Abstract
The Hairy-related transcription-factor (HRT) genes encode three related basic helix–loop–helix transcription factors that show sequence similarity to the Hairy and Enhancer of split family of transcriptional repressors. HRT proteins are expressed in specific regions of the developing heart, vasculature, pharyngeal arches and somites, and the periodicity of their expression in somitic precursors mirrors that of Notch signaling-related molecules. In the present study, we show that the intracellular domain of the Notch1 receptor (Notch1 IC), which is constitutively active, up-regulates HRT2 expression in 10T½ fibroblasts. Luciferase reporter assays using the regulatory regions of the mouse HRT genes revealed that transcription of all three genes is stimulated by Notch1 IC. The promoters of the HRT genes share homology in a binding site for Suppressor of Hairless [Su(H)], a transcriptional mediator of Notch signaling. A dominant-negative Su(H) mutant abolished Notch-activated HRT2 expression, and mutation of the conserved Su(H) consensus site in the HRT2 promoter attenuated transcriptional activation by Notch. Ectopic expression of HRT proteins also blocked activation of HRT2 expression by Notch1 IC through a mechanism requiring the basic region, but not the conserved carboxyl-terminal YQPW-TEVGAF motif of HRT2. These findings identify HRT genes as downstream targets for Notch signaling and reveal a negative autoregulatory loop whereby HRT proteins repress their own expression through interference with Notch signaling.
The transmembrane receptor Notch participates in an evolutionarily conserved cell-interaction system that plays fundamental roles in embryonic patterning and development, including neurogenesis and somitogenesis (1, 2). Notch signaling is especially important in cell-fate specification and boundary formation in which clusters of undifferentiated cells are segregated into different cell lineages. Upon activation by ligands such as Delta or Jagged on neighboring cells, the intracellular domain of the Notch receptor (Notch IC) is cleaved and translocated to the nucleus together with Suppressor of Hairless [Su(H)] or related molecules. Su(H) provides DNA-binding specificity through recognition of the consensus sequence, whereas Notch IC functions as an activation domain (1–3). In response to Notch signaling, Su(H) activates transcription of the Hairy/Enhancer of split genes [H/E(spl)], which encode a family of basic helix–loop–helix (bHLH) transcriptional repressors (1, 2, 4). H/E(spl) proteins then inhibit transcription of their target genes, thereby preventing undifferentiated precursors from achieving differentiated phenotypes. The actions of Notch can also be mediated in Su(H)-independent pathways by the interaction of the Notch receptor with various molecules (1, 2).
We and others recently identified a subclass of Hairy-related transcription factors (HRTs), also called Hesr, Hey, CHF, and Gridlock (5–9). The HRT family consists of three proteins, HRT1, -2, and -3, which share structural similarity in their bHLH regions and contain a unique carboxyl-terminal domain similar to, but distinct from, the region of H/E(spl) proteins responsible for transcriptional repression (5). During embryogenesis, HRT genes show characteristic expression patterns that demarcate regions of the developing heart, vasculature, pharyngeal arches, and somites (5). Within the segmental-plate mesoderm, HRT gene expression exhibits a periodicity reminiscent of H/E(spl) and other components of Notch-signaling pathways.
Based on their embryonic expression patterns and on the importance of bHLH proteins for Notch signaling (4, 10), we investigated whether HRT genes might be downstream targets for Notch signaling. Here, we show that HRT gene expression in cultured cells is activated by Notch signaling and that HRT proteins interfere with Notch-dependent activation of HRT2 expression, thereby fulfilling a negative autoregulatory loop that tightly regulates HRT expression.
Materials and Methods
Expression Constructs.
Plasmid expression constructs for Notch1 IC, mouse HRT (mHRT), and human HRT (hHRT) were prepared by the insertion of PCR fragments into pcDNA3.1 (Invitrogen, Carlsbad, CA) with an amino-terminal Myc-tag. The pCS2+mN1 IC(V1744)wt provided by R. Kopan (Washington University, St. Louis) was used as a template for Notch1 IC. All wild-type HRT constructs were designed to contain the entire coding region without the first methionine residue. The mHRT2 C(−) mutant construct was designed to delete the carboxyl terminus of mHRT2, KPYQPWGTEVGAF, by introducing a premature stop codon. The mHRT2 B(−) construct was prepared by the ligation of two PCR fragments so that the basic domain of mHRT2 (RKKRRGIIEKRRR) was replaced with two amino acids, LE. The plasmids pCMX-RBP-J for wild-type RBPJ expression and pEF-BOSneo-R218H for dominant-negative RBPJ (11) were provided by T. Honjo (Kyoto University).
