Abstract
The bacteriophage P1 Cre/loxP system has become a powerful tool for in vivo manipulation of the genomes of transgenic mice. Although in vitro studies have shown that Cre can catalyze recombination between cryptic “pseudo-loxP” sites in mammalian genomes, to date there have been no reports of loxP-site infidelity in transgenic animals. We produced lines of transgenic mice that use the mouse Protamine 1 (Prm1) gene promoter to express Cre recombinase in postmeiotic spermatids. All male founders and all Cre-bearing male descendents of female founders were sterile; females were unaffected. Sperm counts, sperm motility, and sperm morphology were normal, as was the mating behavior of the transgenic males and the production of two-celled embryos after mating. Mice that expressed similar levels of a derivative transgene that carries an inactive Cre exhibited normal male fertility. Analyses of embryos from matings between sterile Cre-expressing males and wild-type females indicated that Cre-catalyzed chromosome rearrangements in the spermatids that lead to abortive pregnancies with 100% penetrance. Similar Cre-mediated, but loxP-independent, genomic alterations may also occur in somatic tissues that express Cre, but, because of the greater difficulty of assessing deleterious effects of somatic mutations, these may go undetected. This study indicates that, following the use of the Cre/loxP site-specific recombination systems in vivo, it is prudent to eliminate or inactivate the Cre recombinase gene as rapidly as possible.
The bacteriophage P1 recombinase, Cre, is a member of the integrase family of site-specific recombinases that catalyzes recombination between loxP DNA elements as a part of the normal viral life cycle (1, 2). All members of the integrase family share a similar mechanism of action (3). In Cre-mediated recombination, individual loxP sites are bound by Cre homodimers (4). Two Cre-bound loxP sites interact in the recombination reaction. The loxP site is a 34-bp element consisting of two 13-bp inverted repeats separated by a directional 8-bp core (1, 5–7). During cleavage, Cre Tyr324 becomes covalently linked to the 3′-phosphate, forming a DNA-phosphotyrosine intermediate (8, 9). Interaction with a second loxP-bound Cre dimer allows nucleophilic attack of the 3′-phosphotyrosine in one loxP site by the free 5′-hydroxyl of the other. This ligation reaction induces a structural rearrangement of the tetrameric complex, allowing the same reaction to occur on the second strand of each loxP site, thus completing the strand exchange reaction (8, 10–12).
Cre has proven to be a valuable tool for manipulating the genomes of mammalian cells and of mice in situ (13–19). As an example, Cre-mediated recombination has all but eliminated the problem of “selection cassette effects” associated with some targeted mutations (15). Although Cre is most commonly used to generate short deletions in transgenic mice (13), it has also proven useful for recombination between sites up to several centimorgans apart on individual chromosome arms (17) and for catalyzing unequal sister-chromatid exchanges in cultured cells and in mice (17, 18).
In vitro studies have shown that Cre recombinase is capable of catalyzing recombination between DNA sequences found naturally in yeast (20, 21) and mammalian (22) genomes, termed “pseudo-loxP sites.” These illegitimate sites often bear little primary sequence similarity to the phage P1 loxP element (22). Nonetheless, there have been, as yet, no reports of Cre-site infidelity in transgenic animals, suggesting that illegitimate Cre recombination might not occur in vivo. The apparent fidelity of Cre for bona fide loxP sites in vivo has led to numerous proposals and pilot studies that employ the Cre/loxP system in human gene therapy protocols as a means for assembling genetic elements in situ (14, 23, 24), for targeting foreign elements into the genome (22, 25), or for removing unwanted exogenous genetic elements posttherapy (26–29). In this study, we show that chronic high-level expression of Cre in the spermatids of transgenic mice leads to male sterility with 100% penetrance. These results indicate that Cre can catalyze illegitimate recombination having overt pathological consequences in animals.
Materials and Methods
Design of Expression Vectors and Production of Transgenic Mice.
The mouse protamine 1 (Prm1)-Cre-human growth hormone (hGH) vector (30) contains mouse prm1 sequences extending from 4.1-kb pairs upstream of the cap site to a synthetic linker inserted 91 bp downstream of the cap site (31) fused to a version of the bacteriophage P1 Cre cistron, which had been modified for expression in mammalian cell nuclei (13), followed by 0.8 kb of genomic 3′ flanking sequences from the hGH gene extending from a BglII site near the 3′ end of protein-coding region of the last exon, through the polyadenylation site and signals, to the 3′ EcoRI site (Fig. 1a; ref. 32). To create the CreYI-VD cassette, Tyr324 and Ile325 were converted to Val and Asp, respectively (sequence TAT ATC converted to GTC GAC, creating an SalI site) by PCR-mediated site-directed mutagenesis. All constructs were confirmed by sequencing. CreYI-VD-hGH was inserted into both the Prm1 transgenic mouse expression vector and a cytomegalovirus (CMV) expression vector (33). In both cases, the 91-bp 5′-untranslated region from the prm1 gene was retained.
