Abstract
Peptide aptamers are proteins selected from combinatorial libraries that display conformationally constrained variable regions. Peptide aptamers can disrupt specific protein interactions and thus represent a useful method for manipulating protein function in vivo. Here, we describe aptamer derivatives that extend the range of functional manipulations. We isolated an aptamer with increased affinity for its Cdk2 target by mutagenizing an existing aptamer and identifying tighter binding mutants with calibrated two-hybrid reporter genes. We used this and other anti-Cdk2 aptamers as recognition domains in chimeric proteins that contained other functional moieties. Aptamers fused to the catalytic domain of a ubiquitin ligase specifically decorated LexA-Cdk2 with ubiquitin moieties in vivo. Aptamers against Cdk2 and another protein, Ste5, that carried a nuclear localization sequence transported their targets into the nucleus. These experiments indicate that fusion proteins containing aptameric recognition moieties will be useful for specific modification of protein function in vivo.
Keywords: peptide aptamers, combinatorial peptide libraries, protein interactions, cellular nanotechnology, protein design
Peptide aptamers are recognition reagents that embody some features of antibodies (1). They consist of a conformationally constrained variable region (here, the V region is 20 amino acids) displayed by a platform protein (here, Escherichia coli thioredoxin A). We currently select peptide aptamers from combinatorial libraries by two-hybrid methods, using aptamer derivatives that also bear acidic activation domains, and using LexA derivatives of the desired target proteins as baits; such selection ensures that the aptamers function in vivo. Peptide aptamers typically exhibit Kd values for their target of about 10−7 to 10−8 M (1). These molecules can discriminate between closely related members of protein families (1) and even between different allelic forms of proteins (ref. 2 and this work). Anti-Cdk2 aptamers competitively inhibit the interaction of Cdk2 with one of its substrates and, when expressed in human cells, delay progress through the cell cycle (3). Similarly, anti-E2F, anti-HPV E6, and anti-Ras aptamers disrupt the function of their protein targets in mammalian cells (refs. 4 and 5; C. W. Xu, Z. Luo, and R.B., unpublished work), and anti-Drosophila Cdc2 and Cdc2c aptamers inhibit the function of their targets in imaginal disks (6). Finally, we and others have shown that aptamers can be used as dominant genetic agents to cause a phenotype and to identify, in subsequent two-hybrid assays, the proteins and interactions they target (7–9). These results demonstrate that peptide aptamers can disrupt specific protein interactions in vivo and thus allow their precise manipulation (reviewed in ref. 10).
Here, we describe derivative peptide aptamers that covalently modify or change the subcellular localization of their target proteins. We first describe selection of a peptide aptamer with an improved affinity for its target. We use this improved aptamer with others to construct two types of chimeric proteins: “modifiers,” which ubiquitinate their target proteins, and “transporters,” which change the subcellular localization of their targets.
Materials and Methods
Identification of a Higher Affinity Variant.
We amplified the V region of anti-Cdk2 aptamer 10 from the original library vector (1) following a mutagenic PCR protocol as described (11). We ligated the purified amplified products into the RsrII-cut library vector pJM-1 (1), which directs their conditional expression under the control of the Gal1 promoter, and introduced the ligation mix into E. coli DH5α. We prepared plasmid DNA from a pool of 15,000 independent colonies. We transformed (12) this pool into EGY48 that contained LexA-Cdk2 (1) and pRB1840 (13) to obtain 40,000 transformants on Ura−His−Trp−glucose plates. We replica plated these transformants onto Ura−His−Trp− glucose/Xgal and Ura−His−Trp−galactose/Xgal plates. After 2 days at 30°C, we transferred the 16 colonies that showed a blue color onto Ura−His−Trp−glucose plates. We replica plated these master plates onto Ura−His−Trp−glucose/Xgal and Ura−His−Trp−galactose/Xgal plates and confirmed that 12 of the initial 16 colonies again displayed galactose-dependent blue color. We rescued the aptamer-encoding plasmids from these strains (14) and reintroduced the plasmids into EGY48. Seven of these 12 plasmids conferred an interaction phenotype on galactose-containing medium but not on glucose-containing medium.
Construction of Fusion Proteins.
