) with IFNCon1. After 3 days, the cells were enumerated in a Coulter Counter (20). For determining apoptosis, cell cultures (5 × 105 cells per ml) were preincubated overnight with IFNCon1, followed by treatment with either anti-Ig (goat anti-μ antibody at 35 μg/ml) (21) or VSV infection at a high multiplicity of infection (10 plaque-forming units per cell). After an overnight incubation, the cells were deposited onto glass slides in a cytocentrifuge, fixed with 4% formaldehyde, permeabilized with 0.2% Triton X-100, and processed for terminal deoxynucleotidyltransferase-mediated UTP end labeling (TUNEL) according to the manufacturer's recommendations (Roche Molecular Biochemicals). Alternatively, lysates of control and IFN-treated (1,000 units/ml; 24 h) cells were analyzed for apoptotic DNA by modification of a chemiluminescence-based assay (22). In brief, cells (5 × 106) were lysed in hypotonic buffer, and sequentially digested with RNase and proteinase K. Low molecular weight DNA was extracted and subjected to nonisotopic labeling of 3′ ends with digoxigenin-11-dUTP and Taq DNA polymerase. Labeled DNA was separated by electrophoresis on 1.6% agarose, transferred to nitrocellulose, and fragmented DNA visualized by chemiluminescent detection with alkaline phosphatase-conjugated antidigoxigenin and CDP-Star substrate (Roche Molecular Biochemicals).
Results
NF-κB Activation by IFNα/β.
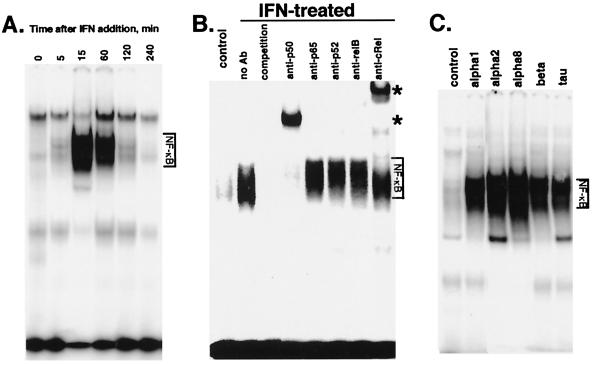
We considered the possibility that IFN may stimulate NF-κB activity because IFN-induced genes, including HLA class I, IFN regulatory factor-1, double-stranded RNA-dependent protein kinase, intercellular adhesion molecule-1, IFNβ, and oligoadenylate synthetase contain NF-κB-binding sites in their 5′ regulatory regions. To determine whether IFNα/β promotes NF-κB activation, highly IFN-sensitive Daudi lymphoblastoid cells were stimulated with IFNα, and NF-κB activation was examined by EMSA. Nuclear extracts from untreated Daudi cells show low constitutive binding to a consensus κB oligonucleotide probe. However, IFN increased κB binding in Daudi cells within 5 min. IFN-dependent κB binding reached a maximal induction after 15 min, and then decreased to basal levels by 4 h (Fig. 1A).
Figure 1.
IFN induces NF-κB activation. (A) EMSA on nuclear extracts from Daudi cells treated with IFNCon1 (5,000 units/ml) for varying times. (B) Nuclear extracts from Daudi cells treated with IFNCon1 (5,000 units/ml; 30 min) were subjected to EMSA in the absence or presence of a 50-fold excess of unlabeled κB oligonucleotide probe (competition). *, Positions of supershifted complexes. (C) EMSA on nuclear extracts from Daudi cells treated with various type 1 IFNs (5,000 units/ml) for 30 min.
Active NF-κB consists of dimers of the Rel/NF-κB family of polypeptides, which include p50, p52, c-Rel, v-Rel, RelA (p65), and RelB. To determine which of these proteins are present in the IFN-induced NF-κB complexes, nuclear extracts prepared from IFN-treated Daudi cells were incubated with antisera directed against specific Rel proteins and NF-κB activation was examined by EMSA. These assays showed the presence of p50 and c-Rel in the IFN-induced NF-κB complexes, because the complexes were supershifted by antisera to either p50 or c-Rel (Fig. 1B). In contrast, antisera to p65, p52, or RelB did not supershift the IFN-induced NF-κB complexes. Moreover, immunofluorescence microscopy confirmed that IFN induced the rapid translocation of p50–c-Rel complexes from the cytoplasm into the nucleus.