Figure 1.
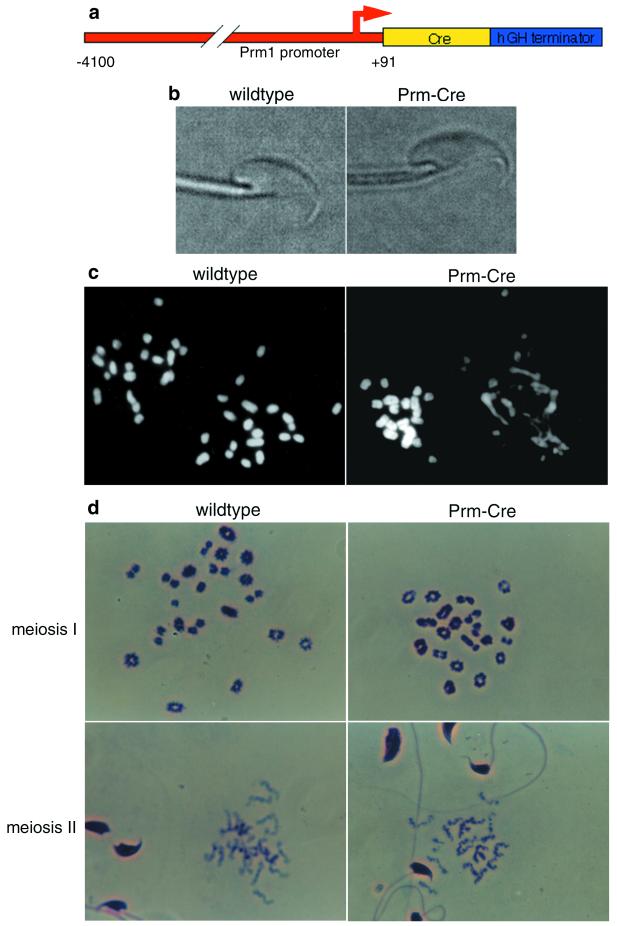
Postmeiotic chromatin disorganization in Prm1-Cre-hGH male mice. (a) Transgene structure. Red indicates sequences derived from the mouse Prm1 gene; yellow denotes the bacteriophage P1 Cre gene; and blue denotes 3′ flanking sequences from the hGH gene. (b) Sperm morphology. High magnification of wild-type and Prm1-Cre-hGH spermatozoa showed no visible differences. The focal plane was set to show the sperm head shape; differences in tail-shaft appearance are because of focal plane. (c) DAPI-stained chromosome spreads of fertilized eggs from matings between wild-type females and either wild-type (Left) or Prm-Cre-hGH males (Right). (d) Meiotic chromosome spreads from wild-type and Prm1-Cre-hGH testes. Testicular sperm at various stages of condensation appear in meiosis II spread fields for stochastic reasons.
Transgene vectors were linearized, DNA was microinjected into one pronucleus of fertilized eggs from matings between wild-type C57BL/6 mice, and pups were reared in surrogate mothers (34). At weaning, tail snips were harvested and DNA was prepared for PCR-based genotyping. Transgene copy was estimated by quantitative PCR and by Southern blotting (30). Founders bearing the transgene were mated with wild-type C57BL/6 mice.
Karyotyping and Embryo Counts.
For examining the chromosomal spreads of fertilized eggs, wild-type C57BL/6 females were mated with wild-type or heterozygous Prm1-Cre-hGH transgenic males, and fertilized eggs were harvested from the oviducts the next morning. Cumulus cells were removed from egg clusters by using 0.3 mg/ml hyaluronidase in M2 medium (Sigma). Eggs were incubated overnight at 37°C in M16 medium (Sigma) containing 0.5 μg/ml colcemid. Zona pelucidae were removed by using acidic Tyrode's solution (Sigma). Eggs were swollen in 0.1 mM KCl, fixed in methanol/acetic acid (3:1), and metaphase chromosome spreads were attached to slides. Slides were stained with 4′,6-diamidino-2-phenylindole (DAPI) and photographed by fluorescence microscopy.
For egg and embryo progression counts, eggs were harvested from the matings described in Table 1 as described above, and these were cultured in M16 media in a humidified, 5% CO2, 37°C incubator.
Table 1.
