Abstract
Mnn9p is a component of two distinct multiprotein complexes in the Saccharomyces cerevisiae cis-Golgi that have both been shown to have α-1,6-mannosyltransferase activity in vitro. In one of these complexes, Mnn9p associates with four other membrane proteins, Anp1p, Mnn10p, Mnn11p, and Hoc1p, whereas the other complex consists of Mnn9p and Van1p. Members of the Mnn9p-containing complexes were incorporated into COPII vesicles made in vitro from endoplasmic reticulum (ER) membranes isolated from cycloheximide-treated cells. This behavior is consistent with an active Golgi to ER recycling process. To examine this path in vivo, we monitored retrograde transport of subunits of the complex in cells blocked in anterograde transport from the ER. In this situation, specific relocation of the proteins from the Golgi to the ER was observed in the absence of new protein synthesis. Conversely, when retrograde transport was blocked in vivo, subunits of the mannosyltransferase complex accumulated in the vacuole. Packaging of Mnn9p in COPI-coated vesicles from purified Golgi membranes was also investigated using a coatomer-dependent vesicle budding assay. Gradient fractionation experiments showed that Mnn9p and the retrograde v-SNARE, Sec22p, were incorporated into COPI-coated vesicles. These observations indicate that the Mnn9p-containing mannosyltransferase complexes cycle back and forth between the ER and Golgi.
The biogenesis of the Golgi complex and the mechanism of intra-Golgi protein transport have long been the subject of intense interest in membrane cell biology. The origin of the different Golgi subcompartments, with their unique set of resident proteins and specialized functions, is a matter of much debate. Two models inform this debate (1, 2). The vesicular transport model posits that Golgi cisternae are stable structures that receive and create vesicles carrying cargo molecules in the anterograde direction and retrograde vesicles returning escaped proteins to their resident cisternae or the endoplasmic reticulum (ER) (3). Alternatively, in the cisternal maturation model, each Golgi cisterna translates through a stack of membranes acquiring new functional capabilities by fusing with retrograde vesicles that carry enzymes from a distal cisterna (2). According to this view, the cis-Golgi can then be considered an outgrowth of the ER, formed by the homotypic fusion of ER-derived transport vesicles. A possible resolution is that the two mechanisms operate in parallel to achieve rapid (vesicular) and slow (cisternal) transport of molecules and large particles, respectively (4).
One strong piece of evidence for cisternal maturation emerged from studies in which procollagen helical fibers were tracked as they moved across the Golgi stack in collagen-secreting fibroblasts (5). Using a combination of biochemical and morphological methods, Bonfanti et al. (5) demonstrated that procollagen traverses the stack while staying enclosed in the lumen of Golgi cisternae. Independent evidence supporting this view came with the observation that the yeast cis-Golgi t-SNARE, Sed5p, cycles between the Golgi and ER (6). Thus, the steady-state localization of Sed5p may be maintained by continuous vesiculation and retrograde transport of cis-Golgi proteins followed by their reappearance in the cis-cisterna that results from the homotypic fusion of ER-derived transport vesicles. Yet another example of this itinerary is the flow of mammalian Golgi proteins through the ER, possibly during mitosis or as a means by which misfolded Golgi proteins may be retrieved and degraded in the ER quality control system (7, 8).
In the course of identifying proteins that are packaged into ER-derived transport (COPII) vesicles, we detected Mnn9p, a subunit of an oligomeric mannosyltransferase. Mnn9p and its partners are type II membrane proteins that form two separate complexes in the cis-Golgi (9). The first comprises Van1p and Mnn9p, and the second consists of Mnn9p, Anp1p, Mnn11p, Hoc1p, and a recently characterized protein Mnn10p (9, 10). These complexes have α-1,6-mannosyltransferase activity in vitro (9) and act in succession to form and extend the mannan backbone of glycosylated proteins traversing the yeast Golgi.
In the present study, we have addressed the intracellular dynamics of the two mannosyltransferase complexes. Despite the normal cis-Golgi localization and function of these proteins, we find their itinerary includes retrieval to the ER.
Materials and Methods
Yeast Strains, Growth Conditions, and Reagents.
Saccharomyces cerevisiae strain RSY445 (Matα, ura3–52,leu2–3,-112, trp1–289, prb1, pep4∷URA3, gal2, his4–579) was used for preparation of microsomes that served as a donor source in the analytical and preparative budding reactions. Strains SEY6210 (Matα, leu2–3,-112, ura3–52, his3Δ200, trp-Δ901, lys2–801, suc2-Δ9) (11), RSY263 (Matα, sec12–4, leu2–3,-112, ura3–52), MLY101 (Matα, Δufe1∷TRP1, leu2–3,-112, ura3–52, trp1–1, his3–11,-15, pUT-1), and MLY103 (ura3–52, trp1–1, his3–11,-15, Δufe1∷TRP1, Δpep4∷URA3, Matα, pUT-1) (12) and derivatives were used in all other experiments. A strain harboring the sec12–4 mutation and a hemagglutinin (HA)-tagged version of OCH1 was a meiotic product from the sporulation of a diploid obtained by crossing RSY264 (Mata, leu2–3, -112, ura3–52) with YAS 31 (Matα, leu2–3, -112, ura3–52, trp1–289, pep4∷URA3, prb1, his4–579,TRP1-OCH1-HA) (this laboratory). Mating, sporulation, and tetrad dissection were performed using standard techniques (13). Yeast strains were grown in rich media at 30°C or at 24°C for temperature-sensitive strains.
