A reduced risk of primary malignant adult brain tumors is observed among people reporting asthma, hayfever and other allergic conditions; however, findings may be attributable to prediagnostic effects of tumors or recall bias. To determine whether asthma and allergic condition polymorphisms are inversely related to glioblastoma multiforme (GBM) risk, we conducted a population-based case-control study of 111 GBM patients and 422 controls. We identified five single nucleotide polymorphisms (SNPS) on three genes previously associated with asthma (interleukin (IL) 4RA, IL-13, ADAM33) and one gene associated with inflammation (COX-2). Confirming previous literature, we found that self-reported asthma, eczema, and fever are inversely related to GBM (e.g., asthma odds ratio (OR)=0.64, 95% confidence interval (CI), 0.33, 1.25). In addition, IL-4RA ser478pro TC, CC and IL-4RA gln551arg AG, AA are positively associated with GBM (OR=1.64, 95% CI=1.05, 2.55; 1.61, 95% CI 1.05, 2.47) while IL-13-1112 CT, TT is negatively associated with GBM (0.56, 95% CI=0.33, 0.96). Each of these polymorphism-GBM associations is in the opposite direction of a corresponding polymorphism-asthma association, consistent with previous findings that self-reported asthmatics and people with allergic conditions are less likely to have GBM than are people who do not report these conditions. Because we used germline polymorphisms as biomarkers of susceptibility to asthma and allergic conditions, our results cannot be attributed to recall bias or effects of GBM on the immune system. However, our findings are also consistent with associations between IL-4RA, IL-13 and GBM that are independent of their role in allergic conditions.
Glioblastoma multiforme (GBM) is the most common primary malignant brain tumor accounting for 23.0% of all primary brain and central nervous system tumors and 51.9% of all gliomas. The median age at diagnosis is 65 years and the age-adjusted incidence rate is 3.24/100,000 although it increases to 13.74/100,000 for ages 65–74. The average five year relative survival rate from time of diagnosis for GBM is only 3.3 % and is lower for people over age 65 years at diagnosis (0.3%) (1).
Although the etiology of this tumor is unknown, there is evidence for a role of the immune system in GBM growth and development (2). In particular, a consistent inverse association between self-reported allergic conditions and glioma has been recently reported in a cohort and several case-control studies (3–7). While the cohort study had rather small numbers of cases, case-control results can be subject to preclinical effects of the tumor on the immune system or to errors in recall among brain tumor patients who may have cognitive deficits (depending on tumor lateralization and type of therapy (8)). Wiemels et al. take another approach to the question of whether allergic disease reduces brain tumor risk by comparing serum IgE levels of cases and controls (9). Although they find that glioma patients have lower serum IgE levels than do controls, the possibility that the tumor itself or its treatment may affect serum IgE levels cannot be excluded. In general, definitive evidence of associations between immunologic biomarkers and glioma risk must be based on measurements taken well before the time of tumor diagnosis. To avoid the influence of the brain tumor itself on an indicator of susceptibility to asthma or allergic conditions, we used germline polymorphisms previously associated with asthma and other allergic conditions as biomarkers of susceptibility. Clearly these genetic variants cannot be influenced by the presence of a brain tumor.
The rationale for using polymorphisms to test the validity of asthma and allergy self-reports is not their superior sensitivity or specificity to self-report because most individual polymorphisms are neither sensitive nor specific indicators of complex diseases (10). Furthermore, asthma, and to a lesser extent allergy, self-reports are relatively sensitive indicators of these conditions (11, 12). Rather, the advantage of using germline polymorphisms as biomarkers of susceptibility is that, unlike asthma self-report or possibly IgE levels, they cannot be influenced by presence of glioblastoma multiforme (GBM) and therefore case and control asthma and allergy measurements are subject to the same degree of error. While this error may reduce the size of measures of association between asthma or allergic conditions and GBM, it will not cause the measure of association to change direction, as may happen when case and control allergic conditions are each measured with a different degree of error (13).
From the extensive literature on asthma and allergic condition polymorphisms, we selected genetic variants that are consistently associated with asthma or allergic conditions in at least two populations and whose functions relate to either glioma development or normal brain physiology. Two of the polymorphisms that meet these criteria are ser478pro and gln551arg on the interleukin (IL)-4 receptor alpha gene (IL-4RA) (14). These IL-4RA polymorphisms are associated with asthma and allergies in two and eleven studies respectively, each study based on different populations (15). Two additional polymorphisms consistently associated with asthma and other allergic conditions are arg130gln (16) and -1112 C/T (17) on the IL-13 gene. Associations between these two IL-13 polymorphisms and asthma or allergic conditions have been identified in nine and four studies respectively (15). IL-4 and IL-13 are cytokines that share immunoregulatory functions and a common IL-4RA chain on their receptors. They both play a central role in allergy by inducing IgE synthesis and both can inhibit inflammatory cytokines (18, 19). Importantly for the present research, IL-4 and IL-13 show strong antitumor activity in mice and inhibit proliferation of astrocytoma and low grade glioma cell lines (20, 21).
We also looked at the T1 polymorphism of a newly identified asthma gene, ADAM33 (22), that was found to be associated with asthma in three different populations (22–24). This gene is a member of a family of matrix metalloproteases, extracellular proteases that participate in matrix degradation and glioblastoma invasion (25).
