Abstract
The DNA single-strand break repair protein XRCC1 contains a BRCT domain that binds and stabilizes intracellular DNA ligase III protein. We recently demonstrated that this domain is largely dispensable for single-strand break repair and cellular resistance to DNA base damage in cycling cells. Here, we report that the BRCT domain is required for single-strand break repair in noncycling cells. Mutations that disrupt the BRCT domain and prevent DNA ligase III interaction abolished XRCC1-dependent repair in serum-starved Chinese hamster ovary cells, and reentry into cell cycle induced by readdition of serum restored repair. Elevating DNA ligase III levels in XRCC1 mutant cells using proteosome inhibitors or by expressing XRCC1 protein in which the BRCT domain is disrupted but can still bind DNA ligase III failed to restore repair in noncycling cells. The requirement for the BRCT domain for DNA strand break repair is thus for more than simply binding and stabilizing DNA ligase III. These data provide evidence in support of a selective role for a DNA repair protein or protein domain in noncycling cells. We propose that the XRCC1 C-terminal BRCT domain may be important for genetic stability in postmitotic cells in vivo.
Thousands of DNA single-strand breaks arise in cells every day from a variety of sources including endogenous reactive oxygen species and the enzymatic excision of damaged DNA bases or abasic sites (1, 2). The threat posed by single-strand breaks is illustrated by the genetic instability of mutant rodent cells in which single-strand break repair (SSBR) is defective. For example, mutations in the SSBR gene XRCC1 result in increased frequencies of spontaneous sister chromatid exchange and chromosomal aberration as well as hypersensitivity to alkylating agents and ionizing radiation (see ref. 1 for review). XRCC1 protein interacts with DNA ligase III (Lig-3) and maintains normal cellular levels of this polypeptide (3, 4). The interaction between XRCC1 and Lig-3 is mediated via BRCT domains located at the C terminus of both polypeptides (5, 6). Intriguingly, there is a cell-cycle stage-specific requirement in mammalian cells for the C-terminal XRCC1 BRCT domain (denoted BRCT II), as it is required for XRCC1-dependent SSBR during G1 but is dispensable for this process in S phase (7). However, the role in G1 is largely dispensable for survival in cycling cells because XRCC1-dependent SSBR in S phase can largely compensate for an absence of repair in G1 (7). If the BRCT II domain is dispensable in cycling cells, what role does it play? Here, we have examined the possibility that the major role for this motif is during SSBR in noncycling cells, which lack S phase and may thus be dependent on this BRCT domain for SSBR.
Materials and Methods
Expression Constructs and Cell Lines.
EM9-V cells harbor empty vector and EM9-XpmBRCT cells express human XRCC1 possessing the BRCT II domain mutations W611D and VI584/585DD (7). EM9-XW611D and EM9-XVI584/585DD cells express human XRCC1 possessing either of these mutations, individually. EM9-XΔ529–633 cells express human XRCC1 lacking the C-terminal 105 amino acids including the entire BRCT II domain (5). These cell lines are pooled populations of >50 independent transfectants and were cultured routinely in α-MEM (GIBCO/BRL) containing 10% FBS. For serum starvation, cells were plated (≈5 × 105 cells/plate) in complete medium for 24 h followed by incubation for a further 2–4 days in α-MEM containing 0.1% FBS. Where indicated, cells were incubated in the presence of the 20S proteosome inhibitors lactacystin (1 μM) and MG-132 (10 μM) (Calbiochem) for 4 h before analysis. For immunoblotting, samples of ≈5 × 105 cells were lysed in 20 mM Tris⋅Cl, pH 7.5/10 mM EDTA/1 mM EGTA/100 mM NaCl/1% Triton X-100/1 mM PMSF, and ≈15 μg of soluble protein fractionated by SDS/PAGE and immunoblotted with anti-XRCC1 mAb (33-2-5), anti-Lig-3 polyclonal antibody (TL25), or anti-pRb mAb (PharMingen).
BrdUrd Pulse Labeling and Flow Cytometry.