Disruption of genes was accomplished by replacement of ORFs with the S. cerevisiae HIS3 gene using homologous recombination. PCR products were generated from plasmid pRS303 (14) containing the HIS3 gene and introduced into yeast cells by transformation. Gene disruption was verified by PCR.
For c-myc epitope tagging, PCR products were amplified from plasmid pSBB13 (this laboratory), which encodes the S. cerevisiae URA3 gene and six copies of the c-myc tag (SRSMEQKKLISEEDLN), and used to transform yeast strains by the lithium acetate method (13). ORFs were tagged at the exact C terminus by homologous recombination, and integrants were evaluated by PCR (9). Cells containing tagged, integrated constructs of VAN1, MNN9, and ANP1 displayed normal growth characteristics, consistent with functional mannosyltransferase complexes expressed at normal levels for each gene product.
Budding and Isolation of ER-Derived Transport Vesicles.
Microsomes were prepared from strain RSY455 as described (15). In vitro vesicle-budding reactions were performed essentially as described by Barlowe et al. (16). Preparative scale incubations (0.8 ml) contained 2 μg/ml Sar1p, 8 μg/ml Sec13p complex, 4 μg/ml Sec23p complex, and ≈400 μg/ml membrane protein. After incubation at 25°C for 30 min, budding reactions were chilled at 4°C for 3 min, and vesicles were separated from donor membranes by centrifugation at 14,000 × g for 5 min. The supernatant (0.5 ml) containing the vesicles was mixed with an equal volume of 70% Nycodenz in buffer 88 and placed at the bottom of a TLS-55 ultracentrifuge tube (Beckman Coulter). Vesicles were overlayed with three Nycodenz solutions of decreasing density (0.5 ml each of 30%, 25%, and 15%) and then floated by centrifugation at 50,000 rpm for 2 h.
Analytical budding assays were performed at one-quarter scale. A peak of 35S-gpαF was detected at the 25%–15% interface that represented an enriched vesicle fraction. Gradient material (0.5 ml) in this area was removed, diluted threefold to 0.2 M Na2CO3, pH 11, and centrifuged in a TLA 100.3 ultracentrifuge rotor (100,000 × g for 30 min) to concentrate the vesicles. Pellets were dissolved in sample buffer and heated at 95°C for 2 min; vesicle membrane proteins were then analyzed on silver-stained acrylamide gels.
Protein Identification by Mass Spectrometry.
Individual protein bands were excised and subjected to in-gel trypsin digestion (17). Samples were resolved on microspray HPLC columns, and mass spectra were recorded on a liquid chromatographic quadropole ion trap mass spectrometer (Finnigan-MAT, San Jose, CA) equipped with a microelectrospray ionization source (18). Electrospray was performed at a voltage of 1.6 kV. Tandem mass spectra were acquired automatically during the entire gradient run as previously described (19).
An S. cerevisiae protein sequence database was searched directly with the tandem mass spectra using the computer program SEQUEST (20). The S. cerevisiae sequence database was obtained from the Saccharomyces Genome Database (Stanford University) and represented the complete genome sequence.
Semi-Intact Spheroplast-Based Cargo Packaging Assay.
Yeast cells grown to an A600 of 1.5 at 24°C were collected, washed in MVD (6.7 g/liter yeast nitrogen base without amino acids, Difco) twice and resuspended in supplemented MVD (2% glucose and auxotrophic supplements) to 5 A600/ml. Before labeling, cultures were shaken at 30°C for 15 min and then labeled with 35S-promix (1,200 Ci/mol, Amersham Pharmacia) at 1 mCi/60 A600 cells for 3 min at 30°C. After addition of NaN3 (20 mM final) to stop metabolic activity, cells were chilled on ice for 15 min and washed twice with cold 20 mM NaN3. For vesicle budding, semi-intact cells (10 μl of 1.5–2.0 A600) were mixed with purified COPII components in a 50-μl reaction in the presence or absence of nucleotide (21). Following incubation at 25°C for 30 min, reactions were chilled on ice and centrifuged (12,000 × g at 4°C for 5 min) to separate vesicles from donor membranes. Before centrifugation, 10 μl of the reaction was removed for analysis (total). Radiolabeled donor membranes and supernatant (medium-speed supernatant) were analyzed by immunoprecipitation.
Indirect Immunofluorescence Microscopy.
Cells were prepared for indirect immunofluorescence (22). Mouse 9E10 anti-myc and anti-HA (HA.11) antibodies (Covance, Richmond, CA) were used at 1:500 dilution. Incubation of the fixed, permeabilized yeast spheroplasts with the primary antibodies proceeded for 2 h at room temperature. Cells were washed in PBS and incubated with Cy3 (indocarbocyanine)-conjugated goat anti-mouse secondary antibodies (Jackson ImmunoResearch) at 1:500 dilution. Nuclei were stained with 30 ng/ml 4′,6-diamidino-2-phenylindole.
Fluorescence was observed with a Zeiss Axioskop microscope. Yeast vacuoles were visualized using differential interference contrast microscopy. Images were collected with 200-E CCD camera (Sony, Tokyo).
Golgi Budding Assay.
Preparation of Golgi membranes and the Golgi vesicle budding assay were performed as described by Spang and Schekman (23).
Preparation of Total Cell Lysate.
Yeast cells were grown to early mid-log phase (A600 = 1.0), and 1.5 A600 were collected by centrifugation and resuspended in 40 μl of sample buffer. Glass beads were added to the cell suspension (3/4 of the volume), and the mix was vigorously vortexed for 5–10 min to disrupt the cells. Lysates were heated at 95°C for 10 min, and fractions were used for SDS/PAGE immunoblot.
Immunoblotting and Immunoprecipitation.