The previous series of experiments were performed with IFNCon1, which is a synthetic human IFN designed as a consensus of several IFNα subtypes. Although all type 1 IFNs bind to a common cell surface receptor, differences between IFNα/β subtypes in receptor interaction and signal transduction have been observed (23–25). Therefore, we tested whether various type 1 IFNs could induce NF-κB activity by treating Daudi cells with the type 1 IFNs for 15 min and preparing nuclear extracts. Fig. 1C shows that a wide variety of IFNα/β subtypes (IFNCon1, IFNα1, IFNα2, IFNα8, IFNβ, and IFNτ) induce NF-κB DNA-binding activity.
IFN Protects Cells Against Apoptotic Cell Death.
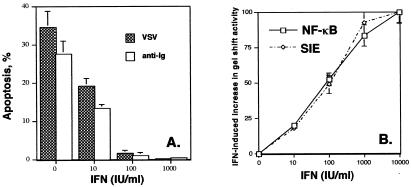
NF-κB activation protects cells from apoptosis induced by cytokines, UV irradiation, and chemotherapeutic agents (10–13). NF-κB knockout mice die early in embryonic development from extensive apoptosis (10). Inhibition of NF-κB enhances sensitivity to apoptotic cell death by various agents (11–13). Because we found that IFN induced NF-κB activation, we tested whether IFN protected Daudi cells against proapoptotic stimuli. Infection by most viruses triggers apoptosis of the infected cell (26). Activation of antigen receptors in B cells (Daudi cells are B-lymphoblastoid cells) induced by antibody-mediated crosslinking results in apoptosis (21). Specifically, we tested whether viral infection (by VSV) or antibody-mediated crosslinking (by anti-Ig addition) induced the programmed death of Daudi cells as determined by TUNEL assays. Infection with VSV or addition of anti-Ig efficiently induced Daudi cell apoptosis (34.6 or 27.5%, respectively). In contrast, as shown in Fig. 2A, overnight pretreatment with IFN markedly protected Daudi cells against apoptosis induced by VSV infection or anti-Ig addition with nearly complete protection observed at an IFN concentration of 100 units/ml. The antiapoptotic action of IFN in Daudi cells had an IC50 of only ≈10 units/ml, which is similar to the IC50 (<25 units/ml) of the antiproliferative and antiviral actions of IFN on Daudi cells. Thus, Daudi cells are highly sensitive to the antiapoptotic action of IFN, and most importantly, IFN is a cell survival factor.
Figure 2.
IFN promotes cell survival. (A) Daudi cells were pretreated overnight with various concentrations of IFNCon1 and analyzed for apoptosis by TUNEL assays at 1 day after either infection by VSV or anti-Ig treatment. The data shown in the graph are the average of two experiments performed in duplicate and are expressed in terms of apoptosis induced by VSV or anti-Ig. (B) Dose-dependent NF-κB activation and sis-inducible element gel shift activity by IFN. Nuclear extracts from Daudi cells treated with varying concentrations of IFNCon1 (0–5,000 units/ml) for 30 min were incubated with a 32P-endlabeled promoter probe for the consensus κB site or the high affinity sis-inducible element (SIE) from the c-fos gene (5′-AGCTTCATTTCCCGTAATCCCTAAAGCT-3′). EMSA results were quantitated on a PhosphorImager (Molecular Dynamics) by using quantity one software (Bio-Rad). The results of three experiments were averaged (SEM < 20%) and expressed relative to the increased gel shift activity observed at an IFN concentration of 10,000 units/ml.
These results led us to examine whether Daudi cells are also highly sensitive to the induction of NF-κB activation by IFN, i.e., NF-κB activation was observed at IFN concentrations that promote cell survival. Daudi cells were treated for 15 min with IFN at concentrations varying from 10 to 10,000 units/ml, nuclear extracts prepared, and NF-κB activation was examined by EMSA. As illustrated in Fig. 2B, increased κB binding in Daudi cells was observed at an IFN concentration of only 10 units/ml and IFN-dependent κB binding reached a maximal induction at 5,000 units/ml. The dose-response relationship for IFN-mediated induction of NF-κB activity in Daudi cells is similar to that for the induction of sis-inducible element gel shift activity.