Modifiers.
We isolated DNA encoding the hect domain of yeast RSP5 by PCR using the oligonucleotides 5′-ATATCTCGAGATTAAAGTACGTAGAAAGAAC-3′ and 5′-ATATGTCGACGGATCCTCATTCTTGACCAAACCCTATG-3′, which contained, respectively, an XhoI site and BamHI and SalI sites. We subcloned the PCR product into XhoI-cut pJG4–4, which contains a Trp1 marker, a 2-micron replication origin, and that directs the expression of native proteins under the control of the GAL1 promoter to create pJG4–4(hect). We amplified TrxA and peptide aptamers using the oligonucleotides 5′-GGAGGCGAATTCGCCGCCACCCATGGCCGATAAAATTATTCACCTGACTGACG-3′ and 5′-ATATCTCGAGCGCCAGGTTAGCGTCGAGGAAC-3′, which contained, respectively, an EcoRI site followed by an initiator codon in a Kozak context and an XhoI site, and inserted the PCR products into EcoRI/XhoI-cut pJG4–4 (hect). To express the hect domain only, we used the 5′ oligonucleotide 5′-ATATGAATTCGCCGCCACCATGGCCATTAAAGTACGTAGAAAGAACATTTTTGAG-3′, which contained an EcoRI site and an initiator codon in a Kozak context with the above-described 3′ oligonucleotide to PCR the hect domain from RSP5, and introduced this fragment into EcoRI/XhoI-cut pJG4–4. We constructed the mutant hect domain using the transformer kit from CLONTECH, according to the manufacturer's instructions, using the mutagenic oligonucleotide 5′-GCCAAAATCTCACACAGCTTTTAACAGAGTTG-3′ to change the hect active site cysteine to alanine, and the selection oligonucleotide 5′-CGCTAACCTGGCGCCTAGGATTAAAGTACGTAG-3′ to change to the XhoI site on the vector to an AvrII site.
For experiments with Myc-tagged ubiquitin, we expressed the modifiers from another vector. To this end, we amplified 5-, 8-, and 10M-hect fusions described above using oligonucleotides 5′-ATATGTCGACGGATCCTCATTCTTGACCAAACCCTATG-3′ and 5′-GGAGGCGAATTCGCCGCCACCCATGGCCGATAAAATTATTCACCTGACTGACG-3′. We introduced the amplified products into EcoRI/XhoI-cut pBC103, a plasmid that carries a Ura3 marker, a 2-micron replication origin, and the GAL1 promoter.
Myc-Ubiquitin.
Yep105 contains a Trp1 marker and a 2-micron replication origin and directs the expression of a Myc-tagged synthetic yeast ubiquitin gene under the control of the copper-inducible CUP1 promoter (15).
LexA-7Lys-Cdk2.
We began with the bait plasmid encoding LexA-Cdk2 (1). We constructed DNA that encoded a stretch of seven lysines by annealing the oligonucleotides 5′-AATTGAAGAAGAAAAAAAAGAAAAAGC-3′ and 5′-AATTGCTTTTTCTTTTTTTTCTTCTTC-3′ and introduced this duplex into the EcoRI site of the bait plasmid. We treated the ligation mixture with EcoRI, introduced it into E. coli XL-1 blue, and identified by PCR cells that bore plasmids containing the insert.
Transporters.
TrxA and anti-Cdk2 peptide aptamers were amplified as described above. We introduced the PCR products into EcoRI/XhoI-cut pBC103 (mentioned above) and pBC104, which also carried a Ura3 marker, a 2-micron replication origin, and a Gal1 promoter, and that directed the synthesis of nuclear localization sequence (NLS) aptamers. pJM-C6, pJM-C7, pJM-N2, and pJM-N3, the plasmids that direct the synthesis of non-NLS- and NLS-tagged anti-Ste5 aptamers, are described elsewhere (7).
Yeast Manipulations.
We performed interaction mating assays as described (16) using the EGY48 strain for the preys and the EGY42 strain for the baits and reporter plasmids (17). We used pSH18–34, pJK103, and pRB1840 (carrying 8, 2, and 1 LexA operators upstream of a Gal1-lacZ fusion gene, respectively) in Fig. 1A, the pSH18–34-derived LexAop-green fluorescent protein (GFP) reporter (Display Systems Biotech, Copenhagen) in Fig. 1C, and pSH18–34 in Fig. 2.