Finally, the last polymorphism that we evaluated is found on the cyclooxygenase-2 (COX-2) gene (−765GC) and is associated with post-surgical C-reactive protein levels (26). (C-reactive protein is produced in response to inflammatory cytokines during the acute phase response.) Although IL-4 and IL-13 function as pro-inflammatory mediators in asthma, allergy and helminth infection, these cytokines also have anti-inflammatory properties resulting, in part, from their inhibition of both cell-mediated immune responses (27, 28) and COX-2 expression (29). In addition, our selection of the COX-2 gene was based on our previous findings of an inverse association between non-steroidal anti-inflammatory drug use and GBM (30).
Methods
Glioma and meningioma cases that occurred in Sweden between September 1, 2000 and August 31, 2002 were identified in collaboration with brain tumor treatment centers. Regional cancer registries were searched approximately every third month for additional case identification, to make sure that no cases had been missed. This system was effective in reducing the time between diagnosis and interview as indicated by the fact that proxy interviews were necessary for only 9% of glioma and 3% of meningioma cases (a low proportion of proxy interviews when compared to other brain tumor studies (7) ). We restricted our study to the most common type of adult glioma, GBM, to reduce genetic heterogeneity
Controls were randomly selected within strata defined by glioma or meningioma patients’ age, sex, and geographic region from a continuously updated population registry. Computer-assisted interviews were conducted by research nurses. Information collected on allergies included questions about whether the participant had been diagnosed with asthma, hayfever, or eczema and the length of time these conditions were present. In addition, data were collected on allergy medication including type of medication and frequency of use. Although information concerning type of glioma of individuals who refused to participate was not available to investigators (to protect nonrespondents’ privacy), we know that of the 499 glioma patients, 73.9% agreed to be interviewed and of the 956 potential controls identified, 66.2% agreed to be interviewed. However, once interviewed, slightly more controls (66.7%) than GBM cases (63.8%) consented to having their blood drawn.
Statistical Analysis
We used unconditional logistic regression to compare case and control polymorphism prevalence adjusted for age and sex. The variable geographic region had no influence on our findings so we eliminated it from our regression models.
Genotyping
Dynamic Allele-Specific Hybridization (DASH) was performed as previously described (31–35). For this, two PCR primers and one DASH probe per target mutation/SNP were designed by means of custom software (36) provided by DynaMetrix Ltd (UK). These oligonucleotides were provided and HPLC purified, by Biomers GmbH (Germany). The DASH PCRs entailed amplifying 50–90 bp genomic fragments spanning the variant of interest, with one of the primers carrying a 5′-biotin label. Amplifications were performed in 5uL volume, containing 1–2ng genomic DNA, 0.38uM biotinylated primer, 0.75uM non-biotinylated primer, 0.03 units AmpliTaq Gold (PE Biosystems, CA), 10% dimethylsulphoxide, 1 × AmpliTaq Gold Buffer including 1.5mM of MgCl2 (Applied Biosystems, CA) and 0.2mM each dNTP. Thermal cycling was conducted on an MBS 384 device (Thermo-Hybaid, Ashford, UK) as follows; 1 × (10 minutes at 94 C), 35× (15 seconds at 94 C, 30 seconds at annealing temperature). To verify successful amplification, 0.5ul of several randomly chosen samples were examined on a 3.0% low-melt agarose gel.
DASH analysis of the PCR product was conduced on membrane macro-arrays, using the DASH-2 protocol (34). Briefly, this entailed transferring samples to the membrane array by centrifugation or robotic gridding (37). Resulting individual arrays with up to 9,600 distinct samples/features were rinsed in 0.1 M NaOH to denature the PCR products, and then exposed to 2ml HE buffer (0.1 M HEPES, 10mM EDTA, pH 7.9) containing 4nmol of suitable probe, itself end-labelled with ROX. After heating to 85 C and cooling to room temperature, the membrane was briefly rinsed in HE buffer. The array was then soaked in 40ml HE-buffer containing SYBR GreenI dye at 1:20,000 dilution for up to 3 hours. Using a DASH-2 device (DynaMetrix Ltd, UK), the membrane was taken through a DASH heating ramp (heating at 3 C/minute from room temperature to 85 C) whilst fluorescence from the ROX acceptor dye on the probe was monitored. Data were collected at interval of 0.5 C. Fluorescence changes with temperature (DNA melting profiles) were used to distinguish different alleles, and this was done by means of the DASH-2 device software which uses negative derivatives of fluorescence against temperature to reveal peaks of denaturation rate (target-probe melting temperatures; Tm) and thereby automatically assign DNA samples into genotype groups. Finally, a random sample of 15% of all DNA samples was reassayed and the genotype assignment confirmed. An error rate of less than one percent was observed.
No samples were sequenced since DASH is an extensively validated method (31–35). Quality control data included in the preceding references confirms the error rate of the method across many different SNPs is <0.1%.
Results
In Table 1 the distributions of age, sex, and allergic condition variables are shown for interviewed cases and controls who did and did not consent to having their blood drawn. Overall, results are similar except when the sample size for a category is small, as is the case for the variable food allergy among people who did not have their blood drawn. For both groups there are consistent inverse relationships between self-reported asthma, eczema, fever during the ten years prior to the interview and GBM. The reason that cases and controls differ with respect to age (median age for all participants: cases 56 years, controls 53 years) and sex (for total participants: male cases 59.18 %, male controls 48.02 %) is that we include controls who were initially matched to age and sex of all glioma and meningioma cases (see Methods Section).
Table 1.