IFN Promotes the Serine Phosphorylation and Degradation of IκBα.
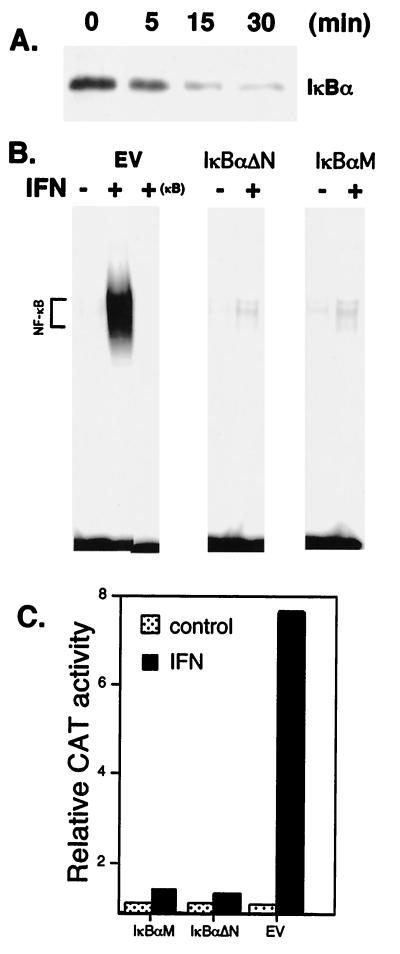
The activity of NF-κB is tightly controlled by inhibitory IκB proteins that bind to NF-κB complexes and thus sequester NF-κB in the cytoplasm. Cytokines, such as IL-1 and tumor necrosis factor, promote the serine phosphorylation of IκB, its polyubiquitination, and proteosome-mediated degradation, and thereby induce NF-κB translocation to the nucleus. To determine whether IFN-dependent NF-κB activation reflects IκBα degradation, IκBα levels were determined at various times after IFN addition by immunoblotting with anti-IκBα antisera. As shown in Fig. 3A, IFN induced a progressive decrease in cellular levels of IκBα, indicating that IFN stimulated NF-κB activation by promoting IκBα degradation. The kinetics of induction of NF-κB activation in Daudi cells is consistent with that of IκBα degradation, with IκB degradation observed within 5 min of IFN addition, at which time NF-κB activation is detectable. Moreover, although similar amounts of c-Rel were immunoprecipitated from control or IFN-treated cells, precipitates from IFN-treated cells showed a progressive decrease in associated IκBα (data not shown). Our results indicate that IFN promotes the dissociation of IκBα/NF-κB complexes and IκBα degradation.
Figure 3.
The role of IκBα in NF-κB activation by IFNα/β. (A) Cell lysates from Daudi cells treated with IFNCon1 (5,000 units/ml) were resolved by SDS/PAGE, blotted onto poly(vinylidene difluoride) membranes, probed with anti-IκBα, and visualized by enhanced chemiluminescence. (B) EMSA with a 32P-labeled κB probe on nuclear extracts from control and IFN-treated Daudi cells transiently transfected for 48 h with IκBαM, IκBαΔN, or with empty vector (EV). Binding to the κB probe was blocked by a 50-fold excess of unlabeled probe (+ κB). (C) NF-κB-dependent reporter gene activity in IFN-treated COS cells transiently cotransfected with the pUX-CAT 3XHLAκB construct and IκBαM, IκBαΔN, or empty vector. At 2 days after transfection, cells were treated in the presence or absence of IFN for 30 min, lysed, and assayed for CAT activity as determined by phosphorimaging. Data shown are the average of three experiments (SEM < 15%), expressed relative to CAT activity in cells transfected with empty vector.
Because serine phosphorylation of IκBα leads to its degradation, IκBα proteins with mutations or deletions of serine phosphorylation sites function as superrepressors of cytokine-induced NF-κB activation (12, 15). We transiently transfected Daudi cells with such IκBα mutant proteins. As shown in Fig. 3B, IκBαM or IκBαΔN expression blocked NF-κB activation by IFN. These results indicate that NF-κB activation results from IκBα serine phosphorylation.