For immunoblotting, we heated samples in loading buffer at 65°C for 15 min; proteins were separated on SDS/PAGE and transferred to a 0.45-μm nitrocellulose membrane (Millipore) at 25 V for 2 h. Blots were blocked in 3% nonfat dry milk in TBST (50 mM Tris-Cl, pH 7.4/150 mM NaCl/0.1% Tween 20); strips were excised and incubated overnight with anti-Sec22p polyclonal antibodies (1:5000), anti-Sec61p antibodies (1:7,500), anti-Sed5p antibodies (1:10,000), anti-Sec23p antibodies (1:2,000), anti-Anp1p antibodies (1:5,000), and anti-myc 9E10 monoclonal antibodies (1:5,000). Immunoreactive proteins were detected using an ECL kit or a Vistra kit (Vistra Systems, Amersham), and signals were quantified using a Storm 860 PhosphorImager (Molecular Dynamics).
Immunoprecipitation reactions were carried out under denaturing and native conditions. Proteins were solubilized either by heating at 65°C for 15 min in the presence of 1% SDS or by treatment with Triton X-100 (1% final) at 4°C for 1 h. Samples were adjusted to 1 ml with IP buffer (150 mM NaCl/1% Triton X-100/15 mM Tris-Cl, pH 7.5), 100 μl (5 μg) of anti-myc antibody, and 40 μl of 20% protein A-Sepharose in PBS. Reactions were incubated at 4°C overnight, and immunoprecipitated material was analyzed by SDS/PAGE and quantified using a PhosphorImager.
Results
Packaging of Yeast Golgi Proteins into COPII Vesicles Derived from ER Membranes of Cycloheximide-Treated Cells.
To better understand the molecular events involved in ER to Golgi transport, we sought to identify membrane protein components of ER-derived COPII vesicles. A number of such proteins have been previously characterized, and their function as it pertains to secretion has been studied. Yeung et al. (24) showed that the v-SNARE Sec22p and Bet1p are actively packaged in vesicles budding from the ER, even when biosynthetic cargo has been depleted from the lumen by treatment of the cells with cycloheximide. Erv14p, Emp24p, and Erv25p, proteins that actively cycle between the ER and Golgi apparatus, have also been shown to be abundant components of COPII vesicles formed from membranes of cycloheximide-treated cells (25, 26). Analysis of the vesicle protein composition by gel electrophoresis demonstrated that the vesicles contain a set of tightly associated polypeptides that can be solubilized by detergents but resist extraction at high pH (27).
To prepare sufficient quantities of vesicle membrane proteins, we performed a large-scale budding reaction using membranes depleted of biosynthetic cargo and purified COPII components as previously described (14). Microsomes were prepared from cells in mid-log phase (A600 1.0) of strain RSY455, following treatment with cycloheximide (final concentration 50 μg/ml) at 24°C for 1 h to stop protein synthesis and purge biosynthetic cargo from the ER membrane and lumen. After such treatment, α-factor precursor could no longer be detected in microsomes (data not shown). Membranes were incubated with purified COPII components and a 1 × ATP regeneration mix in the presence or absence of GTP to generate COPII vesicles. We monitored vesicle budding by performing a parallel budding assay where radioactive α-factor was loaded into the ER lumen as a cargo marker. The vesicular fraction from the budding reaction was purified on Nycodenz density gradients by flotation. Gradient material from the 15–25% Nycodenz interface was withdrawn and diluted fourfold with 0.2 M Na2CO3, pH 11, to remove contaminant proteins nonspecifically associated with the vesicles. Stripped COPII vesicles were collected by ultracentrifugation and solubilized in SDS, and samples were subjected to SDS/PAGE to examine the membrane protein profile.
Silver staining of the gel revealed an array of protein bands ranging in molecular weight from 10 to 90 kDa (not shown). Initially, we decided to focus on the proteins in the 30 to 50-kDa range. About 14 individual bands were manually excised from the acrylamide gel and subjected to mass spectrometry analysis (17) to determine the identity of the proteins. A band that was taken from the 40 to 50-kDa range yielded a fragment with amino acid sequence ELIELGFITPR that was found to correspond to the S. cerevisiae MNN9 gene product.
Mnn9p is a member of a family of proteins that has been shown to be involved in the formation and extension of mannan chains in the yeast Golgi (9). All representatives of this family are type II membrane proteins that share limited sequence homologies with each other (10). Interestingly, they form two separate complexes that have α-1,6-mannosyltransferase activity in the cis-Golgi of which Mnn9p is a shared member. The first complex consists of Van1p and Mnn9p, whereas the second includes five proteins: Mnn9p, Anp1p, Mnn10p, Mnn11p, and Hoc1p. Based on the phenotypes of mutations in individual subunits, Jungmann et al. (10) proposed a model for the sequential action of the two complexes in the synthesis of mannan structures.