In vitro embryo development
| Number of embryos at various times after mating
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.5 day
|
1.5 days
|
2.5
days
|
3.5 days
|
|||||||||||||
| Male | (Egg harvest) | 1 | 2 | 3 | 4 | M | 1 | 2 | 3 | 4 | M | 1 | 2 | 3 | 4 | M |
| wt | (66 eggs)* | 27 | 36 | 3 | — | — | 26 | 3 | 14 | 12 | 11 | 26 | 3 | 3 | 2 | 32 |
| Cre | (60 eggs)* | 40 | 19 | 1 | — | — | 33 | 16 | 7 | 4 | — | 33 | 16 | 7 | 4 | — |
Three wild-type (wt) and three heterozygous Prm1-Cre-hGH (Cre) males were mated with wild-type females (all males were 12–24 wk old, females were 6–8 wk old), and eggs were harvested from oviducts of sacrificed females on the following morning (0.5 day postmating). Zygotes were cultured for 3 days, and the number of 1-, 2-, 3-, 4-, and >4-celled (morulae and blastocysts) embryos (1–4 and M, respectively) were counted each day.
Some of the one-celled embryos may have been unfertilized eggs or may have been damaged during harvest, accounting for the embryos that did not progress beyond the one-celled stage in matings with either wild-type or Prm1-hGH-Cre males. Embryos harvested at 2.5 days after fertilization from matings with either wild-type or Prm1-Cre-hGH males contained only 10% one-celled embryos (see text), which is consistent with this interpretation.
To examine meiotic chromosomal spreads, cells were squeezed from seminiferous tubules of adult wild-type or Prm1-Cre-hGH mice and swollen in 1% sodium citrate solution. After fixation (see above), metaphase chromosomes were attached to slides, stained with Geimsa solution, and photographed by using phase contrast microscopy.
RNase Protection Analyses.
RNase protection analyses were performed as described previously (30). The Cre probe, which hybridizes to 193 bases of either Cre or CreYI-VD mRNA, and the Prm1 probe, have been described previously (30). All hybridizations contained the amount of total testis RNA indicated in the figure, supplemented with yeast RNA to a total of 50 μg. For Cre analyses, 20 fmol of probe was used in each hybridization; for Prm analyses, probe-specific activity was reduced 30-fold, and 100 fmol of probe was used.
Recombination Assay.
For an in vivo recombination substrate, a 1.25-kb loxP-flanked fragment of arbitrary bacterial sequence was inserted into a BamHI site near the middle of an arbitrarily chosen 5.4-kb EcoRI fragment of mouse genomic sequence in a Bluescript SK− vector (Stratagene). Primers flanking the BamHI site yield a 108-bp PCR product on mouse genomic sequence (data not shown), but no product with human genomic DNA (Fig. 2d, lanes 1 and 2). Nonrecombined vector yields a 1.4-kb-pair PCR product, whereas recombination excises the 1.25-kb-pair loxP-flanked insert yielding a 180-bp loxP-containing PCR product (Fig. 2d, lanes 3–9).
Figure 2.
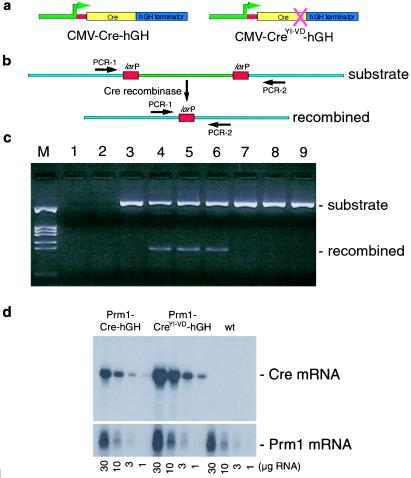
CreYI-VD mutant. (a) Somatic cell CreYI-VD-hGH expression vectors. Colors as in Fig. 1a; green denotes CMV promoter sequences. (b) In vivo recombination assay. A fragment of arbitrary extragenic mouse genomic DNA (light blue) containing two loxP sites (red) flanking a 1.2-kb insert (green) was transiently cotransfected into human 293 cells with either CMV-Cre-hGH (lanes 4–6) or CMV-CreYI-VD-hGH. After 48 h, DNA was extracted and analyzed for recombination by using the indicated PCR primers. Lane M, DNA size markers. Lanes 1 and 2, Cells transfected with 2.0 μg CMV-Cre-hGH or CMV-CreYI-VD-hGH, respectively. Lane 3, Cells transfected with 1.0 μg substrate plasmid only. Lanes 4–6, Cells cotransfected with 1.0 μg substrate plasmid and 2.0 μg, 0.2 μg, or 0.02 μg of CMV-Cre-hGH expression plasmid, respectively. The similar yield of recombined product in all three lanes indicates that even the lowest amount of CMV-Cre-hGH plasmid yielded 100% recombination in cotransfected cells. Unrecombined substrate in lanes 4–6 likely stems from substrate plasmid that did not enter cells and therefore was not exposed to Cre. Lanes 7–9, Like lanes 4–6, except that CMV-CreYI-VD-hGH was used in place of CMV-Cre-hGH. In all cases, Bluescript plasmid vector DNA was used to supplement each transfection to a total of 3.0 μg DNA. (d) Expression of Prm1-Cre-hGH and Prm1-CreYI-VD-hGH mRNAs in transgenic mouse testes. RNase protection analyses were performed on the indicated amounts of total RNA harvested from testes of 12- to 14-wk-old wild-type (wt), Prm1-Cre-hGH, or Prm1-CreYI-VD-hGH mice. The Cre probe hybridizes to 193 bases of either Cre or CreYI-VD mRNA. The endogenous Prm1 mRNA signal served as a normalization control.