To determine the functional importance of NF-κB activation, COS cells were cotransfected with a NF-κB-CAT reporter plasmid and IκBα mutant proteins or an empty vector, and assayed for CAT activity 2 days after transfection. COS cells were used because they are IFN responsive in reporter assays, whereas Daudi cells are not. IFN stimulated κB-dependent transcription by about 8-fold in cells transfected with empty vector (Fig. 3C). In contrast, expression of IκBαM or IκBαΔN suppressed IFN-activated κB-dependent transcription by >90%. Neither IκBα mutant affected IFN-stimulated response element-dependent transcription (data not shown). These results show that the IFN-activated κB-dependent mechanism is distinct from the well-established IFN-stimulated response element-dependent mechanism in regulating gene expression.
Blockage of NF-κB Activation Sensitizes Cells to the Proapoptotic Activity of IFN.
NF-κB antagonizes apoptosis induced by tumor necrosis factor, UV irradiation, and chemotherapeutic agents (11–13). Paradoxically, IFN induces the apoptosis of some tumor cells (27), whereas protecting other cells against proapoptotic stimuli. This led us to hypothesize that the IFN-induced NF-κB pathway counterbalances proapoptotic signals generated by IFN itself.
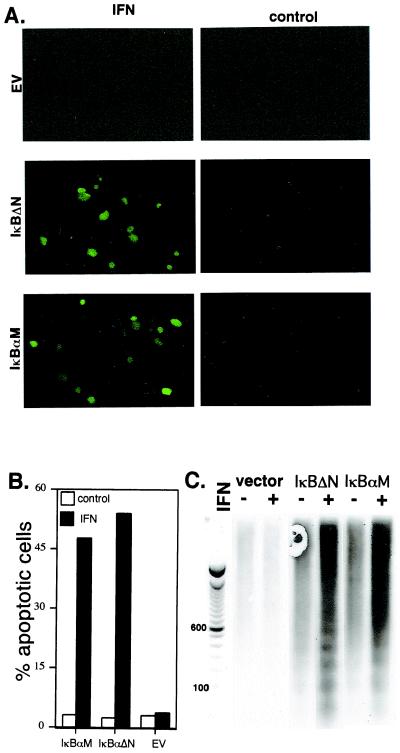
Thus, we examined whether expression of superrepressor IκBα mutants, which block IFN-induced NF-κB activation, would sensitize Daudi cells to IFN-induced apoptosis. In empty vector-transfected cells, IFN induced a very modest increase in apoptosis (from 0.1 to 0.6%) as determined by TUNEL assays. However, as shown in Fig. 4 A and B, expression of superrepressor IκBα constructs markedly sensitized Daudi cells to IFN-induced death (≈50%). A prominent feature of apoptosis is the intranucleosomal cleavage of DNA in a discontinuous ladder of discrete multimers of ≈200 bp. When cell lysates were examined by a highly sensitive chemiluminiscent-based DNA fragmentation assay, the formation of the telltale DNA ladder is clearly evident in IFN-treated Daudi cells expressing either superrepressor IκBα construct (Fig. 4C). These results indicate that a pathway leading to NF-κB activation protects cells against the proapoptotic action of IFN. Thus, IFN generates a strong cell survival signal, which suppresses the death-promoting activity of various stimuli (VSV infection, antigen crosslinking, and even IFN).
Figure 4.
The role of IκBα in IFNα/β promoted cell survival. IFN-treated (1,000 units/ml; 24 h) Daudi cells transiently transfected for 48 h with IκBαM, IκBαΔN, or empty vector were analyzed for apoptosis by TUNEL assays (A and B) or apoptotic DNA by a chemiluminescent assay with a DNA ladder provided for reference (C). Apoptotic cells are fluorescent and thus appear green in the photomicrograph. The TUNEL data (B) represent the mean of three independent experiments in which at least 500 cells were scored for each variable (SEM < 10%).
NF-κB Activation by IFNα/β in a Variety of Cell Types.