Figure 1.
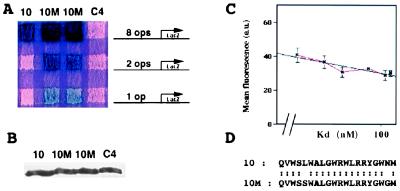
A mutant aptamer with enhanced affinity for its target. (A) Interaction mating assay between LexA-Cdk2 and aptamer 10, two strains carrying aptamer 10M, and a noninteracting aptamer C4, using three different sensitivity LexAop-lacZ reporters. (B) Western blot assay using an anti-TrxA antibody. Diploid exconjugates were grown in galactose-containing medium, and proteins were extracted and subjected to a Western blot analysis with anti-TrxA antibody. (C) Strength of interaction phenotypes as determined by fluorescence from a LexAop-GFP reporter plotted against Kd values measured in evanescent wave experiments. Fluorescence values are in arbitrary units (a.u.). (D) Sequence of the variable regions of aptamers 10 and 10M.
Figure 2.
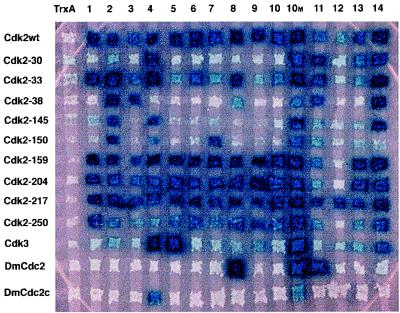
Mapping sites on Cdk2 bound by the original aptamers and by aptamer 10M. We collected the Cdk2 mutant bait proteins, described elsewhere (3), Cdk3, and Drosophila Cdc2 and Cdc2c (16). In this experiment, we expressed TrxA, the 14 different aptamers originally selected (1), and aptamer 10M as preys. We mated yeast to generate an interaction matrix (16).
To measure interaction phenotypes with the LexAop-GFP reporter gene, we grew overnight liquid cultures from diploid exconjugants in Ura−His−Trp− galactose liquid medium. We measured fluorescence with a FACStar Plus (Becton Dickinson) illuminated with two argon lasers tuned to 488 nm and to multiline UV. We recorded with a 530−/+15-nm filter to measure yeast fluorescence. We set the FL3–2 voltage (background) using yeast that did not show an interaction phenotype. We analyzed 30,000 cells for each interaction and determined mean fluorescence of the yeast population above background using the cellquest software package (Becton Dickinson).
To perform the modifier assays, we transformed the EGY48 yeast strain with different combinations of targets and aptamer-hect fusions (12). We plated transformants onto His−Trp−glucose plates, then grew colonies overnight in 4 ml of His−Trp−galactose medium. For experiments with Myc-ubiquitin, we transformed EGY48 with aptamer-hect fusions and Yep105, and EGY42 was transformed with LexA-Cdk2 and pSH18–34. We mated the transformants and selected diploid exconjugants on Ura−His−Trp−Leu−glucose medium. We inoculated exconjugants into liquid Ura−His−Trp−Leu−galactose medium that contained 100 μM CuSO4 and into control medium that lacked copper.
For Western analysis (below), we pelleted equal amounts of yeast in logarithmic growth phase and treated the pelleted yeast with zymolase (Seikagaku America, Rockville, MD) at 1 mg/ml in 50 μl of 1 M sorbitol/0.5 M sodium citrate/0.5 m EDTA/1 M DTT/1 M potassium phosphate/0.1 M PMSF for 1 h at 30°C and lysed them with SDS/PAGE sample buffer.
Immunoassays.