Cycling or serum-starved cells were incubated in the presence of BrdUrd (100 μM) for 1 h, harvested, washed with PBS, and stored in 70% ethanol/30% PBS at 4°C until required. Pelleted cells were resuspended in 2 M HCl at room temperature for 30 min, washed twice with PBS, once with PBT (PBS + 0.5% BSA/0.1% Tween 20), and resuspended in 100-μl PBT + anti-BrdUrd primary antibody (Dako; 1/80 dilution) for 1 h. After two washes with PBS, cells were incubated with 100 μl of PBT + FITC-conjugated goat anti-mouse secondary antibody (Dako; 1/40 dilution in PBS). Cells were washed with PBT, PBS, and incubated with RNase A in PBS at 4°C overnight. After the addition of propidium iodide (10 μg/ml in PBS), cells were analyzed by flow cytometry (FACScan; Becton Dickinson).
Survival Assays.
Cells plated in duplicate in 6-well dishes (150–200 cells/well) in Opti-MEM (5% FBS) were treated with ethyl methanesulfonate (EMS) for 1 h at the concentrations indicated. The surviving fraction was calculated after 10 days by dividing the number of colonies in treated wells by those in untreated wells. Each experiment was conducted at least three times.
DNA Strand-Break Repair Assays.
Strand-break repair was assayed using alkaline single-cell agarose-gel electrophoresis, which quantifies strand breakage via the ability of the latter to increase DNA migration from the cell nucleus during electrophoresis (8). DNA strand breakage measured by this assay is expressed as the “tail moment,” which is the product of the fraction of DNA that has exited the nucleus multiplied by the distance migrated. To measure initial damage, cells (≈2 × 105 per 10-cm plate) were treated with EMS (10 mg/ml) for 15 min, washed in ice-cold PBS, and harvested with a rubber policeman. EMS was used because this agent was used in the original characterization of the repair defect in XRCC1 mutant cells (9). A dose of 10 mg/ml was chosen from a standard curve of EMS concentration versus mean tail moment (data not shown). This dose provides a level of damage that is easily distinguished from background levels but which is still within the linear portion of the standard curve. To measure DNA strand break-repair capacity, drug-treated cells were washed with PBS (37°C) and incubated in drug-free medium for 3 h before harvest. To calculate SSBR proficiency, the fraction (%) of EMS-induced DNA strand breakage remaining after incubation in drug-free medium was calculated by the following equation; [(mean tail momentafter repair − mean tail momentuntreated cells)/(mean tail momentinitial damage − mean tail momentuntreated cells)] × 100. Measurement of the mean tail moment was from 100 cells per slide, present in 10–20 randomly selected fields representing the whole area of the slide, with each data point in each experiment calculated from duplicate slides. Calculation of the mean tail moment was automated using the Comet Assay II software (Perceptive Instruments; Suffolk, U.K.).
Results
We wished to examine the role of the XRCC1 BRCT II domain in DNA SSBR in quiescent cells. However, because primary cell lines or differentiated cells in which XRCC1 is mutated are not available, we chose to measure SSBR in XRCC1 mutant Chinese hamster ovary (CHO) cells in which noncycling status was induced by serum starvation. Serum starvation has been shown to arrest cultured rodent cell lines before the restriction point that defines commitment to proliferation (10). Thus, although it is unlikely to completely reflect a quiescent status in vivo, it provides an approximation of such in cultured cell lines. The CHO cell lines used for this study were transfected derivatives of XRCC1 mutant EM9 cells expressing either wild-type human XRCC1 (EM9-X cells) or XRCC1pmBRCT (EM9-XpmBRCT cells). The BRCT II domain in XRCC1pmBRCT harbors the double mutation VI584/585DD and the single mutation W611D and cannot interact with DNA Lig-3 (6, 7). In addition, in later experiments, EM9 cells expressing XRCC1 that harbors these mutations individually (denoted EM9-XVI584/585DD and EM9-XW611D cells, respectively), or XRCC1 protein in which the entire BRCT II domain was deleted (denoted EM9-XΔ529–633 cells), were used (5).