Cell Culture and Plasmid Transfection.
C3H10T½ (10T½) fibroblasts, COS7 cells, and C2C12 myoblasts were maintained as described (12). Plasmid transfection was performed by using Fugene 6 (Roche Diagnostics) or Lipofectamine Plus reagent (Life Technologies, Grand Island, NY). The total amount of plasmids was adjusted by using vector plasmids in each assay.
Northern Blot Analysis.
Two days after transfection, RNA was purified from 10T½ cells and Northern blot analysis was performed by using 30 μg of total RNA as described (5). In experiments with transfection of mHRT2 expression plasmids, a 0.9-kb RsaI–RsaI fragment of the 3′ noncoding region of mHRT2 cDNA was used as a probe.
Isolation of Mouse HRT Genes.
A mouse genomic DNA bacterial artificial chromosome library (Children's Hospital Oakland Research Institute, Oakland, CA) was screened by using mHRT1, -2, and -3 cDNA fragments. Positive genomic DNA fragments were analyzed by restriction-enzyme digestion, Southern blot analysis, and sequencing.
Luciferase Reporter Analysis.
A BamHI–MfuI partial-digestion fragment of mHRT1, an EcoRI–NruI fragment of mHRT2, and a BamHI–BamHI fragment of mHRT3 were cloned into the pGL3 basic luciferase vector which lacks a promoter (Promega; Fig. 2A), and a series of 5′-deletion constructs was made for mHRT2. The mHES-1-luciferase construct provided by R. Kopan (13) was used as a positive control.
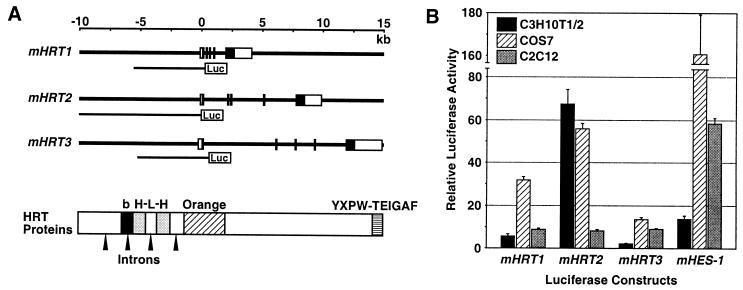
Figure 2.
Genomic structures of mHRT1, mHRT2, and mHRT3 and their transcriptional activation by Notch signaling. (A) Exon–intron structures of the mHRT genes. The filled boxes represent exons for protein-coding regions and open boxes represent exons for 5′ and 3′ noncoding regions. The genomic DNA fragments used for luciferase assays are shown below the gene structure. A schematic of HRT protein functional domains is shown with the positions of introns in the corresponding genomic sequences. Luc, luciferase; b, basic domain; H-L-H, helix–loop–helix motif; Orange, orange domain; YXPW-TEIGAF, carboxyl-terminal YXPW-TE(I/V)GAF motif. (B) Luciferase reporter assays were performed by using the mHRT1, -2, and -3 genomic DNA fragments as shown in A. The indicated cell lines were transfected with luciferase constructs and Notch1 IC expression construct or empty vector (450 ng/35-mm well each). The mHES-1-luciferase construct was used as a positive control. Fold-increase in normalized luciferase activity with Notch1 IC expression is shown.
Cultured cells were transfected with a luciferase reporter construct and various expression constructs. The CMV-LacZ expression construct (100 ng/35-mm well; ref. 12) was cotransfected for normalization. Two days after transfection, luciferase and LacZ activities in the cell extracts were assayed as described (12). All experiments were repeated at least twice, and the results from a representative experiment (n ≥ 3) are shown with standard deviations.
Site-Directed Mutagenesis.
Mutation of the Su(H)-consensus site was introduced in the NheI–NruI fragment of mHRT2 by PCR (CGTGGGAAA to CGTGGCAAA; ref. 14) and the mutated fragment was ligated into several mHRT2-luciferase constructs.
Electrophoretic Mobility Shift Assay.
DNA-binding assays were performed as described (15) using in vitro-translated proteins and oligonucleotides containing a CACGTG motif (5′-TCGAGGGTGGCACGTGCCATTG-3′, 5′-TCGAGCAATGGCACGTGCCACC-3′; ref. 16).