Demographic variables and age- and sex-adjusted self-reported physician-diagnosed asthma and allergic condition and inflammation variables from population-based Swedish case-control study of glioblastoma multiforme (GBM, 2000–2002).
| All study particpants | Interviewed only | Interviewed and collected blood sample | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Demographic variables | Cases n=174 | Controls n=633 | Cases n=63 | Controls n=211 | Cases n=111 | Controls n=422 | |||
| Median Age (inter-quartile range) | 56 (49, 62) | 53 (43, 60) | 57 (53, 63) | 52 (41, 58) | 55 (46, 61) | 54 (44, 61) | |||
| Sex (% male) (95 % CI) | 59.18 (51.89, 66.49) | 48.02 (44.14, 51.90) | 58.73 (46.48, 70.78) | 53.08 (46.34, 59.82) | 59.46 (50.29, 68.63) | 45.50 (40.75, 50.23) | |||
| Allergic and Inflammatory variables | Cases 1 n | Controls 1 n | Odds ratios (95% CI) | Cases 1 n | Controls 1 n | Odds ratios (95% CI) | Cases 1 n | Controls 1 n | Odds ratios (95% CI) |
| None | 163 | 570 | 1.00 | 57 | 186 | 1.00 | 106 | 384 | 1.00 |
| Asthma | 11 | 63 | 0.64 (0.33, 1.25) | 6 | 25 | 0.76 (0.29, 2.00) | 5 | 38 | 0.53 (0.20, 1.38) |
| None | 147 | 532 | 1.00 | 52 | 176 | 1.00 | 95 | 356 | 1.00 |
| Hayfever | 27 | 101 | 0.98 (0.62, 1.57) | 11 | 35 | 1.16 (0.54, 2.50) | 16 | 66 | 0.82 (0.47, 1.60) |
| None | 141 | 490 | 1.00 | 49 | 159 | 1.00 | 92 | 331 | 1.00 |
| Asthma or hayfever | 33 | 143 | 0.82 (0.54, 1.26) | 14 | 52 | 0.88 (0.44, 1.77) | 19 | 91 | 0.77 (0.45, 1.34) |
| None | 144 | 485 | 1.00 | 47 | 172 | 1.00 | 97 | 313 | 1.00 |
| Eczema | 29 | 148 | 0.67 (0.43, 1.05) | 16 | 39 | 1.54 (0.97, 3.08) | 13 | 109 | 0.40 (0.21, 0.74) |
| None | 160 | 573 | 1.00 | 59 | 157 | 1.00 | 101 | 378 | 1.00 |
| Contact allergy | 12 | 46 | 1.02 (0.53, 2.00) | 2 | 9 | 1.06 (0.20, 5.57) | 10 | 37 | 1.09 (0.52, 2.29) |
| None | 161 | 583 | 1.00 | 60 | 196 | 1.00 | 101 | 387 | 1.00 |
| Food allergy | 11 | 37 | 1.23 (0.61, 2.49) | 1 | 10 | 0.63 (0.07, 5.31) | 10 | 27 | 1.55 (0.72, 3.30) |
| None | 151 | 523 | 1.00 | 55 | 182 | 1.00 | 96 | 341 | 1.00 |
| Allergy medication | 23 | 110 | 0.78 (0.48, 1.27) | 8 | 29 | 1.04 (0.43, 2.50) | 15 | 81 | 0.70 (0.38, 1.28) |
| Never (last 10 years) | 43 | 102 | 1.00 | 15 | 35 | 1.00 | 28 | 67 | 1.00 |
| Fever once or less per year | 83 | 410 | 0.54 (0.35, 0.84) | 32 | 132 | 0.68 (0.37, 1.45) | 51 | 278 | 0.46 (0.27, 0.79) |
| Fever 2 or more times per year | 24 | 92 | 0.78 (0.43, 1.40) | 11 | 25 | 1.37 (0.51, 3.68) | 13 | 67 | 0.53 (0.25, 1.14) |
Differences in numbers of cases and controls among variables are attributable to missing responses.
The second column of Table 2 shows previously reported associations between five polymorphisms and asthma (14, 16, 17, 22). Column 3 contains results from the present study for associations of these same polymorphisms with GBM. For each SNP, odds ratios for asthma that are greater than or less than the null value (OR= 1.0) correspond to odds ratios for GBM that are greater than or less than the null value. However, because reference categories that we use for the two diseases differ, our findings indicate that these asthma susceptibility polymorphisms have opposite relationships with asthma and GBM. For example, individuals with the TT polymorphism for the IL-4RA ser478pro SNP appear to be more susceptible to asthma and less susceptible to GBM than are individuals with the TC or CC variants. Also reported at the bottom of the second column are maximum levels of C-reactive protein after coronary by-pass surgery stratified on alleles of the COX-2 -765GC polymorphism (26). The odds ratio characterizing the association between the COX-2 polymorphism and GBM indicates that higher post-surgical levels of C-reactive protein, perhaps indicating a stronger acute phase inflammatory response, are associated with a greater risk of GBM.
Table 2.