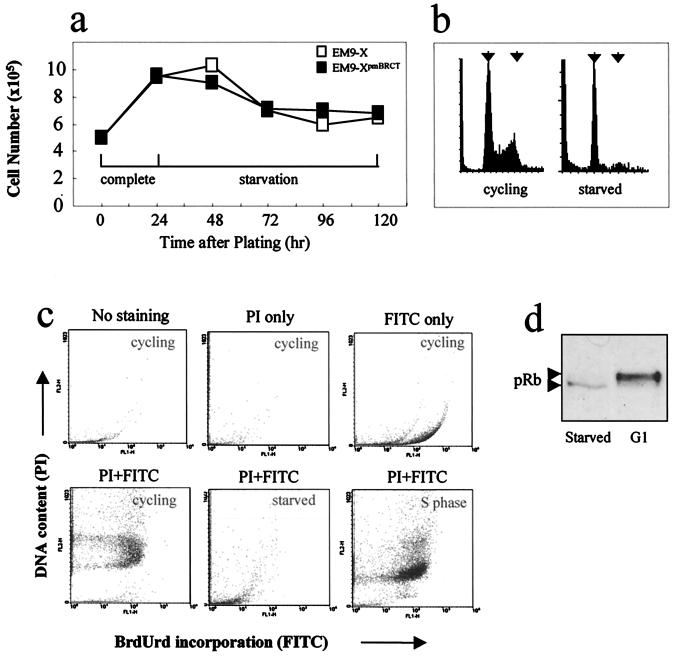
EM9-X and EM9-XpmBRCT cells doubled in number within 24 h after plating in complete medium, but no further increase in cell number was observed after serum withdrawal (Fig. 1a). Indeed, an initial reduction in cell number of ≈30% was observed during the first 48 h of serum starvation, presumably reflecting a subset of cells that failed to survive the synchronization protocol. An analysis of the DNA content of cells that were serum starved for 48 h by flow cytometry suggested that S phase and G2 cells were absent from this population (Fig.1b). Indeed, this cell population did not incorporate BrdUrd during a 1-h pulse-labeling experiment, confirming the absence of S phase cells (Fig. 1c, compare bottom-left and bottom-middle panels). In contrast, >90% of this cell population incorporated BrdUrd during a 1-h pulse label if first reincubated in complete medium, indicating that the cells that were serum starved for 48 h were competent to reenter cell cycle upon readdition of serum (Fig. 1c, bottom-right panel). Finally, to obtain biochemical evidence that serum-starved CHO cells were arrested in a noncycling state, we examined cellular pRb protein for its phosphorylation status. pRb is hypophosphorylated in cells that are located before the restriction point that defines commitment to proliferation and consequently migrates faster during SDS/PAGE than does the hyperphosphorylated form of pRb that is present in cycling cells that have passed this point (11). The pRb present in serum-starved CHO cells migrated more rapidly than did hyperphosphorylated pRb controls, which were derived by arresting cycling cells in late G1 with mimosine (Fig. 1d and data not shown), supporting the presence of noncycling status.
Figure 1.
Induction of noncycling status in CHO cells by serum starvation. (a) EM9-X cells (□) or EM9-XpmBRCT cells (■) were plated at 5 × 105 cells/plate and incubated for 24 h in complete medium followed by a further 4 days in medium containing 0.1% FBS. Cells from duplicate plates were counted at daily intervals. (b) EM9-X cells plated as in A were harvested after 24 h in complete medium (left; cycling) and after a subsequent 3 days in starvation medium (right; starved). Fixed cells were stained with propidium iodide to stain DNA and analyzed by flow cytometry. Left and right arrows indicate the position of cells with G1 and G2 DNA content, respectively. Similar results were observed with EM9-XpmBRCT cells (data not shown). (c) Asynchronous cycling cells (cycling), cells serum starved for 48 h (starved), or cells serum starved for 48 h and then reincubated in complete medium for 5 h (S-phase), were pulse labeled with BrdUrd for 1 h. The pulse-labeled cells were analyzed by flow cytometry either without further processing (no staining) or after staining with propidium iodide (PI only), anti-BrdUrd antibody (FITC), or both (PI + FITC). (d) pRb immunoblot of cell extract from EM9-XpmBRCT after serum starvation (left lane; starved) or in cells arrested in late G1 by incubation for 24 h in complete medium containing mimosine (right lane; G1). Arrows depict the position of hypophosphorylated (lower arrow) and hyperphosphorylated (upper arrow) pRb.