Results
Activation of mHRT2 mRNA Expression by Notch Signaling.
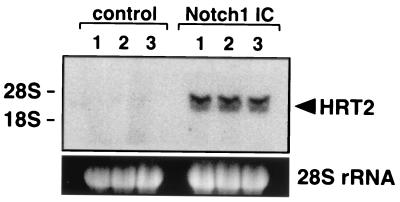
We initially investigated whether Notch activation was sufficient to stimulate HRT gene expression in 10T½ fibroblasts by using expression of Notch1 IC, which mimics activation of Notch signaling triggered by ligand binding (1, 2). As shown in Fig. 1, Notch1 IC significantly induced endogenous HRT2 mRNA expression in 10T½ cells, indicating that mHRT2 is a target gene of Notch signaling.
Figure 1.
Activation of mHRT2 mRNA expression by Notch signaling. 10T½ cells were transfected with a Notch1 IC expression construct or an empty vector (control; 2.7 μg/10-cm dish) and HRT2 transcripts were detected by Northern blot analysis. The blot was exposed to x-ray film for 8 h. Results from three independent transfections with each plasmid are shown. Ethidium bromide staining of 28S rRNA is shown at the bottom and positions of 28S and 18S rRNA are shown on the left.
Structure of Mouse HRT Genes.
To determine whether the HRT genes are direct transcriptional targets for Notch signaling, we isolated genomic DNA fragments encompassing mHRT1, -2, and -3 and their 5′-flanking regions. Each gene comprised at least five exons and intervening introns (Fig. 2A) and all the introns had boundary sequences matching the GT-AG rule. Comparing the protein functional domains with the genomic structure, an intron was present in the middle of the basic domain and in the HLH domain, which is uncommon for other bHLH proteins.
Notch-Activated Transcription of HRT Genes.
We then analyzed the effects of Notch signaling on expression of luciferase reporters linked to the 5′-flanking regions of the HRT genes, using a 6.5-kb mHRT1 fragment, a 10-kb mHRT2 fragment, and a 6.5-kb mHRT3 fragment (Fig. 2A). As shown in Fig. 2B, overexpression of Notch1 IC significantly increased luciferase activity of all three HRT genes in different cell lines. Luciferase activity of an HES-1 construct was also stimulated by Notch1 IC as described (13). The fold-increase in activation for each gene varied among cell lines and was most prominent with mHRT2 in 10T½ cells. We, therefore, further characterized the regulatory region of mHRT2 by using these cells.
Notch Activation of mHRT2 Is Mediated by the Su(H) Pathway.
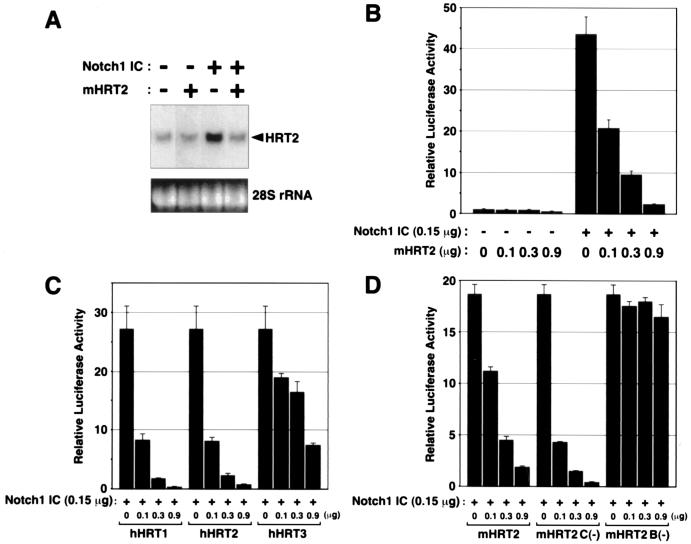
Because the effects of Notch signaling can be fulfilled through Su(H)-dependent or -independent pathways (1–3), we next examined the involvement of RBPJ, a mammalian Su(H) orthologue, in regulation of HRT gene expression. As shown in Fig. 3A, coexpression of the RBPJ dominant-negative mutant, RBPJR218H (11), clearly interfered with mHRT2 mRNA expression in response to Notch1 IC. Furthermore, expression of RBPJR218H abolished Notch-dependent activation of mHRT2-luciferase expression (Fig. 3B).
Figure 3.