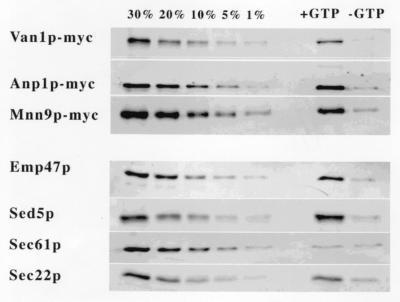
Detection of Mnn9p as a membrane protein component of COPII vesicles raised the question of whether the protein was a bona fide vesicle constituent or was simply a contaminating fragment of Golgi membranes that copurified with the vesicle fraction. Strains that had three members of the protein family, Mnn9p, Anp1p, and Van1p, epitope-tagged with the c-myc antigen and integrated at the corresponding locus to ensure a normal level of expression, were used for the preparation of microsomes from cells treated with cycloheximide. The microsomes were used as donor membranes for the in vitro generation of COPII vesicles. Quantitative immunoblot analysis showed that Mnn9p-myc, Anp1p-myc, and Van1p-myc were released in the vesicular fraction in amounts comparable to that of Sec22p, an ER to Golgi v-SNARE, and Sed5p and Emp47p, two Golgi resident proteins that recycle through the ER (refs. 6 and 7, Fig. 1 and Table 1). Furthermore, release of each protein was dependent on GTP, indicative of the normal requirement for COPII vesicle budding. In contrast, Sec61p, a resident ER membrane protein, was not detected in significant amounts in the vesicles. We consistently obtained between 20 and 30% release of Mnn9p, Anp1p, and Van1p in the vesicles, numbers that compared favorably to the percent release of Sec22p. The fact that the SNAREs and the subunits of the Mnn9p complex are packaged equally efficiently into COPII vesicles from cycloheximide-treated cells suggests that these proteins are subject to the same itinerary of anterograde and retrograde vesicular transport between the ER and Golgi.
Figure 1.
Van1p-myc, Anp1-myc, and Mnn9p-myc are enriched in COPII vesicles generated from microsomes of cycloheximide-treated cells. Vesicles (right two lanes) were generated by incubation of microsomal membranes with purified COPII components, and proteins were resolved on SDS/PAGE. For quantitation, 30%, 20%, 10%, 5%, and 1% fractions from the total membrane content were compared with vesicle proteins by immunoblot (left five lanes).
Table 1.
Quantitative release of proteins into COPII vesicles
| Proteins | % release (+GTP) | % release (−GTP) |
|---|---|---|
| Van1p-myc | 25.4 | >1 |
| Anp1p-myc | 28.6 | 1.3 |
| Mnn9-pmyc | 22.1 | 2.2 |
| Emp47p | 24.1 | 2.7 |
| Sed5p | 28.0 | 3.1 |
| Sec61p | >1 | >1 |
| Sec22p | 23.9 | 4.5 |
Two Distinct Mnn9p-Containing Protein Complexes Are Packaged into COPII Vesicles.
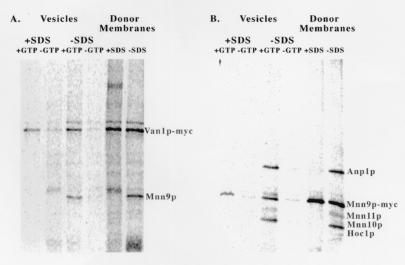
We next investigated the subunit associations of Mnn9p packaged into COPII vesicles using a modified version of the budding assay described earlier. Permeabilized spheroplasts were prepared from 35S-labeled (3 min) mid-log cells (A600 = 1.0) and used as donor source in an in vitro vesicle budding assay. Van1p-myc and Mnn9p-myc were immunoprecipitated from solubilized vesicles and donor membranes with 9E10 anti-myc monoclonal antibody under nondenaturing (1% Triton X-100) and denaturing (1% SDS) conditions.
Whereas in the denaturing incubations, individual bands corresponding to the size of Mnn9p and Van1p were observed, under nondenaturing conditions the antibodies coprecipitated several additional proteins present in both vesicles and donor membranes (Fig. 2). Native Van1p-myc immunoprecipitation also coprecipitated Mnn9p (Fig. 2A). To examine the presence of Mnn9p in the two known complexes containing this protein, we repeated the nondenaturing immunoprecipitation with membranes isolated from a VAN1 deletion strain. Mnn9p-myc coprecipitated with three new proteins (Fig. 2B).
Figure 2.
Two distinct Mnn9p-containing complexes are packaged in COPII vesicles. A strain with a myc-tagged version of Van1p and a strain with a myc-tagged version of Mnn9p and null for VAN1 were used for the preparation of permeabilized spheroplasts. COPII vesicles and total membrane fractions were solubilized under denaturing (1% SDS) or nondenaturing (1% Triton X-100) conditions. Labeled proteins were immunoprecipitated and visualized with a PhosphorImager. (A) Van1p-myc from solubilized COPII vesicles and total membranes. (B) Mnn9p-myc from solubilized COPII vesicles and total membranes.
Native immunoprecipitation using anti-Anp1p polyclonal antibodies revealed an identical ensemble of proteins associated with Anp1p (data not shown). Although we did not attempt to identify the coprecipitated protein bands, the proteins recovered in the native immunoprecipitation reaction, other than Mnn9p-myc and Anp1p, are most likely Mnn11p (45 kDa), Mnn10p (43 kDa), and Hoc1p (43 kDa). Jungmann et al. (10) found that all five proteins associate together, forming a complex with α-1,6-mannosyltransferase activity. Mnn10p and Hoc1p migrate as a single species because the difference in their molecular mass is only 0.7 kDa, and Mnn11p was seen as a less intense species, possibly because of its low methionine content.
The immunoprecipitation experiments show that Mnn9p is a component of two distinct multiprotein complexes that are preassembled in the ER and exported to the Golgi in COPII vesicles.
Mnn9p-myc, Anp1p-myc, and Van1p-myc Recycle Between ER and Golgi.