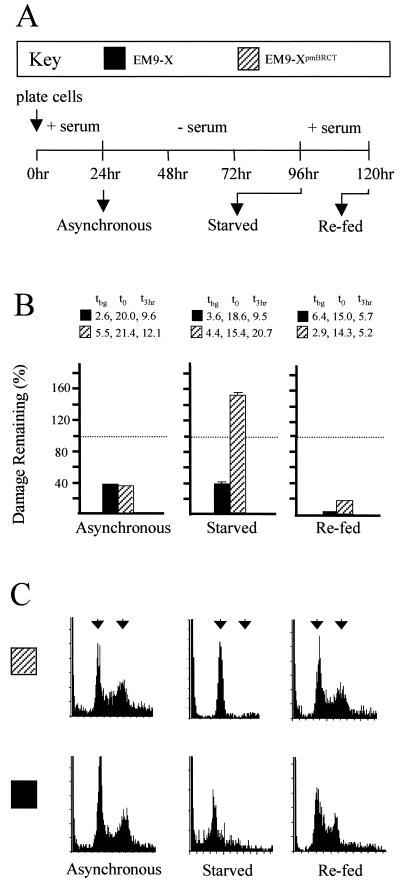
We next measured SSBR before and after serum starvation, and after serum-starved cells were refed with complete medium to induce reentry into cell cycle (see Fig. 2A for experimental design). Repair assays conducted before serum starvation indicated that cycling populations of both EM9-X and EM9-XpmBRCT cells were SSBR proficient during a 3-h incubation in drug-free medium (Fig. 2B Left). Strikingly, however, whereas serum-starved EM9-X cells were also SSBR proficient, reducing strand breakage by ≈65%, EM9-XpmBRCT cells were not (Fig. 2B Center). Indeed, strand breakage appeared to increase in serum-starved EM9-XpmBRCT cells during repair incubation. These data suggest that disruption of the BRCT II domain selectively abolished XRCC1-dependent SSBR specifically in noncycling cells. In agreement with this notion, SSBR proficiency was restored in EM9-XpmBRCT cells by adding back serum for 24 h, suggesting that reentry into cell cycle restored SSBR (Fig. 2C Right). The SSBR measured in these experiments was XRCC1 dependent irrespective of cell-cycle status because EM9 cells harboring empty vectors were SSBR defective before, during, and after serum starvation (data not shown). Flow cytometry confirmed that the cell populations used in these experiments were in the growth phase expected (Fig. 2C).
Figure 2.
Effect of BRCT II mutations on SSBR proficiency in cycling and noncycling CHO cells. (A) Schematic of the experimental design. EM9-X or EM9-XpmBRCT cells were plated in complete medium for 24 h and sampled before serum starvation (asynchronous), after serum starvation for 72 h (starved), or after refeeding for 24 h (re-fed), and analyzed as follows. (B) Aliquots of the indicated cells (see Key) were treated with EMS (10 mg/ml) for 15 min and subsequently incubated in drug-free medium for 3 h to allow repair. The fraction (%) of EMS-induced breakage remaining after the repair incubation is presented graphically, with that present immediately after EMS treatment set at 100% (dotted line). The actual levels of strand breakage present in each cell line (measured as the mean comet tail moment) before exposure to EMS (i.e., background tbg), immediately after EMS treatment (t0), and after a 3-h repair incubation (t3 h) are shown. These data are the means of three independent experiments. (C) DNA content was determined by flow cytometry as a measure of cell-cycle status. Left and right arrows denote the position of cells with G1 and G2 DNA content, respectively.
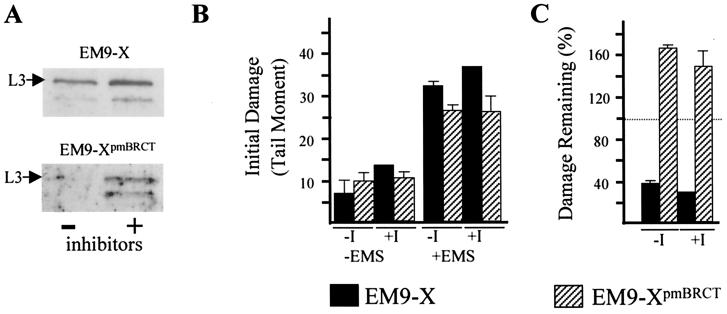
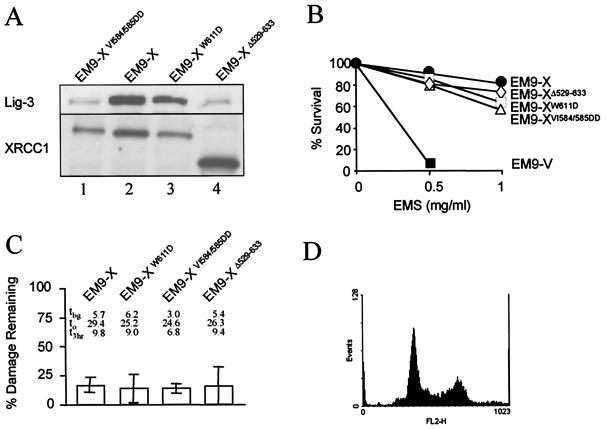
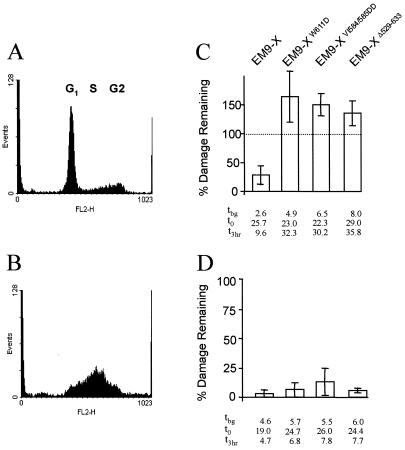