Notch activation of mHRT2 is mediated by the Su(H) pathway. (A) 10T½ cells were transfected with a Notch1 IC expression plasmid (0.9 μg/10-cm dish) and/or 5.4 μg of dominant-negative RBPJ construct (RBPJR218H), and HRT2 mRNA was detected by Northern blot analysis. Note that less Notch1 IC expression plasmid was used compared with Fig. 1 because of the limitation of total plasmid amount in a transfection, and that the blot was exposed to x-ray film for 24 h. (B) The 10-kb mHRT2-luciferase construct (150 ng/35-mm well) was transfected with or without Notch1 IC and RBPJR218H constructs. (C) A series of 5′-deleted mHRT2-luciferase constructs (450 ng) was transfected with or without 450 ng of Notch1 IC plasmid. (D) Notch responsiveness of the luciferase constructs with a mutation in the proximal Su(H) site was examined as in C. Fold-increase in luciferase activity with Notch1 IC expression is shown in C and D.
To identify the cis elements responsible for the Notch responsiveness, we prepared a series of 5′ deletions of the mHRT2-luciferase constructs. As depicted in Fig. 3C, all the deletion constructs showed significant activation of transcription by Notch1 IC. The shortest, a 0.5-kb NheI–NruI fragment, was sufficient for the response to Notch activation, although the longer constructs yielded higher fold-increases.
Sequence analysis of the NheI–NruI fragment revealed two potential Su(H)-binding sites. The proximal motif, 140 bp upstream of the translation-initiation site, had a complete Su(H)-consensus sequence (CGTGGGAAA), whereas the distal one, located 203 bp upstream of the proximal site, was a complementary ATTCCCACG sequence with a single nucleotide difference from the classical Su(H)-consensus motif (T/C-GTG-G/A-GAA-A/C; ref. 3). Binding of RBPJ to the oligonucleotide fragment of the proximal Su(H) site was confirmed by gel mobility shift assay (data not shown). We, therefore, introduced a single nucleotide mutation in the proximal Su(H) site (CGTGGCAAA). Binding of RBPJ to the mutated proximal Su(H)-site fragment was not detectable in the gel mobility shift assay (data not shown). As shown in Fig. 3D, mutation of the proximal Su(H) site abolished transcriptional activation of the NheI-NruI construct (1.5-fold increase versus 19-fold increase of wild type), and drastically decreased transcription of the longer constructs. These results indicated that the transcription of mHRT2 was up-regulated by Notch activation through a Su(H)-dependent pathway.
We also characterized the 5′ regulatory regions of mHRT1 and mHRT3, and found that a proximal Su(H)-binding site was present in a conserved position of the mHRT1 and -3 promoters (data not shown). In the mHRT3 promoter, one nucleotide mismatch to the Su(H)-consensus sequence was present (CCTGGGAAA), which may possibly account for weaker activation of mHRT3 transcription (Fig. 2B).
The 3.6-kb SalI-NruI construct showed significantly higher transcriptional activation by Notch1 IC, compared with shorter constructs (Fig. 3C). In addition, mutation of the proximal Su(H) site did not abolish transcriptional activation with the 10-kb, 5.5-kb, or 3.6-kb construct (Fig. 3D). We, therefore, further sequenced the region between 3.6 kb and 0.5 kb upstream of mHRT2 and found two incomplete complementary Su(H) sites, GTTCACACG and CTTCCCACT. These motifs may provide the Notch responsiveness of mHRT2 in addition to the proximal Su(H) site. mHRT1 also had an additional complete Su(H) site upstream of the conserved proximal Su(H) site within the approximately 1-kb region we sequenced.
Negative Autoregulation of mHRT2 Gene Expression Induced by Notch Signaling.
The structural similarity between HRT and H/E(spl) proteins suggested that HRT proteins might function as negative transcriptional regulators. Because mouse HES-1, a mammalian H/E(spl)-related bHLH protein, negatively autoregulates its gene expression (17), we investigated whether HRT proteins might behave similarly. As shown in Fig. 4A, cotransfection of a mHRT2 expression plasmid significantly down-regulated mHRT2 mRNA expression activated by Notch1 IC. Consistently, expression of mHRT2 markedly down-regulated Notch-stimulated activation of the mHRT2 promoter in luciferase assays (Fig. 4B). We also examined the effects of other HRT family members on Notch-activated HRT2 promoter activity. Interestingly, similar negative regulation was observed with hHRT1 and hHRT2, whereas hHRT3, a structurally divergent member of the family (5), showed a significantly weaker potency of negative regulation (Fig. 4C).
Figure 4.