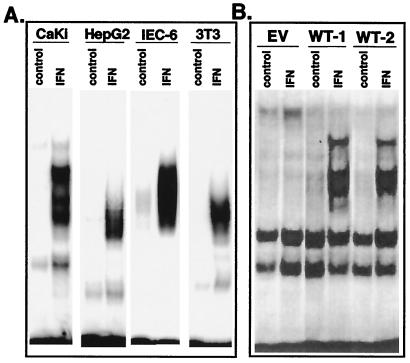
IFN has biological activity in a broad variety of cell types. IFNs are, in general, highly species specific, and therefore human IFN has limited biologic activity in nonprimate cells. Therefore, we examined whether IFN induced NF-κB activity in various human, murine, and rodent cell lines that respond to the antiviral or antiproliferative activity of IFN. As shown in Fig. 5A, rat IFN induced NF-κB activity in rat intestinal epithelial cells (IEC)-6 and murine 3T3 fibroblasts. Human IFN treatment induced NF-κB activity in human HepG2 hepatocarcinoma cells and CaKi renal carcinoma cells (Fig. 5A). IFNα/β promoted NF-κB DNA-binding activity in diverse cell types, including primary rat hepatocytes, primary mouse fibroblasts, as well as transformed human cell lines such as T cell lymphoma, renal carcinoma, epithelioid carcinoma, hepatocarcinoma, and fibrosarcoma (data not shown). Supershift assays with antisera directed against specific Rel proteins showed cell type-specific differences in the composition of the IFN-induced Rel/NF-κB complexes. For example, whereas IFN-induced p50–RelA complexes were found in 3T3, IEC-6, CaKi, HepG2, and HT1080 cells, p50–c-Rel complexes were found in MOLT-16 and Daudi cells.
Figure 5.
IFN induces NF-κB activation in diverse cell types. (A) EMSA on nuclear extracts from IFN-treated (5,000 units/ml rat IFNβ for 30 min) murine 3T3 fibroblasts and rat intestinal IEC-6 cells, or IFN-treated (5,000 units/ml IFNCon1 for 30 min) human CaKi and HepG2 cells. (B) Murine L929 cell transfectants expressing empty vector (EV) or two independent wild-type huIFNAR-1 (WT) transfectants (29) were treated with IFNCon1 (5,000 units/ml) for 30 min before gel shift analysis with a NF-κB probe.
We previously demonstrated that the IFNAR-1 chain of the human IFNα/β receptor acts as a species-specific transducer for type 1 IFN action when transfected into heterologous mouse cells (25, 28, 29). Thus, whereas murine cells are unresponsive to human IFN, murine transfectants expressing the human IFNAR-1 chain respond to human IFN as determined by induction of STAT-containing DNA-binding complexes, IFN-stimulated gene expression, and antiviral activity. We examined whether murine transfectants expressing the human IFNAR-1 chain respond to human IFN as determined by induction of NF-κB activity. As shown in Fig. 5B, murine cells transfected with empty vector do not show detectable NF-κB activity on human IFN addition. However, in two independent murine tranfectants expressing the human IFNAR-1, NF-κB activity was clearly evident after human IFN addition. These results not only demonstrate that NF-κB activation is a common cellular response to IFNα/β, but also suggest a role for the human IFNAR-1 chain in signaling events leading to NF-κB activation.
Discussion
IFNα/β has antiproliferative, antiviral, antibacterial, antiprotozoal, and immunomodulatory functions. In addition, IFN has anticancer activity in vivo and is clinically useful for the treatment of laryngeal and genital papillomas, chronic viral hepatitis, and multiple sclerosis. Understanding the molecular basis of IFN action is very important when one considers the therapeutic potential of IFN as well as its role as a model for understanding the function of many cytokines.
IFN elicits pleiotropic biological effects by regulating gene expression through signals generated on its binding to a distinct surface receptor on target cells. Type 1 IFNs bind to a ubiquitously expressed cell-surface receptor, composed of the IFNAR-1 and IFNAR-2 subunits (2, 4, 30, 31). Whereas IFNAR-2 is the ligand-binding subunit, IFNAR-1 acts as a species-specific transducer for the actions of type 1 IFN (28, 32). These receptor subunits have no inherent enzyme activity, but generate cytoplasmic signals in combination with the nonreceptor Janus protein tyrosine kinases and STAT proteins. On their tyrosine phosphorylation, IFN-activated STATs (STAT1–STAT3) dimerize and translocate into the nucleus to induce gene transcription. STAT proteins function in the gene activation pathway induced by many other cytokines (33, 34). Thus, the IFN-activated Janus kinase/STAT tyrosine phosphorylation pathway serves as a paradigm for understanding cytokine signal transduction.