Sex- and age-adjusted associations between asthma polymorphisms and glioblastoma multiforme (GBM) from population based Swedish case-control study (2000–2002), and comparisons with previously published associations between polymorphisms and asthma or inflammatory conditions.
| Gene (SNP) | Previous asthma literature | GBM (present study) | Previous asthma literature | ||||
|---|---|---|---|---|---|---|---|
| Asthma OR (95% CI) | GBM OR (95% CI) | GBM OR (trend test) | GBM Cases1, 2 n (%) | GBM Controls1, 2 n (%) | Asthma Cases (%) | Asthma Controls (%) | |
| IL-4RA (rs1805015, ser478pro) | Asthma (14) | Asthma (14 ) | |||||
| TT | 1.96 (1.13, 3.44) | 1.00 | 1.00 | 64 (59) | 288 (70) | 77 | 63 |
| TC | 1.00 | 1.64 (1.05, 2.55) | 1.73 | 40 (37) | 107 (26) | 20 | 33 |
| CC | 1.09 | 4 (4) | 16 (4) | 3 | 4 | ||
| p = 0.07 (trend) | |||||||
| IL-4RA (rs1801275, gln551arg) | Asthma (14) | Asthma (14) | |||||
| AA | 1.25 (0.72, 2.01) | 1.00 | 1.00 | 53 (49) | 243 (60) | 68 | 64 |
| AG | 1.00 | 1.61(1.05, 2.47) | 1.52 | 45 (41) | 236 (34) | 26 | 32 |
| GG | 2.09 | 11 (10) | 24 (6) | 5 | 4 | ||
| p = 0.02 (trend) | |||||||
| IL-13 (rs20541, arg130gln) | Asthma (16) | Asthma (16) | |||||
| GG | 0.48 (0.30, 0.76) | 1.00 | 1.00 | 74 (67) | 254 (60) | 36 | 54 |
| AG | 1.00 | 0.75 (0.48, 1.17) | 0.81 | 33 (30) | 144 (33) | 48 | 39 |
| AA | 0.41 | 3 (3) | 24 (7) | 16 | 7 | ||
| p= 0.12 (trend) | |||||||
| IL-13 (rs1800925, -1112C/T) | Asthma (17) | Asthma (17) | |||||
| CC | 0.51 (0.31, 0.84) | 1.00 | 1.00 | 84 (80) | 277 (69) | 58 | 73 |
| CT | 1.00 | 0.56 (0.33, 0.96) | 0.55 | 18 (17) | 111 (28) | 37 | 25 |
| TT | 0.64 | 3 (3) | 15 (4) | 5 | 2 | ||
| p= 0.05 (trend) | |||||||
| ADAM33 (rs2280091, T1) | Asthma (22) | Asthma (22) | |||||
| TT | 0.24 (0.15, 0.39) | 1.00 | 1.00 | 82 (77) | 297 (71) | 58 | 85 |
| TC | 1.00 | 0.82 (0.50, 1.33) | 0.85 | 24 (22) | 106 (26) | 37 | 14 |
| CC | 0.56 | 2 (1) | 13 (3) | 6 | 1 | ||
| p= 0.35 (trend) | |||||||
| COX-2 -765GC | C-reactive protein (CRP) levels (26)) 1 | CRP (26)) 3 | |||||
| GG | 173.64 | 1.00 | 1.00 | 80 (74) | 286 (72) | 75 | |
| GC | 149.57 | 0.84 (0.45, 1.57) | 0.90 | 27 (25) | 105 (26) | 24 | |
| CC | 0.52 | 1 (1) | 8 (2) | 1 | |||
| p= 0.53 (trend) | |||||||
Differences in numbers of cases and controls among polymorphisms reflect failure to identify genotype for some DNA samples.
Numbers in parentheses are percentages of genotypes for GBM cases and controls excluding people reporting physician-diagnosed asthma or hayfever.
Papafili et al. (26) do not report C-reactive protein distribution of case group.
Further details describing associations between each of the six polymorphisms and GBM are reported in columns 4–8 of Table 2. The genotype distribution of GBM cases and controls is shown together with the same distributions for asthma cases and controls from previous literature. For each of the first five polymorphisms, if GBM and asthma cases and controls are ranked by the percentage having the most common polymorphism, GBM and asthma cases are at the extremes and controls lie between them. For example, for the IL-13 -1112 C/T polymorphism, 84% of GBM cases have the CC genotype, while only 58% of asthma cases are in this category. This pattern is reversed for IL-4RA ser478pro where a gradient from GBM (59% TT genotype) to asthma cases (77% TT genotype) can be seen. The order of these percentages does not change when people reporting asthma or other allergic conditions are excluded from the GBM case group (not shown). GBM asthma and control genotype distributions are similar, except those for the ADAM33, T1 SNP (22). These distributions remain similar when people with asthma or other allergic conditions are excluded from the GBM control group (not shown).
In Figure 1 the previously reported effect of the interaction between IL-4RA ser478pro and IL-13 -1112 C/T on asthma risk is shown (14). Consistent with findings in Table 2, GBM odds ratios are inversely associated with previously reported asthma odds ratios. Although we find no similar interaction in the present study among GBM controls (the middle bar labeled 4 is not higher than the sum of middle bars labeled 2 and 3), asthma genotype odds ratios in our sample are similar to those previously reported. However, there is little additional evidence of associations between individual asthma susceptibility polymorphisms reported in Table 2 and self-reported asthma, hayfever, fever, or any of the allergic conditions shown in Table 1 among controls in our data (not shown). Nor did our findings change when asthma or allergic condition variables were stratified on age at onset or duration of disease.
Figure 1.