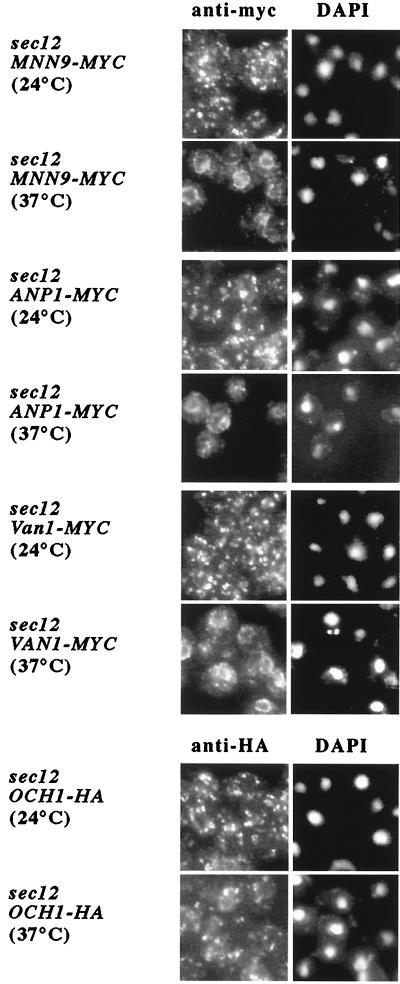
We sought a visual means of testing the prediction that members of the Mnn9p complex recycle between the ER and Golgi. We adopted the procedure of Schroder et al. (28) who documented recycling of the Golgi resident protein, Emp47p, through the ER. The basis of this strategy is to block anterograde transport to the Golgi and then to examine the intracellular distribution of a protein as it recycles to the ER. For this assay, we constructed strains that contained a sec12–1 mutation and a myc- or HA epitope-tagged version of the proteins to be examined. At a restrictive temperature (37°C), budding of COPII vesicles from the ER is blocked in a sec12 strain, but retrieval of proteins from the Golgi to the ER is not affected. To eliminate the possibility that newly synthesized proteins may contribute to the signal in the assay, we treated cells with cycloheximide before the restrictive temperature shift.
As seen in Fig. 3, at permissive temperature (24°C) Mnn9p-myc, Anp1p-myc, and Van1p-myc are distributed throughout the cell in a punctate staining pattern that is characteristic of the Golgi. However, a shift to 37°C for 30 min caused the proteins to redistribute to a ring around the nucleus (perinuclear), consistent with the ER. This redistribution is consistent with the biochemical detection of the Mnn9p subunits in COPII vesicles. In contrast, as previously reported (12, 28), the distribution of Och1p-HA in a sec12 mutant remained unchanged after the shift. Attempts to quantify the packaging or exclusion of Och1p in COPII vesicles were frustrated by a high background of Och1p in slowly sedimenting membranes released from microsomes incubated without GTP.
Figure 3.
Van1p, Anp1p, and Mnn9p, but not Och1p, recycle between ER and Golgi. Strains harboring a sec12 mutation and a myc-tagged copy of Van1p, Anp1p, or Mnn9p, or an HA-tagged version of Och1p were treated with cycloheximide (50 μg/ml) for 1 h before being shifted to 37°C for 30 min. Cultures were taken before and after the temperature shift, fixed with formaldehyde (4%), and processed for indirect immunofluorescence (22).
Wooding and Pelham (6) detected recycling of the cis-Golgi t-SNARE, Sed5p, but not of Anp1p in sec12 mutant cells incubated for 15 min at 37°C without cycloheximide. Our protocol involved cycloheximide pretreatment of sec12 cells for 1 h at 24°C and a 30-min shift to 37°C. Sed5p may have a more rapid cycling time that is revealed by 15 min at 37°C, whereas Anp1p may require longer to return to the ER from its cis-Golgi location.
Anp1p-myc and Van1p-myc Accumulate in the Vacuole When Retrograde Transport to the ER Is Blocked.
The dramatic change in intracellular distribution of the mannosyltransferases upon imposition of a sec12 block suggests that they undergo recycling through the ER, perhaps to maintain their steady-state localization in the Golgi apparatus. If this recycling is used by the cell as a mechanism for maintaining residence in the cis-Golgi, a block of retrograde transport will result in the relocation of the proteins to a distal compartment, or perhaps in their degradation. For example, when the cytosolic di-lysine motif of Emp47p is disrupted by mutation, the protein is transported to the vacuole where it is proteolyzed (28).
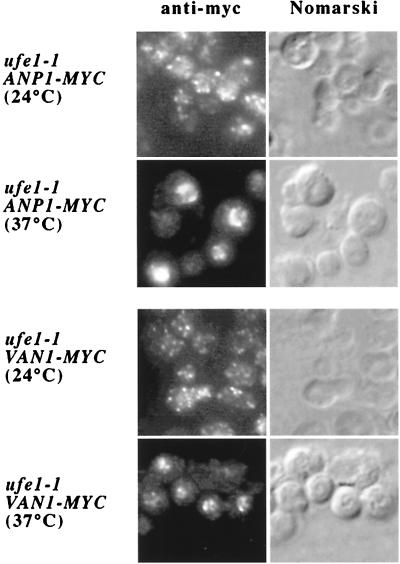
We examined the change in the intracellular localization of Van1p-myc and Anp1p-myc when retrograde transport to the ER was arrested in a ufe1–1 mutant at a restrictive temperature (37°C). Ufe1p is a yeast t-SNARE that resides on the endoplasmic reticulum and mediates retrograde traffic from the Golgi complex (12). If retrograde transport plays a role in Mnn9p mannosyltransferase complexes maintaining a steady-state Golgi distribution, failure of the proteins to undergo recycling may target them to the vacuole. To test this prediction, we used ufe1 mutant cells containing integrated forms of Anp1p or Van1p, components of the two Mnn9p-containing complexes, epitope-tagged with the c-myc antigen. Cells were grown to mid-log phase (A600 = 1.0) and treated with cycloheximide (50 μg/ml) for 1 h before a shift to the restrictive temperature (37°C) for 45 min. At the permissive temperature (24°C) in strains deficient in the vacuolar proteases (Δpep4), both Anp1p-myc and Van1p-myc were localized in a widely distributive punctate pattern, representative of the Golgi. However, a shift to 37°C caused both proteins to relocate to structures that strongly resembled the vacuolar compartment (Fig. 4). Comparison of their immunofluorescence staining pattern with the Nomarski image of the cells, where the vacuoles were seen as indentations, confirmed this identity (Fig. 4).