The interaction of XRCC1 with Lig-3 prevents proteolytic degradation of the latter and is thus required for normal cellular levels of this DNA ligase (4, 7). To examine whether maintaining normal Lig-3 levels is sufficient to account for the role of the BRCT II domain in SSBR, we used proteosome inhibitors to increase levels of Lig-3 in the absence of an interaction with XRCC1 (Fig. 3A). However, increasing the level of Lig-3 in EM9-XpmBRCT cells in this way did not increase SSBR proficiency (Fig. 3C), suggesting that the BRCT II domain fulfilled an additional role in SSBR. To examine this possibility further, we compared the effect on SSBR of the two mutations that comprise XRCC1pmBRCT because, although one of these ablates binding to Lig-3 (VI584/585DD), the other (W611D) does not (6). We also examined the effect on SSBR of a deletion mutation (Δ529–633) that completely removes the BRCT II domain (5). Whereas XRCC1VI584/585DD and XRCC1Δ529–633 were unable to maintain high levels of Lig-3 in transfected EM9 cells, XRCC1W611D was able to, consistent with the respective abilities of these proteins to bind Lig-3 (Fig. 4A, lanes 1, 3, and 4). Indeed, whereas the Lig-3 levels present in EM9-XΔ529–633 and EM9-XVI584/585DD were no higher than those present in EM9 cells transfected with empty vector (data not shown), the Lig-3 level present in EM9-XW611D was similar to that in wild-type EM9-X cells (Fig. 4A, lanes 2 and 3). Although each of the mutant proteins was able to support cellular resistance to alkylating agents and SSBR in cycling cells (Fig. 4 B and C), none of the mutant proteins supported SSBR in serum-starved cells, irrespective of their ability to bind and stabilize Lig-3 (Fig. 5C). SSBR proficiency was restored if the serum-starved cells were subsequently released back into cell cycle, confirming that the SSBR defect was restricted to noncycling status (Fig. 5D). These data further suggest that the stabilization of Lig-3 is insufficient to account for the role of the XRCC1 BRCT II domain in SSBR in noncycling cells, supporting the existence of an additional role for this domain.
Figure 3.
Lig-3 protein levels and SSBR proficiency. (A) Cell extract was prepared from EM9-X (Upper) or EM9-XpmBRCT cells (Lower) after serum starvation for 72 h followed by a further 4 h in the presence (+) or absence (−) of 1 μM lactacystin and 10 μM MG-132. Following SDS/PAGE, cell extracts were immunoblotted for Lig-3. (B) DNA strand breakage was quantified in serum-starved (72 h) EM9-X or EM9-XpmBRCT cells before or immediately after EMS treatment (±EMS). Proteosome inhibitors were present (+I) or absent (−I) for 4 h before and during EMS treatment. (C) The fraction of EMS-induced strand breakage remaining in serum-starved cells after a 3-h repair incubation in EMS-free medium in the presence (+I) or absence (−I) of proteosome inhibitors. Strand breakage present immediately after EMS is set at 100% (dotted line). These data are the means (±SD) of three experiments.
Figure 4.