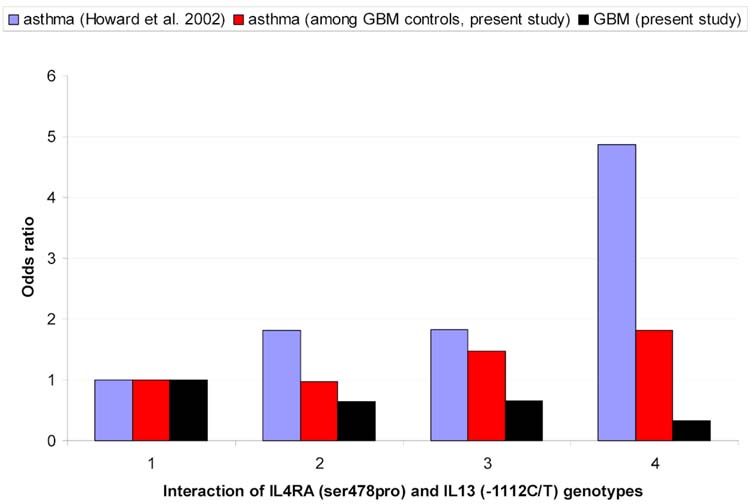
Interaction between IL-4RA (ser478 pro) and IL-13 (-1112C/T) genotypes and : 1) asthma in Dutch population (14); 2) GBM in Swedish Population and 3) asthma among GBM controls in Swedish population.
1= IL-4RA (TC, CT)/IL-13 (CC)
2= IL-4RA (TT)/IL-13 (CC)
3= IL-4RA (TC, CC)/IL-13(CT,TT)
= IL-4RA (TT)/IL-13 (CT,TT)
Discussion
Using previously identified polymorphisms as biomarkers of susceptibility to asthma, we found an association between three of these biomarkers (IL-4RA ser478pro TC, CC, IL-4RA gln551arg AG, AA, and IL-13 -1112 CT, TT) and GBM. In addition, as predicted by both previous epidemiologic and asthma susceptibility polymorphism literature (3–7, 14, 16, 17, 22, 26, 38), odds ratios for GBM were the inverse of those for asthma. Ours is the first study to suggest the validity of the consistently observed inverse association between self-reported asthma and GBM because we used biomarkers that cannot be altered by the presence of GBM. By basing our selection of polymorphisms on strong a-priori epidemiologic and genetic evidence (2), we reduce the risk of false-positive results which so often characterize the genetic polymorphism literature (39). However, because the individual polymorphisms that we identified are not related to self-reported asthma or allergic conditions among controls in our data, our results may reflect a relationship between IL-4RA, IL-13 and GBM that is independent of their role in asthma and other allergic conditions.
Asthma is an inflammatory disease of lung airways that may or may not have an allergic component. Here we define allergy as immune reactions to common environmental proteins characterized by elevated IgE levels and distinguished by allergic symptoms from atopy. Although asthma and other allergic conditions such as hayfever are clinically distinct, they are genetically linked. For example, the AG and AA genotypes of the IL-13 arg130gln polymorphism are associated with atopy, allergen-specific IgE, and asthma (16). A further manifestation of this genetic association among allergic conditions is that within the same family individuals may have allergic diseases of different target organs such as asthma, hayfever, and eczema (40). Because of this genetic link, inferences from our findings can be extended from asthma to other allergic conditions.
Possible reasons for our failure to find expected associations between individual polymorphisms and allergic conditions among controls include both the relatively small numbers of controls reporting asthma or allergic conditions and misclassification of these conditions. In most case-control studies of asthma, all study participants (including controls) are actively screened for asthma by spirometry, reversibility to albuterol or bronchial responsiveness testing to albuterol, e.g., (24). While in the present study asthma and other allergic conditions are identified only by self-report of physician diagnosis. Errors in measurement of allergic conditions may also explain the reason for relatively weak associations between allergic conditions (Table 1, measured with error) and GBM as opposed to relatively strong associations between allergic disease susceptibility polymorphisms (Table 2, measured with little error) and GBM (41).
Additional evidence for an association between allergic conditions and GBM includes the findings of Weimels et al. that glioma cases have lower serum IgE levels than do controls (9), however, their findings need to be validated with prospectively collected data. Perhaps related to the role of allergic disease in the etiology of GBM, are findings from a clinical trial of an IgE blocking drug for asthmatics (Omalizumab) that show a higher rate of solid tumors in the treatment group compared to the control group (rate ratio for solid tumors excluding non-melanoma skin cancer, 3.8, 95% CI 0.9, 34.3 ) (42). Whether brain tumors are included among the solid tumors identified is not known. In addition because the number of tumors found is relatively small (Omalizumab group, n=16, control group, n=5), random allocation of study participants may not have equally distributed individuals with differing prior risks of cancer to the treatment and control groups.
It is also possible that IL-4RA and IL-13 play a role in GBM development that is independent of their roles in asthma and allergic conditions. The function of IL-4 and IL-13 in brain tumor growth has been the subject of several investigations. Barna et al. found that 3 normal astrocytic, 2 low-grade astrocytoma and 3 out of 4 GBM cell lines that she evaluated express IL-4Ralpha receptors. However, IL-4 suppresses DNA synthesis and cell proliferation only in the normal astrocytic and low-grade astrocytoma cell lines but not in the GBM cell lines (21). Their results suggest that IL-4 may interfere with progression from lower to higher grade glioma but may have no role in de novo glioblastomas (43). Saleh et al. attribute the growth-inhibiting properties of mouse IL-4 on implanted C6 glioma cell lines to its ability to promote eosinophil infiltration and to inhibit angiogenesis. Furthermore, Saleh et al. observe that implantation of C6 cell gliomas that produce IL-4 retrovirus are rapidly eradicated in rats (44, 45). Consistent with Saleh et al.’s results, Volpert et al. show that IL-4 blocks corneal neovascularization by fibroblast growth factor in mice as well as inhibiting the migration of cultured bovine and human microvascular cells (46). However, because IL-4 is species-specific the above findings may not be directly applicable to human disease.