Recombination substrate plasmid, CMV-Cre expression vector, and Bluescript carrier DNA were mixed as indicated in the figure and transfected into human kidney H293 cells with Superfect (Qiagen, Chatsworth, CA) using the manufacturer's protocols. After 48 h, cells were rinsed twice with PBS and harvested by scraping in 10 mM Tris, pH 7.5, 5 mM EDTA, 1% SDS, and 200 μg/ml proteinase K. After incubation at 50°C for 2 h, proteins were removed by phenol/chloroform extraction. Samples received 0.5 volumes of 5 M NaCl and two volumes of ethanol, and DNA was spooled out for analysis by PCR.
Results
Production and Phenotype of Prm1-Cre-hGH Mice.
We wished to create a line of transgenic mice that expressed Cre in postmeiotic spermatids to catalyze specific deletions in the germ lines of mouse lines bearing loxP-flanked sequences. We inserted a Cre recombinase transgene, which had been modified for activity in eukaryotic cell nuclei (13), between a 4.1-kb region of the mouse prm1 promoter (31) and 3′-flanking sequences from the hGH gene (32). This vector was injected into the pronuclei of fertilized C57BL/6 mouse eggs (Fig. 1a; ref. 30). We obtained five male and three female founders bearing the Prm1-Cre-hGH transgene. All five males were viable and robust, but infertile. The females were normal and fertile. Curiously, all Cre-bearing male descendents of these females, although exhibiting normal health and viability, were also infertile. Conversely, male descendents lacking the Cre transgene and all female descendents exhibited normal fertility yielding litters with normal sex ratios. We have followed the progeny of one of these founders for seven generations and those of another for three, yet we have obtained no fertile males bearing the transgene. None of these animals carried exogenous loxP sites. RNase protection analyses confirmed that the transgenic males expressed the Cre transgene in their testes (see below).
Because all female and male founder lines resulted in the same male-sterility phenotype, we conclude that the phenotype arose from transgene expression and not from the site of transgene integration into the mouse genome. The same expression cassette has been used to generate mice expressing other transgenes in their haploid spermatids, including the jellyfish green fluorescent protein (GFP) and hGH (30, 32). These transgenic mice generally exhibited normal male fertility, even though they often had transgene copy and mRNA expression levels exceeding those for the Prm1-Cre-hGH transgene in the current study (data not shown; ref. 30).
The Prm1-Cre-hGH males exhibited normal mating behavior based on the presence of semen plugs in wild-type female partners after overnight matings. Semen harvested from the vas deferens of transgenic males showed normal sperm counts. Sperm motility was similar to that of wild-type males, and sperm exhibited no apparent morphological abnormalities (Fig. 1b).
Lethality of Embryos Derived from Matings Between Prm-Cre-hGH Males and Wild-Type Females.
We next harvested 0.5-day zygotes and 2.5-day embryos from the oviducts of wild-type females mated with the transgenic males or with wild-type control males (Table 1). The 0.5-day zygotes exhibited two visible pronuclei, indicating that eggs were successfully bound and fertilized by sperm from both wild-type and Prm1-Cre-hGH males. Overnight culture of the embryos from either transgenic or wild-type males yielded large numbers of two-celled embryos, indicating that sperm from the Prm1-Cre-hGH mice were able to activate embryonic development. Unlike for embryos from wild-type matings, of which most progressed to the four-cell stage or beyond following an additional 48 h in culture, few embryos from transgenic male founders progressed to the four-cell stage. Embryos harvested at 2.5 days from control males were predominantly morulae (85% at 4- to 16-cell stage; 21 embryos from 2 mated females), whereas embryos from matings with the Prm1-Cre-hGH transgenic males were arrested at the 1- (10%), 2- (86%), 3- (2%) , or 4-cell (2%) stage (48 embryos from 4 mated females).