Effect of individual BRCT II mutations on EMS sensitivity and SSBR proficiency in cycling cells. (A) Whole cell extract from the indicated cell lines was fractionated by SDS/PAGE, transferred to nitrocellulose, and immunoblotted with anti-Lig-3 (Upper) or anti-XRCC1 (Lower) antibodies. (B) The cell lines indicated were treated with EMS at the indicated concentration for 1 h and subsequently incubated for 10 days in drug-free medium. The fraction of surviving cells (%) was calculated from the relative number of colonies on treated versus untreated plates. Values are the mean of at least three experiments. Error bars are within 20% of the mean value and are omitted for clarity. (C) The cell lines indicated were treated with EMS (10 mg/ml) for 15 min and subsequently incubated in drug-free medium for 3 h to allow repair. The fraction (%) of EMS-induced breakage remaining after repair incubation is presented graphically. The actual levels of strand breakage present in each cell line (measured as the mean comet tail moment) before exposure to EMS (i.e., background tbg), immediately after EMS treatment (t0), and after a 3-h repair incubation (t3 h) are also shown. Results are the mean (±1 SD) of at least three experiments. (D) DNA content of asynchronous cycling CHO cells determined by flow cytometry following staining with propidium iodide.
Figure 5.
Effect of individual BRCT II domain mutations on SSBR proficiency in serum-starved and S-phase cells. (A and B) DNA content of CHO cells following serum starvation (A) or following serum starvation and subsequent release into S phase by replacement of serum (B). (C and D) The cell lines indicated were treated with EMS (10 mg/ml) for 15 min either during serum starvation (C) or after subsequent release into S phase (D) and then incubated in drug-free medium for 3 h to allow repair. The fraction (%) of EMS-induced breakage remaining after repair incubation is presented graphically. Results are the mean (±1 SD) of at least three experiments. The actual levels of strand breakage present in each cell line (measured as the mean comet tail moment) before exposure to EMS (i.e., background tbg), immediately after EMS treatment (t0), and after a 3-h repair incubation (t3 h) are shown.
Discussion
XRCC1 is required for SSBR and genetic stability in mammalian cells, but the biochemical role of this protein is unclear (1). However, the observation that this polypeptide interacts with several other SSBR proteins suggests that it may function as a chaperone or scaffold protein. One of the proteins with which XRCC1 interacts is DNA Lig-3 (3, 4). This interaction is mediated via BRCT domains located at the C terminus of both proteins (5, 6), and we have reported previously that the XRCC1 BRCT domain (denoted BRCT II) is important for SSBR during G1 (7). However, this requirement is dispensable for cell survival in cycling cells because of an ability of XRCC1 to mediate SSBR independently of this domain in S/G2 (7). Here, we have thus examined the possibility that the major role of the BRCT II domain is to mediate SSBR in noncycling cells, which lack an S/G2 cell-cycle phase and may thus be dependent on this domain for SSBR.
To induce a noncycling status in CHO cell populations, we used serum starvation. The serum-starved populations failed to incorporate BrdUrd during a 1-h pulse label, confirming the absence of S-phase cells. Also, the electrophoretic mobility of pRb protein was increased following serum starvation, suggesting that this protein was hypophosphorylated and thus that the serum-starved cells were synchronized before the commitment point for cell division. This contrasted with CHO cells synchronized in G1 with mimosine, which are beyond this commitment point. A number of mutations within the BRCT II domain were examined for their effect on SSBR. The double mutation VI584/585DD removes two nonpolar amino acids from this domain and ablates its interaction with Lig-3 (6). The mutation W611D removes the tryptophan residue that is largely invariant among BRCT domains and is believed to disrupt proper folding of XRCC1 BRCT II (12). This mutation does not ablate interaction with Lig-3, however (6). Finally, we also examined the effect on SSBR of a deletion mutation that removes the entire BRCT II domain (5). Each of these mutations ablated the ability of XRCC1 to support SSBR in serum-starved EM9 cells when present alone or when combined. However, these mutations did not ablate the ability of XRCC1 to support SSBR in EM9 cells before serum starvation or after serum-starved cells were released into cell cycle. These data strongly suggest that the BRCT II domain is selectively required for SSBR in noncycling cells. What might be the role of the BRCT domain? This structure binds Lig-3 and in so doing maintains normal levels of the DNA ligase (4–7). In cells lacking XRCC1 or expressing XRCC1 that cannot interact with Lig-3, the latter polypeptide is degraded, resulting in 4 to 6-fold reduced levels of the DNA ligase (4, 6, 13). Thus, one role served by the BRCT II domain is to maintain high cellular levels of Lig-3. However, it is unlikely that this is the only role for this domain. This is suggested by two observations. First, the employment of proteosome inhibitors to increase the level of Lig-3 in EM9-XpmBRCT cells possessing a mutated BRCT II domain was not sufficient to restore SSBR. Second, removal of the largely invariant tryptophan from BRCT II abolished SSBR in noncycling cells even though this mutation did not ablate Lig-3 binding or stabilization (ref. 6 and this work). Thus, interaction with Lig-3 is insufficient to account for the role of the XRCC1 BRCT II domain in SSBR in noncycling cells. One additional role for this domain might be to facilitate the translocation of XRCC1–Lig-3 complex to sites of single-strand breakage, hundreds of thousands of which arise throughout the genome per cell, per day. In this scenario, the dispensability of the BRCT domain in cycling cells could be explained if the architectural organization of SSBR differs such that the translocation of protein complexes to different genome sites is not required. For example, perhaps during S phase, DNA repair proteins are arranged into protein factories analogous to those that mediate DNA replication and through which chromosomes can translocate (see ref. 14 for review).