IL-13 shares the IL-4R alpha signaling receptor chain with IL-4, and like IL-4, inhibits astrocyte and low grade astrocytoma proliferation but does not inhibit GBM cell proliferation (20). In addition, Shin et al. found that IL-13 controls brain inflammation by inducing death of activated microglia (major inflammatory cells of the central nervous system) (47). Further evidence for a role of IL-13 in GBM development or growth comes from the overexpression of IL-13Ralpha2 receptors in glioblastoma tissue (48, 49). The role of these receptors in GBM cells is to inhibit IL-13 and IL-4 dependent signal transduction (50).
Not only do IL-4 and Il-13 not inhibit cell growth in GBM (as they appear to do in lower grade gliomas) but also IL-4 or IL-13 stimulation of IL-4RA contributes to the pathogenesis of GBM cells (51). In addition, Madhankumer and Debinski find that IL-13 stimulates a signaling cascade that increases the oncogenic potential of GBM cells (52). There is also extensive evidence indicating that IL-13 downregulates antitumor response by indirectly suppressing production of cytotoxic T cell production e.g., (53). (Although these effects have not been observed for brain tumors, they may be in the future.) As a consequence, Berzofsky et al. have suggested a possible benefit of IL-13 withdrawal as a means of both increasing anti-tumor immunity and the efficiency of vaccines (54). There are also proposals that asthma and allergic conditions be prevented or treated by inhibiting IL-13 production (55, 56).
Our findings of an inverse association between IL-4RA and IL-13 polymorphisms, and GBM suggest that IL-13 withdrawal therapies to treat allergic conditions may interfere with possible tumor-inhibiting effects of this cytokine in the brain. However, if these tumor inhibiting effects occur early in GBM development then perhaps reducing endogenous IL-13 production after GBM development may still be beneficial.
Further research should include investigation of additional allergy susceptibility polymorphisms, use of biomarkers of allergic conditions measured before GBM diagnosis and validation of asthma and allergic condition self-reports. Overall, one of the goals of subsequent research should be to determine whether allergic conditions per se or their related cytokines affect GBM risk.
Acknowledgments
The authors thank Arthur E. Varner, M.D. for his helpful comments. This research was supported by grants from the United States National Cancer Institute (R03CA103379), the Swedish Council for Working Life and Social Research, the Swedish Cancer Society, the Swedish Research Council, the European Union Fifth Framework Program, and the International Union against Cancer (UICC).
References
- 1.CBTRUS. Statistical Report: Primary Brain Tumors in the United States, 1995–1999.: Central Brain Tumor Registry of the United States, 2002.
- 2.Fisher JL, Schwartzbaum JA, Wrensch M, Berger MS. Evaluation of epidemiologic evidence for primary adult brain tumor risk factors using Evidence-Based Medicine. Review of Neurological Surgery by Evidence-Based Medicine Standards: S Karger AG, 2005. [DOI] [PubMed]
- 3.Schlehofer B, Blettner M, Becker N, Martinsohn C, Wahrendorf J. Medical risk factors and the development of brain tumors. Cancer. 1992;69:2541–47. doi: 10.1002/1097-0142(19920515)69:10<2541::aid-cncr2820691025>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 4.Schlehofer B, Blettner M, Preston-Martin S, et al. Role of medical history in brain tumour development. Results from the international adult brain tumour study. Int J Cancer. 1999;82:155–60. doi: 10.1002/(sici)1097-0215(19990719)82:2<155::aid-ijc1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 5.Wiemels JL, Wiencke JK, Sison JD, Miike R, McMillan A, Wrensch M. History of allergies among adults with glioma and controls. Int J Cancer. 2002;98:609–15. doi: 10.1002/ijc.10239. [DOI] [PubMed] [Google Scholar]
- 6.Brenner AV, Linet MS, Fine HA, et al. History of allergies and autoimmune diseases and risk of brain tumors in adults. Int J Cancer. 2002;99:252–9. doi: 10.1002/ijc.10320. [DOI] [PubMed] [Google Scholar]
- 7.Schwartzbaum J, Jonsson F, Ahlbom A, et al. Cohort studies of association between self-reported allergic conditions, immune-related diagnoses and glioma and meningioma risk. Int J Cancer. 2003;106:423–8. doi: 10.1002/ijc.11230. [DOI] [PubMed] [Google Scholar]
- 8.Scheibel RS, Meyers CA, Levin VA. Cognitive dysfunction following surgery for intracerebral glioma: influence of histopathology, lesion location, and treatment. J Neurooncol. 1996;30:61–9. doi: 10.1007/BF00177444. [DOI] [PubMed] [Google Scholar]
- 9.Wiemels JL, Wiencke JK, Patoka J, et al. Reduced immunoglobulin E and allergy among adults with glioma compared with controls. Cancer Res. 2004;64:8468–73. doi: 10.1158/0008-5472.CAN-04-1706. [DOI] [PubMed] [Google Scholar]
- 10.Becker KG. The common variants/multiple disease hypothesis of common complex genetic disorders. Med Hypotheses. 2004;62:309–17. doi: 10.1016/S0306-9877(03)00332-3. [DOI] [PubMed] [Google Scholar]
- 11.Annesi-Maesano I, Didier A, Klossek M, Chanal I, Moreau D, Bousquet J. The score for allergic rhinitis (SFAR): a simple and valid assessment method in population studies. Allergy. 2002;57:107–14. doi: 10.1034/j.1398-9995.2002.1o3170.x. [DOI] [PubMed] [Google Scholar]
- 12.Toren K, Brisman J, Jarvholm B. Asthma and asthma-like symptoms in adults assessed by questionnaires. A literature review Chest. 1993;104:600–8. doi: 10.1378/chest.104.2.600. [DOI] [PubMed] [Google Scholar]
- 13.Rothman K, Greenland S. Modern Epidemiology: Lippincott–Raven Publishers, 1998.