We show that IFNs also transmit nuclear signals through activation of the important transcription factor family, NF-κB. The family of NF-κB transcription factors regulates the expression of genes involved in immune and inflammatory responses and cell growth, as well as viral genes by binding to cis-acting κB sites in the promoters and enhancers of genes bearing the consensus sequence 5′-GGGPuNNPyPyCC-3′. We show that IFN promotes transcription through a NF-κB-dependent reporter and this effect is blocked by two different superrepressor IκB constructs (Fig. 3). Moreover, we found in preliminary studies that IFN appears to up-regulate antiapoptotic genes (Bcl-2 and inhibitor of apoptosis protein-2) that depend on NF-κB (W. J. Valentine and L.M.P., unpublished observations).
In addition, NF-κB has an important antiapoptotic function. We report that IFN promotes cell survival by protecting cells against proapoptotic stimuli, such as virus infection, antibody-mediated crosslinking, and IFN itself. We have also shown that NF-κB activation is a common cellular response to IFNα/β. Recently, IFNα/β has been found to promote the survival of activated T cells in vivo, but has little or no effect on resting T cells (35), observations consistent with those reported here. It will be interesting to determine whether this effect is mediated through the NF-κB pathway.
By using murine transfectants expressing the human IFNAR-1 chain, we found that NF-κB activation involves signals generated through this subunit of the human IFNα/β receptor. The IFNAR-1 chain plays a critical role in the function of the IFNα/β receptor. IFNAR-1 acts as a species-specific transducer for type 1 IFN action undergoing rapid IFN-dependent tyrosine phosphorylation, and its knockout in mice results in the loss of the antiviral response to type 1 IFNs (28, 32, 36, 37). We have previously identified a conserved motif within the intracellular domain of IFNAR-1, which is required for the IFN-dependent activation of STAT3 and phosphatidyinositol 3-kinase (PI-3K) (17, 38). Furthermore, STAT3 is responsible for bringing PI-3K into a complex with the IFNα/β receptor (38). Expression of STAT3 in an IFN-resistant Daudi cell line complemented defective NF-κB activation, as well as several other signaling defects (18). These results suggest that NF-κB activation by IFNα/β may require signals generated through the IFNAR-1 chain involving STAT3 and PI-3K (C.H.Y., A.M., S.R.P., and L.M.P.; unpublished results). Moreover, NF-κB can be activated through a PI-3K-dependent pathway (39, 40), and IFN activates PI-3K (38, 41).
Our findings are important because they are clinically relevant to the use of IFN in the treatment of cancer and viral diseases. The clinical efficacy of IFN is often limited by its inability to efficiently induce cell death (27, 42). Our results suggest that this inability reflects the induction by IFN of potent cell survival signals generated through NF-κB. On the other hand, the therapeutic action of IFN in multiple sclerosis may reflect its ability to protect neuronal cells against proapoptotic cytokines (43). Because we show that the distinct actions of IFN on cell proliferation, viral replication, apoptosis, and cell survival can be modulated, it may now become possible to enhance clinically useful, or alternatively attenuate undesirable, IFN actions in specific pathological conditions.
Acknowledgments
We thank N. Rice (National Cancer Institute, Frederick, MD) for providing a panel of anti-Rel antibodies; A. Martinez (University of Tennessee, Memphis) for help in preparing primary rat hepatocytes; G. Murti (St. Jude Children's Research Hospital, Memphis) for help with confocal microscopy, and I. Verma (Salk Institute, La Jolla, CA), D. Ballard (Vanderbilt University, Nashville), and J. Vilcek (New York University, New York) for providing expression vectors. This work was supported by National Institutes of Health Grant CA73753 (L.M.P.) and by funds from the Department of Pathology, University of Tennessee.