We performed Western blots after SDS/PAGE as described (18) with rabbit anti-LexA serum (19) or rabbit anti-TrxA serum (20), secondary antibodies coupled to alkaline phosphatase, and nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate as substrates. We scanned the blots with an optical scanner. For the Myc-ubiquitin experiments, we used an enhanced chemifluorescent substrate (Amersham Pharmacia) and scanned the blot with a phosphoimager (Molecular Dynamics). For immunofluorescence, we induced aptamer expression by growth in galactose for 3 h and fixed the cells by adding formaldehyde (3.7% final concentration) to the medium for 90 min. We probed samples with a polyclonal rabbit anti-LexA antibody (Upstate Biotechnology), followed by affinity-purified Texas Red- or fluorescein-conjugated secondary antibodies (Jackson ImmunoResearch), essentially as described (21).
Results
Generation and Identification of a Higher Affinity Anti-Cdk2 Aptamer.
We mutagenized an existing anti-Cdk2 aptamer to select one with higher affinity. The starting molecule, aptamer 10, has a measured Kd of 1.05 × 10−7 M (1). We performed random PCR mutagenesis on the aptamer 10 V region, and we reintroduced the PCR products into the library vector, pJM-1. This vector directs the expression of aptamers fused to the SV40 nuclear localization sequence, the B112 activation domain, and the hemagglutinin epitope tag under the yeast GAL1 promoter (1). We created a pool of 15,000 mutants. To obtain a measure of the efficiency of the PCR mutagenesis, we sequenced the V regions of two clones from this pool; sequencing revealed that these carried three and four mutations that resulted in one and three amino acid changes, respectively. To identify tighter-binding variants from this pool, we took advantage of the existence of different LexA operator reporter genes with different sensitivities. We began with a strain that expressed a LexA-Cdk2 bait and that carried pRB1840, a relatively insensitive lacZ reporter (13) with a single synthetic LexA operator. This reporter was not activated by aptamer 10 (Fig. 1A). We introduced the pool of PCR-mutagenized plasmids into this strain and identified transformants that gave rise to blue colonies. The V regions of the seven plasmids thus identified carried identical nucleotide changes and thus were presumably not independent. The nucleotide changes resulted in two amino acid changes: Leu to Ser at V region position 5, and Asn to Gly at position 19 (Fig. 1D). This mutant did not interact with Max and Rb, proteins unrelated to the Cdk family (not shown). We termed the mutant aptamer 10M.
We compared the affinity of 10M and 10 for Cdk2 by interaction mating. The reporters, pSH18–34, pJK103, and pRB1840, contain, respectively, eight high affinity operators, two high affinity operators, and one lower affinity LexA operator (Fig. 1A). Whereas, as judged by blue color from the pSH18–34 reporter, aptamer 10M had only a slightly greater affinity than aptamer 10, blue color from pJK103 and pRB1840 clearly indicated that 10M bound LexA-Cdk2 more strongly. To verify that the apparent higher affinity was not due to increased expression and/or stability of aptamer 10M, we performed Western blotting experiments and showed that the steady-state levels of aptamer 10, 10M, and a control aptamer were identical (Fig. 1B).
We measured the gain in affinity by two different means. First, we used a LexAop-GFP reporter gene to quantify interactions between anti-Cdk2 aptamers and LexA-Cdk2. We plotted mean fluorescence obtained from each interaction against Kd values measured in evanescent wave experiments (1). This plot followed a logarithmic equation (Table 1, Fig. 1C). We used this equation to calculate the Kd of the interaction between LexA-Cdk2 and aptamer 10M from the fluorescence it conferred (Table 1). We also performed evanescent wave experiments with purified aptamer 10M and His6-Cdk2 (1). The measured Kd was 5 nM (not shown), quite close to the 2 nM Kd calculated from the GFP data.
Table 1.
GFP interaction phenotypes and Kds
| Aptamer | Kd, nM | Mean fluorescence, arbitrary units | Standard deviation |
|---|---|---|---|
| 8 | 38 | 40.8 | 4.3 |
| 5 | 52 | 36.8 | 3.7 |
| 2 | 64 | 30.9 | 3.0 |
| 11 | 87 | 33.0 | 1.4 |
| 10 | 105 | 29.0 | 2.8 |
| 3 | 112 | 30.3 | 1.7 |
| 10M | 2 | 67.5 | 3.8 |
Average green fluorescence of yeast above background caused by interactions between LexA-Cdk2 and aptamers 2, 3, 5, 8, 10, and 11 in four independent experiments. Previously measured Kds (1) were plotted against measured fluorescence (Fig. 1c). The plot fit the following equation: Kd = 10 exp (−fluo + 123.7/21.9). We calculated a 2 nM Kd for aptamer 10M by interpolation using this equation.