- 14.Howard TD, Koppelman GH, Xu J, et al. Gene-gene interaction in asthma: IL4RA and IL13 in a Dutch population with asthma. Am J Hum Genet. 2002;70:230–6. doi: 10.1086/338242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffjan S, Nicolae D, Ober C. Association studies for asthma and atopic diseases: a comprehensive review of the literature. Respir Res. 2003;4:14. doi: 10.1186/1465-9921-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinzmann A, Mao XQ, Akaiwa M, et al. Genetic variants of IL-13 signalling and human asthma and atopy. Hum Mol Genet. 2000;9:549–59. doi: 10.1093/hmg/9.4.549. [DOI] [PubMed] [Google Scholar]
- 17.Howard TD, Whittaker PA, Zaiman AL, et al. Identification and association of polymorphisms in the Interleukin-13 gene with asthma and atopy in a Dutch population. Am J Respir Cell Mol Biol. 2001;25:377–384. doi: 10.1165/ajrcmb.25.3.4483. [DOI] [PubMed] [Google Scholar]
- 18.Holgate ST. The epidemic of allergy and asthma. Nature. 1999;402:B2–4. doi: 10.1038/35037000. [DOI] [PubMed] [Google Scholar]
- 19.Wills-Karp M, Luyimbazi J, Xu X, et al. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–61. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Jacobs BS, Liu J, et al. Interleukin-13 sensitivity and receptor phenotypes of human glial cell lines: non-neoplastic glia and low-grade astrocytoma differ from malignant glioma. Cancer Immunol Immunother 2000;49. [DOI] [PMC free article] [PubMed]
- 21.Barna BP, Estes M, Pettay J, Iwasaki K, Zhou P, Barnett GH. Human astrocyte growth regulation: interleukin-4 sensitivity and receptor expression. J Neuroimmunol. 1995;60:75–81. doi: 10.1016/0165-5728(95)00055-7. [DOI] [PubMed] [Google Scholar]
- 22.Van Eerdewegh P, Little RD, Dupuis J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–30. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 23.Werner M, Herbon N, Gohlke H, et al. Asthma is associated with single-nucleotide polymorphisms in ADAM33. Clin Exp Allergy. 2004;34:26–31. doi: 10.1111/j.1365-2222.2004.01846.x. [DOI] [PubMed] [Google Scholar]
- 24.Howard TD, Postma DS, Jongepier H, et al. Association of a disintegrin and metalloprotease 33 (ADAM 33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol. 2003;112:717–22. doi: 10.1016/s0091-6749(03)01939-0. [DOI] [PubMed] [Google Scholar]
- 25.Nakada M, Okada Y, Yamashita J. The role of matrix metalloproteinases in glioma invasion. Front Biosci. 2003;8:e261–9. doi: 10.2741/1016. [DOI] [PubMed] [Google Scholar]
- 26.Papafili A, Hill MR, Brull DJ, et al. Common promoter variant in Cyclooxygenase-2 represses gene expression:Evidence of role in acute-phase inflammatory response. Atheroscler Thromb Vasc Biol. 2002;22:1631–36. doi: 10.1161/01.atv.0000030340.80207.c5. [DOI] [PubMed] [Google Scholar]
- 27.Wynn TA. IL-13 effector functions. Annu Rev Immunol. 2003;21:425–56. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 28.Mentink-Kane MM, Wynn TA. Opposing roles for IL-13 and IL-13 receptor alpha 2 in health and disease. Immunol Rev. 2004;202:191–202. doi: 10.1111/j.0105-2896.2004.00210.x. [DOI] [PubMed] [Google Scholar]
- 29.Saito A, Okazaki H, Sugawara I, Yamamoto K, Takizawa H. Potential action of IL-4 and IL-13 as fibrogenic factors on lung fibroblasts in vitro. Int Arch Allergy Immunol. 2003;132:168–76. doi: 10.1159/000073718. [DOI] [PubMed] [Google Scholar]
- 30.Sivak-Sears N, Schwartzbaum J, Miike R, Moghadassi M, Wrensch M. Case-control study of non-steroidal anti-inflammatory drug use and glioblastoma multiforme. Am J of Epidemiol. 2004;159:1131–1139. doi: 10.1093/aje/kwh153. [DOI] [PubMed] [Google Scholar]
- 31.Howell WM, Jobs M, Gyllensten U, Brookes AJ. Dynamic allele-specific hybridisation: A new method for scoring single nucleotide polymorphisms. Nature Biotech. 1999;17:87–88. doi: 10.1038/5270. [DOI] [PubMed] [Google Scholar]
- 32.Prince JA, Feuk L, Howell WM, et al. Robust and accurate single nucliotide polymorphism genotyping by dynamic allele-specific hybridization (DASH) design. Criteria and assay validation Genome Research. 2001;11:152–62. doi: 10.1101/gr.150201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Howell WM, Jobs M, Brookes AJ. iFRET: an improved fluorescence system for DNA-melting analysis. Genome Res. 2002;12:1401–7. doi: 10.1101/gr.297202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jobs M, Howell WM, Stromqvist L, Mayr T, Brookes AJ. Dash-2: Flexible, low-cost, and high-throughput SNP genotyping by dynamic allele-specific hybridication on membrane arrays. Genome Research. 2003;13:916–924. doi: 10.1101/gr.801103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawyer SL, Howell WM, Brookes AJ. Scoring insertion-deletion polymorphisms by dynamic allele-specific hybridization. Biotechniques 2003;35:292–6, 298. [DOI] [PubMed]
- 36.Fredman D, Jobs M, Stromqvist L, Brookes AJ. DFold: PCR design that minimizes secondary structure and optimizes downstream genotyping applications. Hum Mutation. 2004;24:1–8. doi: 10.1002/humu.20066. [DOI] [PubMed] [Google Scholar]
- 37.Jobs M, Howell WM, Brookes AJ. Creating assays by centrifugation. Biotechniques. 2002;32:1322–1229. doi: 10.2144/02326mt03. [DOI] [PubMed] [Google Scholar]
- 38.Sawaya R, Ramo OJ. Systemic and thromboembolic effects of meningiomas. In: Al-Mefty O, ed. Meningiomas. New York: Raven Press, Ltd., 1991:137–144.