Studies of transgene inheritance indicated that lines from our female founders each had a single independent site of transgene insertion. Because only females were fertile, only heterozygous transgenic animals could be generated. Therefore, half of the sperm from transgenic males were hemizygous-wild-type. However, no sperm were capable of contributing to an embryo that could develop past the four-cell stage. We have previously used highly sensitive techniques to look for spermatid mRNAs in epididymal sperm (Prm1 and TATA box-binding protein mRNA) and were unable to detect any (≤0.1 mRNA molecule per spermatozoan; ref. 35), suggesting that mRNAs synthesized during spermiogenesis are neither packaged into sperm nor transported to the egg. Thus, the phenotype is not likely to stem from Cre expression postfertilization (see below), but rather, from a Cre-dependent modification of the sperm that has no effect on the quantity, motility, morphology, egg-binding, fertilization, or embryogenic activation potential of the sperm.
Transgene-containing and transgene-lacking sperm from heterozygous males appeared to be equally affected. Because the vector that we used directs transgene expression exclusively in postmeiotic spermatids (30–32, 36), the Prm1-Cre-hGH transgene is likely affecting hemizygous-wild-type sperm only after they have been segregated from their hemizygous-transgenic sister nuclei by meiosis II (see below). Postmeiotic sister spermatids are connected by cytoplasmic bridges that allow the transfer of mRNA and/or protein (37–39). We hypothesize, therefore, that, in heterozygous males, a product from hemizygous transgenic spermatids, either Cre mRNA or Cre protein, traversed the cytoplasmic bridges and compromised the hemizygous-wild-type sperm (see below). Because Cre is capable of binding, cleaving, and trans-ligating DNA (8, 9), a likely mechanism of sterility would involve one or more of these activities.
Chromosomal Consequences of Spermatid-Specific Cre Expression.
To examine the transgenic karyotype, transgenic males were mated with wild-type females and fertilized eggs were harvested. These were incubated overnight in colcemid, at which time chromosome spreads were prepared. Colcemid has two effects in this assay: it blocks nuclear migration, such that male and female pronuclei remain separated, and it arrests nuclei in metaphase. As a result, male and female pronuclei can be detected as adjacent but discrete entities. Eggs fertilized by wild-type males gave two adjacent spreads, having 20 normal chromosomes in each. Eggs fertilized by transgenic males gave one apparently normal spread of 20 chromosomes and one with “scrambled DNA,” having amorphous regions of chromatin condensation connected by fine strands of DNA (Fig. 1c). Because pronuclei containing normal haploid spreads never contained a visible Y chromosome, we infer that these were from the female pronuclei, and that the scrambled spreads were from the male pronuclei. In every case, only one pronucleus was visibly affected, further confirming that the effect did not likely stem from expression of transported Cre mRNA or protein in the fertilized egg (see above). The quantity of DAPI fluorescence in each scrambled male pronucleus indicated that they were haploid, which is consistent with the idea that scrambled DNA resulted from rearrangements after the completion of meiosis II (see above), and subsequent to the segregation of the individual haploid nuclei. If scrambling had occurred earlier, we would expect the male pronucleus to contain diploid (if scrambling were before meiosis II), tetraploid (if before meiosis I), or intermediate (if during S phase) DNA content. To further test this hypothesis, we prepared chromosome spreads from wild-type and transgenic mouse testes. Both meiosis I and meiosis II chromosomes from transgenic male testes showed no signs of scrambling and were indistinguishable from those of the wild-type controls (Fig. 1d), which confirmed that chromosome scrambling followed completion of meiosis II.
Cre Enzyme Activity Is Required for Chromatin Scrambling in the Transgenic Mice.
Taken together, the results described above imply strongly that male infertility in the transgenic mice occurs in the testis from some postmeiotic action of a Cre transgene product, expressed independently of the transgene's insertion site, and transmitted through cytoplasmic bridges to wild-type male spermatids. However, the results did not determine whether the chromatin scrambling resulted from Cre-mediated enzymatic activity. Therefore, we produced a second transgene that was identical to the first in all respects except that Tyr324 and Ile325 of Cre were converted to Val and Asp, respectively (CreYI-VD mutant). This mutation was chosen for four reasons: First, Tyr324 forms the covalent DNA-phosphotyrosine intermediate in Cre catalysis (see above) and Val cannot participate in this reaction. Second, these amino acids are near the C terminus of the 343-residue protein and are away from the hydrophobic core (9), reducing the likelihood that they would disrupt overall protein structure. Third, neither of the mutated amino acids is among the 42 nonactive-site amino acids in Cre that contact DNA (≤3.6 angstroms; ref. 9), reducing the likelihood that the mutations would affect DNA-binding or Cre–Cre interactions. Finally, by using this substitution, we could introduce a diagnostic restriction enzyme recognition site into the mutant while making a replacement that was not expected to disrupt the local α-helical nature of this region of the protein.