Negative autoregulation of mHRT2 gene expression induced by Notch signaling. (A) 10T½ cells were transfected with a Notch1 IC construct (0.9 μg/10-cm dish) and/or 5.4 μg of mHRT2 expression plasmid and HRT2 mRNA was detected by Northern blot analysis. The blot was exposed to x-ray film for 24 h. (B) The 10-kb mHRT2-luciferase construct (150 ng/35-mm well) was transfected with or without Notch1 IC and mHRT2 expression constructs. (C and D) 10T½ cells were transfected with a 10-kb mHRT2-luciferase construct and a Notch1 IC plasmid (150 ng each), and the effects of cotransfection of various HRT constructs were observed. mHRT2 C(−), mHRT2 mutant without carboxyl-terminal YQPW-TEVGAF motif; mHRT2 B(−), mHRT2 mutant lacking basic domain. Luciferase activity without Notch1 IC or HRT cotransfection was given a value of 1 in B–D.
We next tested the effects of mHRT2 mutants to identify the domain(s) responsible for transcriptional repression. In the carboxyl-terminal regions, HRT proteins contain a YXPW motif that is structurally similar to the WRPW motif in H/E(spl) proteins, which is essential for their association with the Groucho family of corepressors (18). In addition, mouse and human HRT proteins possess a highly conserved TE(I/V)GAF motif at their carboxyl termini (5). Carboxyl-terminal elongation of Gridlock, a zebrafish orthologue of HRT2, results in impaired vessel formation (9), suggesting that the conserved carboxyl termini of HRT proteins are functionally important in vivo. As shown in Fig. 4D, a mHRT2 mutant lacking the carboxyl-terminal YQPW-TEVGAF sequence [mHRT2 C(−)] repressed Notch-dependent activation of the HRT2 promoter. In contrast, a mutant lacking the basic domain [mHRT2 B(−)] showed no inhibition, indicating that the basic domain is essential for negative regulation (Fig. 4D). Protein expression with each construct was confirmed by Western blot analysis (data not shown).
Transcriptional Inhibition Independent of the HRT-Binding Site.
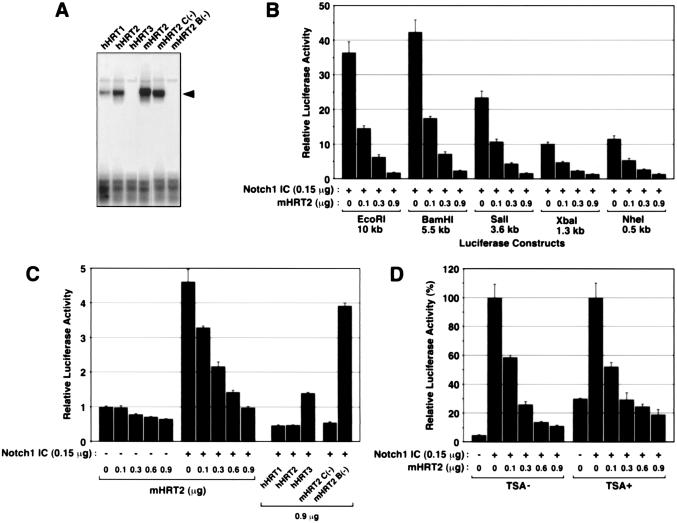
HES-1 inhibits transcription of the HES-1 gene mainly by binding to N boxes (CACNAG) in the 5′ regulatory region of the gene (17). Because the consensus cis elements to which HRT proteins bind had not been determined, we examined the binding of in vitro-translated HRT proteins to several candidate motifs by using gel mobility shift assays. As shown in Fig. 5A, mHRT2, hHRT1, and hHRT2 preferentially bound to an E box motif, CACGTG, which was recently shown to be an additional cis element for E(spl) binding (16). hHRT3 showed no detectable binding to this site. The mHRT2 C(−) mutant also bound to the CACGTG fragment, whereas the mHRT2 B(−) mutant did not show detectable binding. Binding of mHRT2 to the N box (CACAAG) and other E box motifs (CAACTG, CACCTG, CACTTG and CATCTG) was not detectable by gel mobility-shift assays (data not shown).
Figure 5.