Figure 4.
Van1p and Anp1p accumulate in the vacuole when retrograde traffic to the ER is blocked in a ufe1 mutant. Strains harboring a ufe1 mutation and a myc-tagged copy of either Van1p or Anp1p were treated with cycloheximide (50 μg/ml) for 1 h before being shifted to 37°C for 45 min. Vacuoles were visualized by differential interference contrast microscopy.
To confirm the result that Anp1p-myc and Van1p-myc were targeted to the vacuole when their recycling to the ER was abrogated by the ufe1–1 mutation, we followed the fate of Anp1p in a ufe1–1 mutant strain with an intact Pep4p vacuolar protease. ufe1–1 mutant and phenotypically wild-type (ufe1 ts revertant) strains that were wild-type for PEP4 were treated with cycloheximide (50 μg/ml) before the shift at 37°C. A total cell lysate was prepared from cells that did and did not undergo a shift to the restrictive temperature and was used in immunoblot experiments to assess the levels of Anp1p after the different incubation conditions. A shift to 37°C for 1 h resulted in significant reduction of Anp1p levels in the ufe1–1 mutant, whereas the protein was stable in the ufe1–1 ts revertant (data not shown). In both strains and at both temperatures, the levels of Sec61p remained relatively unchanged. These data suggest that the decrease in the amount of Anp1p in the strain that is mutant for UFE1 and wild-type for PEP4 is probably attributable to its transport to the vacuole, where it becomes a substrate for the vacuolar proteases.
The finding that the two mannosyltransferase complexes travel to the vacuole, just as does Emp47p when retrograde traffic is abrogated, gives strong support to the hypothesis that recycling through the ER can be an important mechanism used by the cell to maintain a dynamic Golgi apparatus and steady-state levels of a number of Golgi resident proteins.
Mnn9p-myc Buds Directly in COPI-Coated Vesicles from Golgi-Enriched Membranes.
The results from the indirect immunofluorescence experiments strongly indicate that the two mannosyltransferase complexes undergo continuous cycles of anterograde and retrograde transport between the ER and the Golgi. We used an in vitro Golgi vesicle budding assay to test this possibility directly. Spang and Schekman (23) demonstrated that it is possible to generate COPI-coated vesicles from ER-free Golgi membranes incubated with purified COPI proteins and Arf1p in the presence of guanosine 5′-[γ-thio]triphosphate (GTP[γS]). The vesicles package cargo from the Golgi because the uptake of several proteins, such as Emp47p, Och1p, and Sec22p, was stimulated by coatomer and Arf1p but not by COPII and Sar1p.
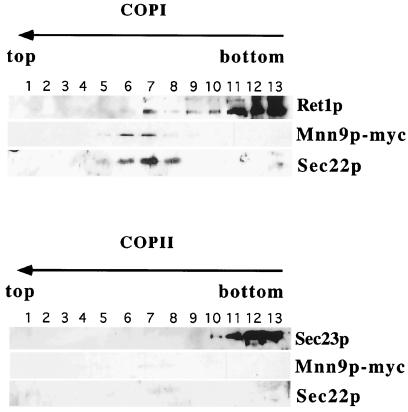
Membranes from a postnuclear pellet of lysed cells (SEY6210, where Mnn9p was tagged with the c-myc epitope) were incubated with COPI (including Arf1p and GTP[γS]) and COPII (including Sar1p and GTP[γS]). After a 30-min incubation at 25°C, the reactions were chilled and subjected to velocity sedimentation on a sucrose density gradient to separate slowly sedimenting vesicles from donor membranes. The vesicle fractions were pooled and further purified by buoyant density sedimentation. We analyzed the gradient by immunoblot examining the levels of Ret1p (αCOP), Sec22p, Sec23p (COPII subunit), and Mnn9p-myc in the fractions. Sec22p was used as the common marker for the formation of coated vesicles.
Mnn9p-myc and Sec22p displayed overlapping distributions, coincident with Ret1p in the incubations with coatomer, suggesting that the proteins were packaged in COPI-coated vesicles derived from the Golgi. In contrast, COPII and Sar1p did not generate Mnn9p or Sec22p signals in the position of coated vesicles. Fractions containing the Golgi-derived coated vesicles were pooled and subjected to a second fractionation based on coated membrane buoyant density. Samples from budding reactions that contained COPII, Sar1p, and GTP[γS] showed no signal for Mnn9p-myc or Sec22p in the fractions corresponding to coated vesicles (Fig. 5, bottom). Fractionation of pools from COPI-containing reactions, on the other hand, revealed coincident sedimentation of Mnn9p-myc, Sec22p, and Ret1p (Fig. 5, top). This pattern suggested that fractions 6–8 represented enriched COPI-coated vesicles emanating from Golgi membranes that contain both Sec22p and Mnn9p-myc as cargo.
Figure 5.
Mnn9p packaged in COPI-coated vesicles from Golgi-enriched membranes. Golgi membranes from a strain with a myc-tagged Mnn9p were incubated with coatomer and Arf1p, or with COPII components in the presence of GTP[γS].