To test the effectiveness of this mutation, both wild-type Cre and CreYI-VD were cloned into a CMV promoter-based expression vector (33), and these were transiently cotransfected into human 293 cells with a loxP-containing recombination substrate (Fig. 2 a and b). Analyses after 48 h revealed efficient recombination of the substrate by wild-type Cre, but no detectable recombination by Cre YI-VD (Fig. 2c).
Next, transgenic mice bearing the Prm1-CreYI-VD-hGH transgene were produced. Males bearing the mutant transgene were found to exhibit normal fertility. RNase protection analyses were used to compare transgene mRNA expression in testes from a sterile Prm1-Cre-hGH transgenic mouse line and a fertile Prm1-CreYI-VD-hGH mouse line (Fig. 2d). The results showed that transgene mRNA expression was similar between the two lines. We conclude that it is the Cre recombinase activity that induces male sterility in the Prm1-Cre-hGH mice. Specifically, the subtle nature of the rescuing mutation in Prm1-CreYI-VD-hGH, affecting only the active-site DNA-cleavage residues of Cre, leads us to posit that infertility in the Prm1-Cre-hGH males stemmed from hydrolysis and/or ligation of DNA mediated by Cre recombinase.
Discussion
The Cre/loxP site-directed recombination system has become an important tool for genetic manipulation of transgenic animals and may have future importance as a tool for human gene therapy. In the current study, we show that persistent high-level Cre expression in the spermatids of transgenic mice can lead to gross chromosomal rearrangements in spermatozoa. The rearrangements are dependent on Cre enzyme activity, but occur in the absence of exogenous loxP sites. Our finding may have important implications for in vivo use of the Cre/loxP system. Specifically, they emphasize the importance of limiting the duration of in vivo Cre expression to only the period required for the desired recombination event, rather than allowing chronic Cre activity in the target cells.
Many lines of transgenic mice that express the Cre recombinase gene have been generated in multiple laboratories. However, this is the first report of an overt effect of Cre in mice that is independent of exogenous (i.e., transgenic) loxP sites. Thus, we may ask why similar effects have not been reported previously. Several factors may have contributed to the outcome of our experiments. First, a very proficient promoter was used to direct the expression of the Cre transgene, which might increase otherwise undetectable rates of Cre-mediated recombination between pseudo-loxP sites. Second, the target tissue for Cre expression was developing spermatids, which may have influenced the outcome in several ways. Thus, only mutations that affect the germ line, as opposed to the somatic genome, are transmitted from generation to generation, which makes them readily apparent. In addition, the spermatid genome undergoes dramatic chromatin reorganization wherein histones are replaced by transition proteins and protamines (40). During this transition, the DNA of spermatids may be particularly vulnerable to Cre-mediated reactions.
Conversely, if the expression of the Cre transgene is restricted to somatic tissues, the consequences of recombination between pseudo-loxP sites in these cells could be missed. If the cells were dividing, a likely consequence of such genomic alterations would be cell death because of the inability to properly segregate chromosomes between daughter cells. Only if the fraction of dying cells in the Cre-expressing tissue were very large would there be a likelihood of detecting an associated defect in the affected tissue. Mammals are remarkably tolerant of the effects of somatic cell death. For example, during normal development, approximately half of the neurons generated by both the central and peripheral nervous systems undergo programmed cell death (41). Other tissues can exhibit similarly high levels of programmed cell death as well (42).
Interestingly, a previous study reported the generation of mice carrying a Cre transgene driven by a promoter similar to that reported here (43). However, those transgenic mice were not reported to exhibit male sterility. Because both the current and former studies produced multiple independent founder lines, the difference in phenotypes is likely because of differences in Cre expression rather than to effects of different transgene insertion sites. The previous study used a truncated form of the Prm1 promoter (i.e., 652 bp as compared with 4100 bp in our vector), used a different untranslated region, which is known to affect the timing of translation of spermatid mRNAs (30, 32), and incorporated an epitope tag. These differences are likely to have resulted in lower levels of Cre transcription, as well as differences in the timing of Cre mRNA translation or accumulation of Cre protein within the nuclei of developing spermatids. Any of these effects could readily account for the differences in phenotype exhibited in the two studies.
It is possible that Cre-mediated recombination reactions similar to those described here in developing spermatids can also occur in somatic cells. However, as discussed above, the frequency of such events may be lower in somatic tissues. The RAG recombinases of vertebrate immune systems are also members of the integrase family, and, like Cre, show high target site fidelity (3). Nevertheless, translocations caused by rare instances of site infidelity by RAG recombinases in somatic cells are involved in many human leukemias (44, 45).