Transcriptional inhibition of mHRT2 independent of the HRT-binding site. (A) Electrophoretic mobility shift assays were performed by using oligonucleotide probes containing a CACGTG motif and in vitro-translated HRT proteins. Specific DNA-protein complexes are indicated by an arrowhead. mHRT2 C(−), mHRT2 mutant without YQPW-TEVGAF motif; mHRT2 B(−), mHRT2 mutant lacking basic domain. (B) 10T½ cells were transfected with 5′-deletion mHRT2-luciferase constructs and Notch1 IC plasmid (150 ng each) and the effects of coexpression of mHRT2 were examined. (C) The 190-bp mHRT2-luciferase construct and Notch1 IC plasmid (150 ng each) were transfected and the effects of cotransfection of various HRT constructs were examined. Luciferase activity without Notch1 IC or HRT cotransfection was given a value of 1 in B and C. (D) The 10-kb mHRT2-luciferase construct (150 ng) and Notch1 IC and/or mHRT2 expression constructs were transfected into 10T½ cells. Trichostatin A (TSA; 300 μM) or vehicle was added to the medium 24 h after transfection and cells were incubated for 20 h. TSA treatment increased luciferase and LacZ activities in control cells with vector transfection, and the luciferase activity of cells with Notch1 IC cotransfection was given a value of 100%.
To determine whether inhibition of mHRT2 transcription was mediated through the CACGTG element, we first examined the effects of mHRT2 on a series of 5′-deletion HRT2 constructs. As shown in Fig. 5B, Notch-activated transcription of all the deletion constructs was clearly down-regulated by coexpression of mHRT2. The shortest NheI–NruI fragment did not contain a CACGTG motif but had two E box motifs (CAGGTG). To eliminate the possibility that repressive effects of HRT proteins occurred via these sites, we prepared a further 5′-deletion construct with a 190-bp ApaI–NruI fragment that contained the proximal Su(H) site but no N box or E box motifs. As shown in Fig. 5C, transcription from the ApaI-NruI construct was significantly activated by Notch1 IC, and cotransfection of various HRT plasmids suppressed it to the same extent as we observed with the 10-kb luciferase construct (Fig. 4 B–D). These results suggested that negative regulation of mHRT2 gene expression was independent of binding of HRT proteins to these consensus elements.
Stra13, a distantly related member of the H/E(spl) family, also negatively autoregulates its gene expression (19). Although the mechanism of negative regulation is unknown, the repressive effects of Stra13 are inhibited by treatment with trichostatin A (TSA), a histone deacetylase (HDAC) inhibitor (19). In contrast, repression of the mHRT2 promoter activity by mHRT2 was maintained in cells treated with TSA (Fig. 5D), suggesting that the effects of mHRT2 did not occur by recruitment of HDACs. These results suggested that negative autoregulation of mHRT2 gene expression may be mediated by a mechanism distinct from those in the HES-1 and Stra13 genes.
Discussion
The present study demonstrates that the HRT genes are downstream targets for transcriptional activation by Notch signaling and that their responsiveness to Notch is mediated by binding of Su(H) to their 5′ regulatory regions. Our results also reveal the existence of a negative-feedback loop in which HRT proteins interfere with Notch-dependent activation of HRT2 expression. This type of negative autoregulation may function to terminate or dampen Notch signaling, thereby resulting in a transient or periodic signal as is characteristic of Notch signaling in the developing paraxial mesoderm.
Within the presomitic mesoderm, the HRT genes show unique expression patterns similar to those of Notch-related molecules (5), suggesting their participation in Notch signaling pathways. Consistent with this notion, mouse embryos lacking the Notch-receptor ligand, Dll1, show decreased expression of HRT1/Hesr1 in the somites (6). Conversely, transgenic overexpression of activated Notch1 in the cortex layer of hair follicles in mice results in misexpression of HRT3/HeyL (20). H/E(spl) proteins are also expressed in presomitic precursors and confer Notch responsiveness to specific target genes (10). The present study shows that HRT and HES proteins exhibit distinct DNA-binding specificities that may reflect unique roles of these transcriptional repressors in regulation of downstream genes in Notch signaling cascades. Intriguingly, coimmunoprecipitation experiments have shown that HRT2 can heterodimerize with HES-1 (D.G.M. and E.N.O., unpublished observations), raising the possibility that these factors may act independently or cooperatively to mediate the effects of Notch on somitic development.