To evaluate the contributions of the V region changes to increased affinity, we mutated both residues individually to wild type and analyzed the single mutants by interaction mating (data not shown). These experiments showed that the mutation of V region residue 5 (Leu to Ser) contributed to the gain of affinity but that the mutation of residue 19 (Asn to Gly) did not.
We then sought to determine whether the gain in affinity in 10M was caused by changes in its contact(s) with Cdk2. To this end, we examined binding of all existing anti-Cdk2 aptamers, including 10M, to a collection of Cdk2 mutants, related members of this protein family, and control proteins (not shown). Fig. 2 shows that, as previously observed, some of these aptamers recognize different epitopes conserved among Cdk proteins (1). However, by contrast with the aptamers isolated in our previous work, aptamer 10M showed distinct crossreactivity to other Cdk proteins. This interaction with Cdk family members is consistent with three ideas. First, the Leu to Ser change might create direct contact(s) between the variable region and residue(s) conserved among the Cdk proteins tested. Second, the change might indirectly create contacts by changing the conformation of the variable region. Third, even though Leu is more dihedrally constrained than Ser, a change to Ser might create new internal contacts within the variable region and thus reduce its conformational entropy.
Targeted Intracellular Protein Modifiers.
We used aptamer 10M and others to make protein derivatives that ubiquitinated target proteins in vivo. We based our design on the structural organization of the hect domain-containing ubiquitin ligases, which conjugate ubiquitin received from an E2 enzyme and transfer it to a protein substrate (22). The amino-terminal substrate-recognizing region of these enzymes varies greatly, whereas the carboxyl-terminal region, the hect domain, which carries the catalytic activity, is conserved between family members and throughout evolution (23). This modular structure suggested that we could fuse a hect domain to peptide aptamers and create ubiquitin ligases with engineered specificities (Fig. 3A).
Figure 3.
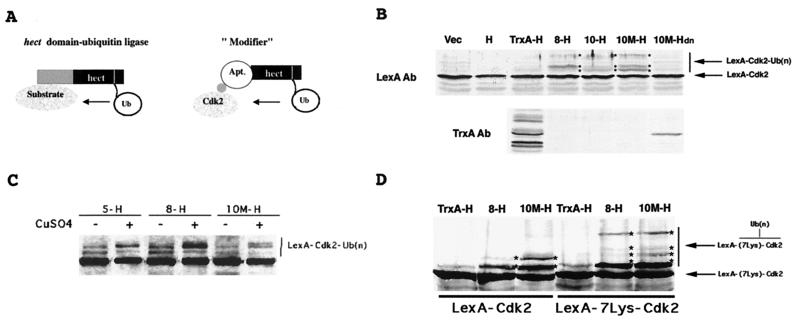
Targeted ubiquitination of LexA-Cdk2 by aptamer-hect fusions. (A) Design of a “modifier,” inspired by the structure of hect domain containing ubiquitin ligases. (B) Western blot analysis of LexA-Cdk2 (Upper) and TrxA-hect or aptamer-hect fusions (Lower) using anti-LexA and anti-TrxA antibodies, respectively. (C) Western blot analysis of LexA-Cdk2 when aptamer-hect fusions are expressed by growth overnight in a medium that does or does not contain CuSO4 and that does or does not express Myc-tagged ubiquitin. (D) Western blot analysis of LexA-Cdk2 and LexA-7Lys-Cdk2 when TrxA-, 8- and 10M-hect fusions are expressed, using the anti-LexA antibody.