Discussion
In this paper, we have examined the intracellular dynamics of two yeast mannosyltransferase complexes that reside in the cis-Golgi compartment. Our data suggest that although at steady state these proteins localize to the cis-Golgi, they undergo continuous recycling through the ER.
Our initial interest in the intracellular trafficking of the complexes arose from the detection of Mnn9p as a membrane protein packaged in COPII vesicles produced from ER membranes depleted of biosynthetic cargo. One possible interpretation of this finding is that Mnn9p and its partners continuously recycle between ER and Golgi. A growing list of proteins, residing at steady state in both Golgi and ER, have been described to undergo such a pattern of trafficking. This group includes the ER to Golgi v-SNAREs (12, 23), members of the p24 family (29), membrane proteins essential to the rapid transport of plasma membrane and vacuolar membrane proteins (26, 30), the mammalian lectin-binding protein ERGIC-53 (31) and its close yeast relative Emp47p (28), and the KDEL/HDEL receptor Erd2p (12). The cycling of some of these proteins between the two compartments is consistent with their functional role in ER to Golgi traffic.
We present evidence that two yeast Golgi resident mannosyltransferase complexes also actively recycle between the ER and cis-Golgi. Why should proteins with a clearly defined enzymatic function in a specific Golgi compartment be engaged in retrograde trafficking to the preceding organelle? It seems unlikely that these mannosyltransferases also act as chaperones or receptors for glycoproteins in the ER on their way to the Golgi, a role proposed for ERGIC-53 (31) and Emp47p (28), because null mutations or mutant combinations do not result in any specific secretory defect. However, a defect in the traffic of nonessential proteins may have no obvious phenotype. It seems likely that the traffic pattern of the Mnn9p complexes reflects an essential aspect of Golgi membrane biogenesis.
Recycling of resident Golgi proteins through the ER has been addressed and documented in several instances, in both yeast and mammalian cells. The yeast t-SNARE Sed5p was reported to move to and be retained in the endoplasmic reticulum in a sec12 strain upon shift to restrictive conditions, suggesting that Sed5p normally cycles between the ER and the Golgi (6). The retrograde trafficking of Sed5p was interpreted in the context of the cisternal maturation model of Golgi biogenesis as a necessary transport event for the maintenance of the cis-cisterna. Cole et al. (7) demonstrated recycling to the ER of fusion proteins that contain the luminal domain of a temperature-sensitive mutant form of VSV-G fused to targeting domains that confer Golgi or plasma membrane localization. Shift to restrictive conditions caused relocation of the chimeric proteins to the ER in the absence of new protein synthesis and apparently not as a result of unfolding of the chimeras in the Golgi. These results lead to the conclusion that transport of Golgi residents to the ER is constitutive, perhaps designed to re-expose them to the quality control machinery of the ER. Data showing slow recycling through the ER of two Golgi enzymes in HeLa cells, N-acetylgalactosaminyltransferase-2 (GalNAc-T2) and the trans/TGN β-1,4-galactosyltransferase, confirm and extend this conclusion (8). Other observations, on the other hand, argue against the generality of this pathway. For example, results from microinjection experiments demonstrated that introduction of a dominant-negative mutant of Sar1 in living cells in the presence of nocodazole caused the accumulation of the intermediate compartment marker ERGIC-53 in the ER but not that of the medial Golgi resident protein giantin (32).
The dynamic pattern of traffic of the Mnn9p-containing complexes must be reconciled with its steady-state localization to the cis-Golgi cisterna. As is the case with Sed5p (6), the recycling of the Mnn9p family of proteins to the ER may be explained by the cisternal progression model for Golgi biogenesis. Mnn9p and biosynthetic cargo molecules would originate in the ER in COPII vesicles that would fuse to generate the cis-cisterna (1). As a cis-Golgi cisterna matures to a medial cisterna, COPI vesicles would retrieve Mnn9p, Sed5p, and other obligatory recycled proteins back to the ER. Indeed, we observed Mnn9p packaged into COPI vesicles budding from Golgi membranes. Although these vesicles probably represent the retrieval event, it is possible that Mnn9p and other Golgi resident proteins are carried forward in COPI vesicles along with anterograde cargo proteins. A chimeric protein containing a cis-Golgi protein, Och1p, fused to a reporter that allows transit of the hybrid protein to distal compartments to be monitored, was shown to pass through the trans-Golgi on its way to a steady-state distribution in the cis-cisterna (33). The role of COPI vesicles or maturing Golgi cisternae in this anterograde movement has not been established.
Transport of Och1p to the trans-Golgi is in contrast to its failure to recycle through the ER. Och1p-HA remained in a punctate distribution typical of Golgi membranes in sec12 mutant cells held at the restrictive temperature for as long as 1 h. The distinct behavior of Mnn9p and Och1p may be explained by a differential recognition by the COPI retrieval mechanism. Those molecules that recycle to the ER may be more avidly captured by the COPI coat, whereas others less favored by the coat may escape recycling until later (34).
Unlike the standard retrieval signals KKXX or HDEL, no familiar motif for retrograde transport is found in the cytoplasmic amino acid sequences of the Mnn9p protein family. A potential signal, … KIKKS at the C terminus of Mnn11p, is positioned on the luminal side of the membrane, unavailable to the cytoplasmic retrieval apparatus. Clearly, other such sequences or structural features, such as transmembrane domains (35), may serve this purpose. Mutations that allow the mannosyltransferase complex to escape to the plasma membrane may identify such sorting features.
Acknowledgments
We thank Sean Munro for providing us with strains and antibodies and Hugh Pelham for the ufe1 mutant strains. This work was supported by the Howard Hughes Medical Institute and National Institutes of Health Grant GM26755.