In summary, we have demonstrated that expression of the Cre recombinase gene in developing spermatids can lead to male sterility with 100% penetrance. The cause of sterility is likely because of Cre-mediated genomic rearrangements, perhaps at pseudo-loxP sites within the mouse genome. Such sites have been identified previously within the mouse and human genomes and have been shown to function in Cre-mediated recombination assays performed in bacteria and cultured mammalian cells (22). Our results suggest that, subsequent to in vivo use of the Cre/loxP system to mediate genomic rearrangements, or to control gene activities in a conditional manner, it would be prudent to remove or inactivate the Cre gene as rapidly as possible. For such purposes, the self-excising Cre vectors described by Bunting et al. (19) may be particularly attractive. The potential consequences of Cre-mediated recombination between cryptic sites within the human genome should also be seriously considered before implementing Cre/loxP-based technologies in human gene therapy protocols.
Acknowledgments
We thank N. Hobbs, T. Frerk, T. Tucker, A. Meloni, R. Shepard, A. Bondareva, and M. Hocking for technical assistance. This work was supported by funding from the Montana Agricultural Experimental Station, National Science Foundation-MONTs, and the National Institutes of Health (to E.E.S.), and by grants from the Howard Hughes Medical Institute and the Mathers Charitable Foundation (to M.R.C.). E.E.S. is a Howard Hughes Fellow of the Life Sciences Research Foundation.
Abbreviations
- hGH
human growth hormone
- Prm1
mouse protamine 1
- DAPI
4′,6-diamidino-2-phenylindole
- CMV
cytomegalovirus
- GFP
green fluorescent protein
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240471297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240471297
References
- 1.Sternberg N, Hamilton D. J Mol Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 2.Sternberg N, Hamilton D, Austin S, Yarmolinsky M, Hoess R. Cold Spring Harb Symp Quant Biol. 1981;45:297–309. doi: 10.1101/sqb.1981.045.01.042. [DOI] [PubMed] [Google Scholar]
- 3.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V, Pierson L S d, et al. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mack A, Sauer B, Abremski K, Hoess R. Nucleic Acids Res. 1992;20:4451–4455. doi: 10.1093/nar/20.17.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoess R H, Ziese M, Sternberg N. Proc Natl Acad Sci USA. 1982;79:3398–3402. doi: 10.1073/pnas.79.11.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoess R, Abremski K, Sternberg N. Cold Spring Harb Symp Quant Biol. 1984;49:761–768. doi: 10.1101/sqb.1984.049.01.086. [DOI] [PubMed] [Google Scholar]
- 7.Hoess R H, Wierzbicki A, Abremski K. Nucleic Acids Res. 1986;14:2287–2300. doi: 10.1093/nar/14.5.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopaul D N, Guo F, Van Duyne G D. EMBO J. 1998;17:4175–4187. doi: 10.1093/emboj/17.14.4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo F, Gopaul D N, van Duyne G D. Nature (London) 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 10.Wierzbicki A, Kendall M, Abremski K, Hoess R. J Mol Biol. 1987;195:785–794. doi: 10.1016/0022-2836(87)90484-0. [DOI] [PubMed] [Google Scholar]
- 11.Sauer B, Henderson N. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoess R, Abremski K, Irwin S, Kendall M, Mack A. J Mol Biol. 1990;216:873–882. doi: 10.1016/S0022-2836(99)80007-2. [DOI] [PubMed] [Google Scholar]
- 13.Gu H, Zou Y R, Rajewsky K. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- 14.Akagi K, Sandig V, Vooijs M, Van der Valk M, Giovannini M, Strauss M, Berns A. Nucleic Acids Res. 1997;25:1766–1773. doi: 10.1093/nar/25.9.1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin D I, Enver T, Ley T J, Groudine M. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 16.Tsien J Z, Chen D F, Gerber D, Tom C, Mercer E H, Anderson D J, Mayford M, Kandel E R, Tonegawa S. Cell. 1996;87:1317–1326. doi: 10.1016/s0092-8674(00)81826-7. [DOI] [PubMed] [Google Scholar]
- 17.Ramirez-Solis R, Liu P, Bradley A. Nature (London) 1995;378:720–724. doi: 10.1038/378720a0. [DOI] [PubMed] [Google Scholar]