Accordingly, we used a hect domain native to the yeast protein Rsp5 to construct various aptamer-hect fusions. We expressed these in yeast together with their putative LexA-Cdk2 targets and examined the fates of these fusion proteins. Fig. 3B shows that although the control TrxA-hect fusion that lacked a variable region was detectable by Western analysis, none of the anti-Cdk2 aptamer-hect fusions could be detected. This result indicates that attachment of a TrxA aptamer to a hect domain destabilizes the aptamer. However, expression of anti-Cdk2 aptamer-hect fusions resulted in the appearance of a ladder of higher molecular weight forms of LexA-Cdk2, suggesting that these chimeric proteins, even expressed at low levels, still caused ubiquitination of the target. The ladder of higher molecular weight LexA-Cdk2 forms was most apparent when the 10M-hect fusion was expressed, suggesting that the affinity of the modifier for the target affected the degree of target modification. Modification did not result in destruction; as determined from Western (Fig. 3B) and pulse-chase experiments (not shown), expression of these modifiers had no effect on LexA-Cdk2 stability or half-life.
To confirm that this ladder of higher molecular weight species corresponded to LexA-Cdk2-ubiquitin conjugates, we performed two different experiments. First, we used a yeast strain that conditionally expressed Myc-tagged ubiquitin, under the control of a copper-inducible promoter (15), and repeated the above experiments. This ubiquitin derivative is larger than native ubiquitin, and conjugates that contain it have higher molecular weights (15). On addition of CuSO4 to the culture medium, the bands on the ladder of higher molecular weights shifted higher than those in the uninduced culture (Fig. 3C). Second, we generated a loss of function anti-Cdk2 aptamer-hect fusion by changing the cysteine residue that forms the thioester bond with ubiquitin to alanine (23). In cells that expressed this mutant fusion, we did not observe the LexA-Cdk2 ladder but did observe the enzymatically dead aptamer-hect derivative (Fig. 3B). This fact suggests that active aptamer-hect derivatives may be sensitive to proteolysis after self-ubiquitination. This idea is supported by two lines of evidence. First, ubiquitination occurs on lysines (24), and, by contrast with aptamer 5-, 8-, 10-, and 10M-hect fusions, aptamer 2- and 11-hect fusions, which contain lysines in their V regions, did not ubiquitinate LexA-Cdk2 (not shown), even though aptamers 2 and 11 bind Cdk2 as tightly as the others (Fig. 2). This fact is consistent with the idea that ubiquitination of their V regions blocks their binding. Second, variant aptamer-hect fusions in which we changed different combinations of the five solvent-exposed lysines (K19, K37, K53, K70, K83) (25) on TrxA to arginines resulted in proteins detectable by anti-TrxA antiserum (data not shown). These observations suggest that the aptamer-hect fusions are vulnerable to ubiquitination by their own hect moieties and subsequently proteolyzed but are able to ubiquitinate their targets even though expressed at very low steady states.
Finally, we tested whether we could sensitize LexA-Cdk2 to ubiquitin-dependent proteolysis by introducing into it extra lysines as ubiquitin acceptors. To this end, we expressed in yeast a LexA-Cdk2 derivative that carried seven lysines between the LexA and the Cdk2 moieties. We coexpressed this LexA-Lys7-Cdk2 target together with TrxA-hect, 8-hect, and 10M-hect modifiers and visualized it with anti-LexA antibody as above. Fig. 3D shows that, as compared with the “native” LexA-Cdk2 target, the additional lysines on the experimental target improved its ubiquitination, both increasing the amount of higher molecular weight LexA-Cdk2 derivatives and causing the appearance of at least one new still higher molecular weight conjugate. However, as judged by its steady-state level, the increased ubiquitination of this LexA-Lys7-Cdk2 target did not destabilize it (Fig. 3D).
Targeted Intracellular Protein Transporters.