In summary, we report that mutations within the C-terminal BRCT domain of human XRCC1 selectively abolish XRCC1-dependent SSBR in serum-starved cells. This is evidence for a role for a DNA repair protein or protein domain in noncycling cells. These results support a model in which the XRCC1 BRCT II domain is selectively required for genetic stability in postmitotic tissues, in vivo.
Acknowledgments
This work was funded by Medical Research Council Grants G9603130 and G9809326.
Abbreviations
- CHO
Chinese hamster ovary
- SSBR
single-strand break repair
- Lig-3
DNA ligase III
- EMS
ethyl methanesulfonate
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250477597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250477597
References
- 1.Thompson L H, West M G. Mutat Res. 2000;459:1–18. doi: 10.1016/s0921-8777(99)00058-0. [DOI] [PubMed] [Google Scholar]
- 2.Lindahl T. Nature (London) 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 3.Caldecott K W, Mckeown C K, Tucker J D, Ljungquist S, Thompson L H. Mol Cell Biol. 1994;14:68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldecott K W, Tucker J D, Stanker L H, Thompson L H. Nucleic Acids Res. 1995;23:4836–4843. doi: 10.1093/nar/23.23.4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nash R A, Caldecott K W, Barnes D E, Lindahl T. Biochemistry. 1997;36:5207–5211. doi: 10.1021/bi962281m. [DOI] [PubMed] [Google Scholar]
- 6.Taylor R M, Wickstead B, Cronin S, Caldecott K W. Curr Biol. 1998;8:877–880. doi: 10.1016/s0960-9822(07)00350-8. [DOI] [PubMed] [Google Scholar]
- 7.Taylor R M, Moore D J, Whitehouse J, Johnson P, Caldecott K W. Mol Cell Biol. 2000;20:735–740. doi: 10.1128/mcb.20.2.735-740.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fairbairn D W, Olive P L, O'Neill K L. Mutat Res. 1995;339:37–59. doi: 10.1016/0165-1110(94)00013-3. [DOI] [PubMed] [Google Scholar]
- 9.Thompson L H, Brookman K W, Dillehay L E, Carrano A V, Mazrimas J A, Mooney C L, Minkler J L. Mutat Res. 1982;95:427–440. doi: 10.1016/0027-5107(82)90276-7. [DOI] [PubMed] [Google Scholar]
- 10.Larsson O, Zetterberg A, Engstrom W. J Cell Sci. 1985;75:259–268. doi: 10.1242/jcs.75.1.259. [DOI] [PubMed] [Google Scholar]
- 11.Mittnacht S. Curr Opin Genet Dev. 1998;8:21–27. doi: 10.1016/s0959-437x(98)80057-9. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X D, Morera S, Bates P A, Whitehead P C, Coffer A I, Hainbucher K, Nash R A, Sternberg M J E, Lindahl T, Freemont P S. EMBO J. 1998;17:6404–6411. doi: 10.1093/emboj/17.21.6404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cappelli E, Taylor R, Cevasco M, Abbondandolo A, Caldecott K, Frosina G. J Biol Chem. 1997;272:23970–23975. doi: 10.1074/jbc.272.38.23970. [DOI] [PubMed] [Google Scholar]
- 14.Cook P R. Science. 1999;284:1790–1795. doi: 10.1126/science.284.5421.1790. [DOI] [PubMed] [Google Scholar]