HRT genes are also highly expressed in the embryonic vasculature, including the outflow tract of the heart and the aortic sac (5), and zebrafish embryos harboring a mutation in Gridlock, an orthologue of HRT2, show impairment of vascular formation (9). Notch4 receptor and Dll4 ligand are specifically expressed in the vasculature (21, 22), and Notch1 or Notch1/Notch4 mutant embryos show embryonic lethality because of vascular defects (23). Moreover, human mutations in Jagged1, which encodes a Notch ligand, cause defects in the outflow tract and aortic arch artery derivatives (24, 25). Because the HES genes have not been reported to be highly expressed in the embryonic vasculature (4, 26), it is conceivable that the HRT genes play primary roles downstream of Notch signaling in vascular development.
There is not an exact correlation between sites of Notch signaling and HRT expression throughout embryogenesis, indicating that HRT proteins are not obligatory downstream mediators of Notch signaling. In the heart, for example, HRT1 and HRT2 are expressed in the atria and ventricles, respectively, in a complementary fashion (5). Notch receptors and their ligands have not been reported to show this type of chamber-restricted expression (1, 2), suggesting that cardiac expression of these genes may be Notch-independent. HRT genes are likely to respond to regulatory inputs in addition to Notch signaling.
HES proteins repress transcription mainly by direct binding to an N box, and by dimerization with E proteins, thereby preventing other bHLH activators from binding to an E box (26). Our results show that HRT proteins can bind to the E box motif, CACGTG. However, negative autoregulation of mHRT2 transcription seemed to be independent of this binding activity, because the repressive effects of HRT proteins were observed with a mHRT2 fragment containing no N box or E box motifs. The finding that the basic region was required for the transcriptional inhibition by HRT2 suggests that DNA binding is important for this effect. The basic domain could also mediate the protein-protein interaction independent of DNA binding, analogous to that between MyoD and its transcriptional cofactor MEF2 (27). HRT2/CHF1 binds to the aryl hydrocarbon receptor nuclear translocator (ARNT) and inhibits ARNT-dependent transcription by dissociating the ARNT complex from DNA (8). HRT proteins may dimerize with other proteins and bind to unrecognized sites in the mHRT2 promoter.
The bHLH protein Stra13 directly associates with the promoter complex and inhibits the promoter activity of c-myc by a histone deacetylase (HDAC)-independent mechanism, like the autoregulation of mHRT2 in this study, whereas the effects of Stra13 on its own gene expression were suppressed by HDAC inhibition (19). It is also conceivable that HRT proteins inhibit mHRT2 expression through association with components of the basal transcriptional machinery.
Notch signaling pathways are characterized by multiple mechanisms of feedback regulation (1, 2). Activation of Notch signaling causes up-regulation of Notch receptor expression, reinforcing the responsiveness to Notch ligands. In contrast, Notch signaling down-regulates the expression of Notch ligands, which specifies the signaling and responding cells. Downstream of Notch signaling, negative autoregulation of HRT or HES gene expression can serve to spatially and temporally restrict the activation of Notch-dependent signaling. In light of the well known roles of Notch signaling in diverse developmental processes, it will be especially interesting to determine which of the activities of Notch rely on HRT proteins as essential downstream mediators and to identify target genes for HRT proteins in different cell types.
Acknowledgments
We thank R. Kopan, U. Lendahl, and T. Honjo for the plasmids; J. Page, S. Johnson, and A. Tizenor for assistance with the manuscript; and J. Johnson, W. Klein, and R. MacDonald for helpful comments. E.N.O. was supported by grants from National Institutes of Health and the D. W. Reynolds Foundation; D.S. was supported by grants from National Institutes of Health, the March of Dimes, and Smile Train, Inc.; O.N. was supported by Japan Heart Foundation & Bayer Yakuhin Research Grant Abroad, the Uehara Memorial Foundation, and the Yamanouchi Foundation for Research on Metabolic Disorders; and D.G.M. was supported by a Medical Scientist Training Program grant from the National Institutes of Health.