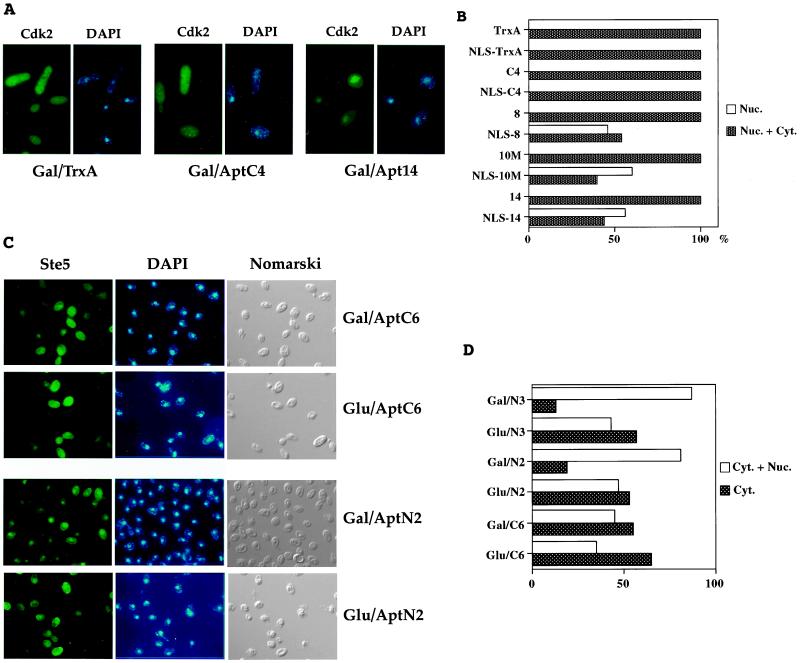
We then investigated the possibility of using derivatized aptamers to change the localization of their protein targets in vivo. In the original library vector, aptamers are expressed in yeast fused to a simian virus 40 NLS. LexA-fusion proteins that lack nuclear localization signals are uniformly distributed within the yeast cell (ref. 26 and this study). We tested whether anti-Cdk2 aptamers addressed to the nucleus would also concentrate LexA-Cdk2 in the nucleus. As shown by immunofluorescence experiments, LexA-Cdk2 is uniformly distributed in cells in which the chimeras B112-NLS-TrxA and B112-NLS-aptamer are not expressed. Similarly, LexA-Cdk2 is evenly distributed inside cells in which the control chimera B112-NLS-TrxA is expressed. However, in cells in which B112-NLS-anti-Cdk2 aptamers are expressed, LexA-Cdk2 is concentrated in the nucleus (Fig. 4A). All of the tested fusions triggered substantial nuclear localization of their LexA-Cdk2 target.
Figure 4.
Nuclear translocation of LexA-Cdk2 and LexA-Ste5 by interacting NLS-aptamer derivatives. (A) Yeast photomicrographs. (Left) Indirect immunofluorescence of LexA-Cdk2 protein using anti-LexA antibody. (Right) DNA staining with DAPI. Gal/TrxA, TrxA is expressed. Gal/AptC4, control aptamer C4 is expressed. Gal/Apt14, anti-Cdk2 aptamer 14 is expressed. (B) Percentage of yeast that displayed clear nuclear immunofluorescence, in presence of aptamers addressed to the nucleus or not. Dark bars, nuclear and cytoplasmic staining. White bars, nuclear staining. At least 50 cells were observed for each assay. (C) Yeast photomicrographs. (Left) Indirect immunofluorescence of LexA-Ste5 fusion protein using anti-LexA antibody. (Center) DNA staining with DAPI. (Right) Yeast observed with Nomarski optics. Gal/AptC6, non-NLS aptamer C6 is expressed. Glu/C6, non-NLS aptamer C6 is not expressed. Gal/AptN2, NLS-aptamer N2 is expressed. Glu/N1, NLS-aptamer N2 is not expressed. (D) Percentage of yeast that displayed cytoplasmic + nuclear or purely cytoplasmic staining, in presence of various aptamers addressed to the nucleus or not. Dark bars, cytoplasmic staining. White bars, cytoplasmic and nuclear staining. At least 100 cells were observed for each assay.
To confirm that the concentration of LexA-Cdk2 in the nucleus was due to the nuclear translocation of peptide aptamers rather than any other aspect of the binding of aptamers to their target, we expressed LexA-Cdk2 together with either aptamers or NLS-aptamers and determined the percentage of yeast in which LexA-Cdk2 was clearly nuclear. The results show that the nuclear localization of LexA-Cdk2 depends on the expression of peptide aptamers that contain NLSs (Fig. 4B). Finally, we used NLS-containing aptamers made against another protein, Ste5 (7), to determine whether NLS-aptamer fusions could cause nuclear localization of a protein that is thought to be predominantly cytoplasmic (27). When no aptamer was expressed or when a non-nuclear localized aptamer was expressed, LexA-Ste5 showed a predominant cytoplasmic localization. However, when NLS-aptamers were expressed, LexA-Ste5 showed a distinct concentration in the nucleus (Fig. 4 C and D).