- 39.Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abbas AK, Lichtman AH, Pober JS. Cellular and molecular immunology. London: W. B. Saunders Company, 1997.
- 41.Vineis P. A self-fulfilling prophecy: are we underestimating the role of the environment in gene-environment interaction research? Int J Epidemiol. 2004;33:945–6. doi: 10.1093/ije/dyh277. [DOI] [PubMed] [Google Scholar]
- 42.Genentech. Briefing Document on Safety (BLA STN 1039760/0): Omalizumab (Xolair) for treatment of allergic asthma., 2003.
- 43.Louis DN. A molecular genetic model of astrocytoma histopathology. Brain Pathol. 1997;7:755–64. doi: 10.1111/j.1750-3639.1997.tb01062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saleh M, Davis ID, Wilks AF. The paracrine role of tumor-derived mIL-4 on tumour-associated endothelium. Int J Cancer. 1997;72:664–72. doi: 10.1002/(sici)1097-0215(19970807)72:4<664::aid-ijc19>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 45.Saleh M, Wiegmans A, Malone Q, Stylli SS, Kaye AH. Effect of in-situ retroviral interleukin-4 transfer on established intracranial tumors. J Natl Cancer Inst. 1999;91:438–45. doi: 10.1093/jnci/91.5.438. [DOI] [PubMed] [Google Scholar]
- 46.Volpert OV, Fong T, Kock AE, et al. Inhibition of angiogenesis by interleukin 4. J Exp Med. 1998;188:1039–46. doi: 10.1084/jem.188.6.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shin WH, Lee D-Y, Keun WP, et al. Microglia expressing interleukin-13 undergo cell death and contribute to neuronal survival in vivo. Glia. 2004;46:142–152. doi: 10.1002/glia.10357. [DOI] [PubMed] [Google Scholar]
- 48.Debinski W, Gibo D, Mintz A. Epigenetics in high-grade astrocytomas: opportunities for prevention and detection of brain tumors. Ann N Y Acad Sci. 2003;983:232–42. doi: 10.1111/j.1749-6632.2003.tb05978.x. [DOI] [PubMed] [Google Scholar]
- 49.Debinski W, Miner R, Leland P, Obiri NI, Puri RK. Receptor for Interleukin (IL) 13Does Not Interact with IL4 but Receptor for IL4 Interacts with IL13 on Human Glioma Cells. J Biol Chem. 1996;271:22428–22433. doi: 10.1074/jbc.271.37.22428. [DOI] [PubMed] [Google Scholar]
- 50.Rahaman SO, Sharma P, Harbor PC, Aman MJ, Vogelbaum MA, Haque SJ. IL-13R(alpha)2, a decoy receptor for IL-13 acts as an inhibitor of IL-4-dependent signal transduction in glioblastoma cells. Cancer Res. 2002;62:1103–9. [PubMed] [Google Scholar]
- 51.Rahaman SO, Vogelbaum MA. IL-4 potentiates persistent activation of STAT3 in glioblastoma multiforme cells: cause and consequence. Neuro-oncol. 2004;6:344. [Google Scholar]
- 52.Madhankumar AB, Debinski W. Structural requirements for interleukin 13 to bind to IL13Ralpha2, a tumor-associated receptor. Neuro-Oncology. 2004;6:343. [Google Scholar]
- 53.Terabe M, Park JM, Berzofsky JA. Role of IL13 in regulation of anti-tumor immunity and tumor growth. Cancer Immunol Immunother. 2004;53:79–85. doi: 10.1007/s00262-003-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berzofsky JA, Terabe M, Oh S, et al. Progress on new vaccine strategies for the immunotherapy and prevention of cancer. J Clin Invest. 2004;113:1515–25. doi: 10.1172/JCI21926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ichinose M, Barnes PJ. Cytokine-directed therapy in asthma. Curr Drug Targets Inflamm Allergy. 2004;3:263–9. doi: 10.2174/1568010043343688. [DOI] [PubMed] [Google Scholar]
- 56.Kumar RK, Herbert C, Webb DC, Li L, Foster PS. Effects of anticytokine therapy in a mouse model of chronic asthma. Am J Respir Crit Care Med. 2004;170:1043–8. doi: 10.1164/rccm.200405-681OC. [DOI] [PubMed] [Google Scholar]