Abbreviations
- bHLH
basic helix–loop–helix
- H/E(spl)
Hairy and Enhancer of split family
- HRT
Hairy-related transcription factor
- hHRT
human HRT
- mHRT
mouse HRT
- Notch1 IC
intracellular domain of Notch1 receptor
- Su(H)
Suppressor of Hairless
Footnotes
Data deposition. The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF311883, AF311884, and AF311885, for human cDNAs HRT1, HRT2, and HRT3, respectively).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250485597.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250485597
References
- 1.Artavanis-Tsakonas S, Rand M D, Lake R J. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Weinmaster G. Curr Opin Genet Dev. 1998;8:436–442. doi: 10.1016/s0959-437x(98)80115-9. [DOI] [PubMed] [Google Scholar]
- 3.Bailey A M, Posakony J W. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 4.Kageyama R, Nakanishi S. Curr Opin Genet Dev. 1997;7:659–665. doi: 10.1016/s0959-437x(97)80014-7. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa O, Nakagawa M, Richardson J A, Olson E N, Srivastava D. Dev Biol. 1999;216:72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- 6.Kokubo H, Lun Y, Johnson R L. Biochem Biophys Res Commun. 1999;260:459–465. doi: 10.1006/bbrc.1999.0880. [DOI] [PubMed] [Google Scholar]
- 7.Leimeister C, Externbrink A, Klamt B, Gessler M. Mech Dev. 1999;85:173–177. doi: 10.1016/s0925-4773(99)00080-5. [DOI] [PubMed] [Google Scholar]
- 8.Chin M T, Maemura K, Fukumoto S, Jain M K, Layne M D, Watanabe M, Hsieh C-M, Lee M-E. J Biol Chem. 2000;275:6381–6387. doi: 10.1074/jbc.275.9.6381. [DOI] [PubMed] [Google Scholar]
- 9.Zhong T P, Rosenberg M, Mohideen M-A P K, Weinstein B, Fishman M C. Science. 2000;287:1820–1824. doi: 10.1126/science.287.5459.1820. [DOI] [PubMed] [Google Scholar]
- 10.Pourquie O. Curr Opin Genet Dev. 1999;9:559–565. doi: 10.1016/s0959-437x(99)00011-8. [DOI] [PubMed] [Google Scholar]
- 11.Kato H, Taniguchi Y, Kurooka H, Minoguchi S, Sakai T, Nomura-Okazaki S, Tamura K, Honjo T. Development (Cambridge, UK) 1997;124:4133–4141. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, McKinsey T A, Nicol R L, Olson E N. Proc Natl Acad Sci USA. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroeter E H, Kisslinger J A, Kopan R. Nature (London) 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 14.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 15.Charité J, Graaff W D, Consten D, Reijnen M J, Korving J, Deschamps J. Development (Cambridge, UK) 1998;125:4349–4358. doi: 10.1242/dev.125.22.4349. [DOI] [PubMed] [Google Scholar]
- 16.Jennings B H, Tyler D M, Bray S J. Mol Cell Biol. 1999;19:4600–4610. doi: 10.1128/mcb.19.7.4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takebayashi K, Sasai Y, Sakai Y, Watanabe T, Nakanishi S, Kageyama R. J Biol Chem. 1994;269:5150–5156. [PubMed] [Google Scholar]
- 18.Fisher A L, Caudy M. Genes Dev. 1998;12:1931–1940. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 19.Sun H, Taneja R. Proc Natl Acad Sci USA. 2000;97:4058–4063. doi: 10.1073/pnas.070526297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin M-H, Leimeister C, Gessler M, Kopan R. Development (Cambridge, UK) 2000;127:2421–2432. doi: 10.1242/dev.127.11.2421. [DOI] [PubMed] [Google Scholar]
- 21.Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Development (Cambridge, UK) 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 22.Shutter J R, Scully S, Fan W, Richards W G, Kitajewski J, Deblandre G A, Kintner C R, Stark K L. Genes Dev. 2000;14:1313–1318. [PMC free article] [PubMed] [Google Scholar]
- 23.Krebs L T, Xue Y, Norton C R, Shutter J R, Maguire M, Sundberg J P, Gallahan D, Closson V, Kitajewski J, Callahan R, et al. Genes Dev. 2000;14:1343–1352. [PMC free article] [PubMed] [Google Scholar]
- 24.Oda T, Elkahloun A G, Pike B L, Okajima K, Krantz I D, Genin A, Piccoli D A, Meltzer P S, Spinner N B, Collins F S, Chandrasekharappa S C. Nat Genet. 1997;16:235–242. doi: 10.1038/ng0797-235. [DOI] [PubMed] [Google Scholar]
- 25.Li L, Krantz I D, Deng Y, Genin A, Banta A B, Collins C C, Qi M, Trask B, Kuo W L, Cochran J, et al. Nat Genet. 1997;16:243–251. doi: 10.1038/ng0797-243. [DOI] [PubMed] [Google Scholar]
- 26.Sasai Y, Kageyama R, Tagawa Y, Shigemoto R, Nakanishi S. Genes Dev. 1992;6:2620–2634. doi: 10.1101/gad.6.12b.2620. [DOI] [PubMed] [Google Scholar]
- 27.Molkentin J D, Black B L, Martin J F, Olson E N. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]