Abbreviations
- ER
endoplasmic reticulum
- HA
hemagglutinin
- GTP[γS]
guanosine 5′-[γ-thio]triphosphate
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250472397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250472397
References
- 1.Pelham H. Trends Cell Biol. 1998;8:45–49. doi: 10.1016/s0962-8924(97)01185-9. [DOI] [PubMed] [Google Scholar]
- 2.Glick B, Malhotra V. Cell. 1998;95:883–889. doi: 10.1016/s0092-8674(00)81713-4. [DOI] [PubMed] [Google Scholar]
- 3.Rothman J, Wieland F. Science. 1996;272:227–234. doi: 10.1126/science.272.5259.227. [DOI] [PubMed] [Google Scholar]
- 4.Pelham H, Rothman J. Cell. 2000;102:713–719. doi: 10.1016/s0092-8674(00)00060-x. [DOI] [PubMed] [Google Scholar]
- 5.Bonfanti L, Mironov A, Jr, Martinez-Menarguez J, Martella O, Fusella A, Baldassarre M, Buccione R, Geuze H, Mironov A, Luini A. Cell. 1998;95:993–1003. doi: 10.1016/s0092-8674(00)81723-7. [DOI] [PubMed] [Google Scholar]
- 6.Wooding S, Pelham H. Mol Biol Cell. 1998;9:2667–2680. doi: 10.1091/mbc.9.9.2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole N, Ellenberg J, Song J, DiEuliis D, Lippincott-Schwartz J. J Cell Biol. 1998;140:1–15. doi: 10.1083/jcb.140.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Storrie B, White J, Rottger S, Stelzer E, Suganuma T, Nilsson T. J Cell Biol. 1998;143:1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jungmann J, Munro S. EMBO J. 1998;17:423–434. doi: 10.1093/emboj/17.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jungmann J, Rayner J C, Munro S. J Biol Chem. 1999;274:6579–6585. doi: 10.1074/jbc.274.10.6579. [DOI] [PubMed] [Google Scholar]
- 11.Raymond C, O'Hara P, Eichinger G, Rothman J, Stevens T. J Cell Biol. 1990;111:877–892. doi: 10.1083/jcb.111.3.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis M, Pelham H. Cell. 1996;85:205–215. doi: 10.1016/s0092-8674(00)81097-1. [DOI] [PubMed] [Google Scholar]
- 13.Guthrie C, Fink G. Methods Enzymol. 1991;194:21–109. [Google Scholar]
- 14.Sikorski R, Hieter P. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wuestehube L J, Schekman R. Methods Enzymol. 1992;219:124–136. doi: 10.1016/0076-6879(92)19015-x. [DOI] [PubMed] [Google Scholar]
- 16.Barlowe C, Orci L, Yeung T, Hosobubuchi M, Hamamoto S, Salama N, Rexach M, Ravazzola M, Amherdt M, Schekman R. Cell. 1994;77:895–707. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 17.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 18.Gatlin T, Kleeman G, Hays L, Yates J. Anal Biochem. 1998;263:93–101. doi: 10.1006/abio.1998.2809. [DOI] [PubMed] [Google Scholar]
- 19.Link A J, Hays L G, Carmack E B, Yates J. Electrophoresis. 1997;18:1314–1334. doi: 10.1002/elps.1150180808. [DOI] [PubMed] [Google Scholar]
- 20.Eng J K, McCormack A L, Yates J. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 21.Doering T L, Schekman R. EMBO J. 1996;15:182–191. [PMC free article] [PubMed] [Google Scholar]
- 22.Chuang J, Schekman R. J Cell Biol. 1996;135:597–610. doi: 10.1083/jcb.135.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spang A, Schekman R. J Cell Biol. 1998;143:589–599. doi: 10.1083/jcb.143.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeung T, Barlowe C, Schekman R. J Biol Chem. 1995;270:30567–30570. doi: 10.1074/jbc.270.51.30567. [DOI] [PubMed] [Google Scholar]
- 25.Belden W, Barlowe C. J Biol Chem. 1996;271:26939–26946. doi: 10.1074/jbc.271.43.26939. [DOI] [PubMed] [Google Scholar]
- 26.Powers J, Barlowe C. J Cell Biol. 1998;142:1209–1222. doi: 10.1083/jcb.142.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rexach M, Latterich M, Schekman R. J Cell Biol. 1994;126:1133–1148. doi: 10.1083/jcb.126.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroder S, Schimmoller F, Singer-Kruger B, Riezman H. J Cell Biol. 1995;131:895–912. doi: 10.1083/jcb.131.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickel W, Sohn K, Bunning C, Wieland F. Proc Natl Acad Sci USA. 1997;94:11393–11398. doi: 10.1073/pnas.94.21.11393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill K, Stevens T. Mol Biol Cell. 1994;5:1039–1050. doi: 10.1091/mbc.5.9.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kappeler F, Klopfenstein D, Foguet M, Paccaud J P, Hauri H P. J Biol Chem. 1997;272:31801–31808. doi: 10.1074/jbc.272.50.31801. [DOI] [PubMed] [Google Scholar]
- 32.Shima D, Cabrera-Poch N, Pepperkok R, Warren G. J Cell Biol. 1998;141:955–966. doi: 10.1083/jcb.141.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris S L, Waters G. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glick B, Elston T, Oster G. FEBS Lett. 1997;414:177–181. doi: 10.1016/s0014-5793(97)00984-8. [DOI] [PubMed] [Google Scholar]
- 35.Letourneur F, Cosson P. J Biol Chem. 1998;273:33273–33278. doi: 10.1074/jbc.273.50.33273. [DOI] [PubMed] [Google Scholar]