Abbreviations
- CAT
chloramphenicol acetyltransferase
- EMSA
electrophoretic mobility-shift assay
- IκB
inhibitor of kappa B
- STAT
signal transducer and activator of transcription
- TUNEL
terminal deoxynucleotidyltransferase-mediated UTP end labeling
- VSV
vesicular stomatitis virus
- IFNAR
IFNα/β receptor
- PI-3K
phosphatidyinositol 3-kinase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250477397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250477397
References
- 1.Branca A A, Faltynek C R, D'Alessandro S B, Baglioni C. J Biol Chem. 1982;257:13291–13296. [PubMed] [Google Scholar]
- 2.Uze G, Lutfalla G, Gresser I. Cell. 1990;60:225–234. doi: 10.1016/0092-8674(90)90738-z. [DOI] [PubMed] [Google Scholar]
- 3.Novick D, Cohen B, Rubinstein M. Cell. 1994;77:391–400. doi: 10.1016/0092-8674(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 4.Domanski P, Witte M, Kellum M, Rubinstein M, Hackett R, Pitha P, Colamonici O R. J Biol Chem. 1995;270:21606–21611. doi: 10.1074/jbc.270.37.21606. [DOI] [PubMed] [Google Scholar]
- 5.Friedman R L, Stark G R. Nature (London) 1985;314:637–639. doi: 10.1038/314637a0. [DOI] [PubMed] [Google Scholar]
- 6.Larner A C, Chaudhuri A, Darnell J E. J Biol Chem. 1986;261:453–459. [PubMed] [Google Scholar]
- 7.Darnell J E, Jr, Kerr I M, Stark G R. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 8.Schindler C, Shuai K, Prezioso V R, Darnell J E. Science. 1992;257:809–813. [PubMed] [Google Scholar]
- 9.Fu X-Y. Cell. 1992;70:323–335. doi: 10.1016/0092-8674(92)90106-m. [DOI] [PubMed] [Google Scholar]
- 10.Beg A A, Sha W C, Bronson R T, Ghosh S, Baltimore D. Nature (London) 1995;376:167–170. doi: 10.1038/376167a0. [DOI] [PubMed] [Google Scholar]
- 11.Beg A A, Baltimore D. Science. 1996;274:782–784. doi: 10.1126/science.274.5288.782. [DOI] [PubMed] [Google Scholar]
- 12.Van Antwerp D J, Martin S J, Kafri T, Green D R, Verma I M. Science. 1996;274:787–789. doi: 10.1126/science.274.5288.787. [DOI] [PubMed] [Google Scholar]
- 13.Wang C-Y, Mayo M W, Baldwin A S., Jr Science. 1996;274:784–787. doi: 10.1126/science.274.5288.784. [DOI] [PubMed] [Google Scholar]
- 14.Baeuerle P A, Henkel T. Annu Rev Immunol. 1994;12:141–179. doi: 10.1146/annurev.iy.12.040194.001041. [DOI] [PubMed] [Google Scholar]
- 15.Brockman J A, Scherer D C, McKinsey T A, Hall S M, Qi X, Lee W Y, Ballard D W. Mol Cell Biol. 1995;15:2809–2818. doi: 10.1128/mcb.15.5.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oliviera I C, Mukaida N, Matsushiam K, Vilcek J. Mol Cell Biol. 1994;14:5300–5308. doi: 10.1128/mcb.14.8.5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang C H, Shi W, Basu L, Murti A, Constantinescu S N, Blatt L, Croze E, Mullersman J E, Pfeffer L M. J Biol Chem. 1996;271:8057–8061. doi: 10.1074/jbc.271.14.8057. [DOI] [PubMed] [Google Scholar]
- 18.Yang C H, Murti A, Pfeffer L M. Proc Natl Acad Sci USA. 1998;95:5568–5572. doi: 10.1073/pnas.95.10.5568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Improta T, Pine R, Pfeffer L M. J Interferon Res. 1992;12:87–94. doi: 10.1089/jir.1992.12.87. [DOI] [PubMed] [Google Scholar]
- 20.Eisenkraft B L, Nanus D M, Albino A P, Pfeffer L M. Cancer Res. 1991;51:5881–5887. [PubMed] [Google Scholar]
- 21.Racila E, Hseuh R, Marches R, Tucker T F, Krammer P H, Scheuermann R H, Uhr J W. Proc Natl Acad Sci USA. 1996;93:2165–2168. doi: 10.1073/pnas.93.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blanco F L, Gonzalez-Reyes J, Fanjul L F, Ruiz de Galarreta C M, Aguiar J Q. BioTechniques. 1998;24:354–358. doi: 10.2144/98243bm04. [DOI] [PubMed] [Google Scholar]