- 18.Herault Y, Rassoulzadegan M, Cuzin F, Duboule D. Nat Genet. 1998;20:381–384. doi: 10.1038/3861. [DOI] [PubMed] [Google Scholar]
- 19.Bunting M, Bernstein K E, Greer J M, Capecchi M R, Thomas K R. Genes Dev. 1999;13:1524–1528. doi: 10.1101/gad.13.12.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauer B. J Mol Biol. 1992;223:911–928. doi: 10.1016/0022-2836(92)90252-f. [DOI] [PubMed] [Google Scholar]
- 21.Sauer B. Nucleic Acids Res. 1996;24:4608–4613. doi: 10.1093/nar/24.23.4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thyagarajan B, Guimaraes M J, Groth A C, Calos M P. Gene. 2000;244:47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 23.Li L P, Schlag P M, Blankenstein T. Hum Gene Ther. 1997;8:1695–1700. doi: 10.1089/hum.1997.8.14-1695. [DOI] [PubMed] [Google Scholar]
- 24.Okuyama T, Fujino M, Li X K, Funeshima N, Kosuga M, Saito I, Suzuki S, Yamada M. Gene Ther. 1998;5:1047–1053. doi: 10.1038/sj.gt.3300704. [DOI] [PubMed] [Google Scholar]
- 25.Feng Y Q, Seibler J, Alami R, Eisen A, Westerman K A, Leboulch P, Fiering S, Bouhassira E E. J Mol Biol. 1999;292:779–785. doi: 10.1006/jmbi.1999.3113. [DOI] [PubMed] [Google Scholar]
- 26.Berghella L, De Angelis L, Coletta M, Berarducci B, Sonnino C, Salvatori G, Anthonissen C, Cooper R, Butler-Browne G S, Mouly V, et al. Hum Gene Ther. 1999;10:1607–1617. doi: 10.1089/10430349950017617. [DOI] [PubMed] [Google Scholar]
- 27.Russ A P, Friedel C, Grez M, von Melchner H. J Virol. 1996;70:4927–4932. doi: 10.1128/jvi.70.8.4927-4932.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Westerman K A, Leboulch P. Proc Natl Acad Sci USA. 1996;93:8971–8976. doi: 10.1073/pnas.93.17.8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Capecchi M R. In: Engineering the Human Germline. Campbell G S J, editor. New York: Oxford Univ. Press; 1999. pp. 31–42. [Google Scholar]
- 30.Schmidt E E, Hanson E S, Capecchi M R. Mol Cell Biol. 1999;19:3904–3915. doi: 10.1128/mcb.19.5.3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zambrowicz B P, Harendza C J, Zimmermann J W, Brinster R L, Palmiter R D. Proc Natl Acad Sci USA. 1993;90:5071–5075. doi: 10.1073/pnas.90.11.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braun R E, Peschon J J, Behringer R R, Brinster R L, Palmiter R D. Genes Dev. 1989;3:793–802. doi: 10.1101/gad.3.6.793. [DOI] [PubMed] [Google Scholar]
- 33.Rusconi S, Severne Y, Georgiev O, Galli I, Wieland S. Gene. 1990;89:211–221. doi: 10.1016/0378-1119(90)90008-f. [DOI] [PubMed] [Google Scholar]
- 34.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 35.Schmidt E E, Schibler U. Dev Biol. 1997;184:138–149. doi: 10.1006/dbio.1997.8514. [DOI] [PubMed] [Google Scholar]
- 36.Peschon J J, Behringer R R, Palmiter R D, Brinster R L. Ann N Y Acad Sci. 1989;564:186–197. doi: 10.1111/j.1749-6632.1989.tb25897.x. [DOI] [PubMed] [Google Scholar]
- 37.Caldwell K A, Handel M A. Proc Natl Acad Sci USA. 1991;88:2407–2411. doi: 10.1073/pnas.88.6.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Braun R E, Behringer R R, Peschon J J, Brinster R L, Palmiter R D. Nature (London) 1989;337:373–376. doi: 10.1038/337373a0. [DOI] [PubMed] [Google Scholar]
- 39.Dym M, Fawcett D W. Biol Reprod. 1971;4:195–215. doi: 10.1093/biolreprod/4.2.195. [DOI] [PubMed] [Google Scholar]
- 40.Hecht N B. Dev Genet. 1995;16:95–103. doi: 10.1002/dvg.1020160202. [DOI] [PubMed] [Google Scholar]
- 41.Kuan C Y, Roth K A, Flavell R A, Rakic P. Trends Neurosci. 2000;23:291–297. doi: 10.1016/s0166-2236(00)01581-2. [DOI] [PubMed] [Google Scholar]
- 42.Vaux D L, Korsmeyer S J. Cell. 1999;96:245–254. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 43.O'Gorman S, Dagenais N A, Qian M, Marchuk Y. Proc Natl Acad Sci USA. 1997;94:14602–7. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fuscoe J C, Knapp G W, Hanley N M, Setzer R W, Sandlund J T, Pui C H, Relling M V. Mutat Res. 1998;419:107–121. doi: 10.1016/s1383-5718(98)00129-6. [DOI] [PubMed] [Google Scholar]
- 45.Greaves M. Eur J Cancer. 1999;35:1941–1953. doi: 10.1016/s0959-8049(99)00296-8. [DOI] [PubMed] [Google Scholar]