- 23.Abramovich C, Shulman L M, Ratovitski E, Harroch S, Tovey M, Eid P, Revel M. EMBO J. 1994;13:5871–5877. doi: 10.1002/j.1460-2075.1994.tb06932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Platanias L C, Uddin S, Colamonici O R. J Biol Chem. 1994;269:17761–17764. [PubMed] [Google Scholar]
- 25.Constantinescu S N, Croze E, Murti A, Wang C, Basu L, Hollander D, Russell-Harde D, Garcia-Martinez V, Mullersman J E, Pfeffer L M. Proc Natl Acad Sci USA. 1995;92:10487–10491. doi: 10.1073/pnas.92.23.10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roulston A, Marcellus R C, Branton P E. Annu Rev Microbiol. 1999;53:577–628. doi: 10.1146/annurev.micro.53.1.577. [DOI] [PubMed] [Google Scholar]
- 27.Einhorn S, Grander D. J Interferon Cytokine Res. 1996;16:275–281. doi: 10.1089/jir.1996.16.275. [DOI] [PubMed] [Google Scholar]
- 28.Colamonici O R, Porterfield B, Domanski P, Constantinescu S N, Pfeffer L M. J Biol Chem. 1994;269:9598–9602. [PubMed] [Google Scholar]
- 29.Basu L, Yang C H, Murti A, Garcia J V, Croze E, Constantinescu S N, Mullersman J E, Pfeffer L M. Virology. 1998;242:14–21. doi: 10.1006/viro.1997.9002. [DOI] [PubMed] [Google Scholar]
- 30.Vanden Broecke C, Pfeffer L M. J Interferon Res. 1988;8:803–811. doi: 10.1089/jir.1988.8.803. [DOI] [PubMed] [Google Scholar]
- 31.Lutfalla G, Holland S J, Cinato E, Monneron D, Reboul J, Rogers N C, Smith J M, Stark G R, Gardiner K, Mogensen K E, Kerr I M, Uze G. EMBO J. 1995;14:5100–5108. doi: 10.1002/j.1460-2075.1995.tb00192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Constantinescu S N, Croze E, Wang C, Murti A, Basu L, Mullersman J E, Pfeffer L M. Proc Natl Acad Sci USA. 1994;91:9602–9606. doi: 10.1073/pnas.91.20.9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadowski H B, Shuai K, Darnell J E, Jr, Gilman M Z. Science. 1993;261:1739–1743. doi: 10.1126/science.8397445. [DOI] [PubMed] [Google Scholar]
- 34.Larner A C, David M, Feldman G M, Igarishi K, Hackett R H, Webb D, Sweitzer S M, Petricoin E F, Finbloom D S. Science. 1993;261:1730–1736. doi: 10.1126/science.8378773. [DOI] [PubMed] [Google Scholar]
- 35.Marrack P, Kappler J, Mitchell T. J Exp Med. 1999;189:521–530. doi: 10.1084/jem.189.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleary C M, Donnelly R J, Soh J, Mariano T M, Pestka S. J Biol Chem. 1994;269:18747–18749. [PubMed] [Google Scholar]
- 37.Muller U, Steinhoff U, Reis L F L, Hemmi S, Pavlovic J, Zinkernagel R M, Aguet M. Science. 1994;264:1918–1921. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 38.Pfeffer L M, Mullersman J E, Pfeffer S R, Murti A, Shi W, Yang C H. Science. 1997;276:1418–1420. doi: 10.1126/science.276.5317.1418. [DOI] [PubMed] [Google Scholar]
- 39.Reddy S A, Huang J H, Liao W S. J Biol Chem. 1997;272:29167–29173. doi: 10.1074/jbc.272.46.29167. [DOI] [PubMed] [Google Scholar]
- 40.Ozes O N, Mayo L D, Gustin J A, Pfeffer S R, Pfeffer L M, Donner D B. Nature (London) 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 41.Uddin S, Yenush L, Sun X-J, Sweet M E, White M F, Platanias L C. J Biol Chem. 1995;270:15938–15941. doi: 10.1074/jbc.270.27.15938. [DOI] [PubMed] [Google Scholar]
- 42.Cremer I, Viellard V, De Maeyer E. Virology. 1999;253:241–249. doi: 10.1006/viro.1998.9470. [DOI] [PubMed] [Google Scholar]
- 43.Benveniste E N. In: Human Cytokines: Their Role in Research and Therapy. Aggarwal B, Puri R, editors. Berkeley, CA: Blackwell Scientific; 1995. pp. 195–216. [Google Scholar]