Discussion
We have described peptide aptamer derivatives that covalently modify and change the localization of target proteins in vivo. To make them, we first generated a higher affinity mutant anti-Cdk2 aptamer, mutagenizing the variable region of an existing aptamer and screening for tighter binding mutants using relatively insensitive two-hybrid reporter genes. This variant had a significant increase in affinity and exhibited a Kd between 2 and 5 nM. We imagine that use of still less-sensitive reporter genes (28), LexA mutants with a decreased affinity for their operators (29), and/or substitution of weaker activation domains on the aptamer library should allow us to identify mutant aptamers with subnanomolar affinity in one step.
To construct modifiers, proteins that covalently alter target proteins in vivo, we exploited ubiquitination, the coupling of ubiquitin molecules to lysine residues on proteins. We showed that these modifiers ubiquitinated the LexA-Cdk2 target. Moreover, our results suggested that the modifiers destroyed themselves by self-ubiquitination on lysine residues exposed at the surface of the thioredoxin platform. This observation is consistent with the fact that the native Rsp5 protein ubiquitinates itself in vitro, probably on lysine residues lying in the amino-terminal part of the protein, outside of its hect domain (23, 30).
Our work provides an in vivo demonstration of targeted protein modifications by enzymes of redirected specificity. However, these aptamer-hect fusions did not destroy their Cdk2 targets, even those that contained extra lysine residues. It is possible that, for Cdk2, ubiquitination mediated by a more active effector domain would result in destabilization. Consistent with this idea, Gosink et al. (31) redirected the specificity of two plant E2 ubiquitin-conjugating enzymes, Ubc1 and Ubc4, in vitro by fusing different protein-binding peptides, including the Ig binding domains of Staphylococcus aureus A protein, to Ubc carboxyl termini. In vitro, those authors observed ubiquitination of the cognate substrates and a partial degradation of the targeted IgG. However, we believe the most likely explanation for the stability of ubiquitinated LexA-Cdk2 fusions is that Cdk2 is simply refractory to ubiquitin-mediated proteolysis. In fact, no Cdk is known to undergo ubiquitin-dependant proteolysis, although some of its binding partners are degraded by this means (32). A number of other proteins are also ubiquitinated without being degraded, including H2A (24), cyclins in certain cell-cycle phases (33), and some membrane receptors whose ubiquitination signals endocytosis without involving the proteasome (34).
We also used peptide aptamers fused to an NLS as “transporters.” Anti-Cdk2 and anti-Ste5 aptamers that carried nuclear localization signals caused their targets to accumulate in the nucleus. These results are similar to those found by Schneider et al. (35), who showed that NLS-containing chimeric proteins, which contain PDZ domains that bind different target peptides, direct their target peptides to the nucleus of mammalian 283T cells (35). Our results suggest that we should be able to build transporters with other protein moieties that readdress their protein targets to other subcellular compartments: the endoplasmic reticulum (36), the mitochondrial membrane (37), or the plasma membrane (38). The aptamers in these transporters all blocked the activity of their target proteins, but, as we have noted in the case of the NLS containing anti-Ste5 aptamers, inactivation may in fact be caused by enforced nuclear localization rather than by blocking interaction with partners (7). Because transporting aptamers allow mislocalization of targeted proteins that are expressed under the control of their own promoters, the perturbations that transporters induce in cell function should be less severe than those caused by overexpression of target proteins fused to addressing sequences.
The construction of new proteins from different functional modules has been reported for many types of proteins (28, 39), and the success of these methods can be taken to support the current picture in which exons are shuffled among proteins during evolution (40). We imagine that peptide aptamers will be useful general purpose recognition moieties in chimeric proteins with other effector domains that perform other functions. The ability to control the spatial and temporal expression of such “modifiers” and “transporters” in cells and whole organisms should facilitate a finer control of protein modification, inactivation, and localization. Furthermore, the ability to select aptamers that distinguish among allelic variants of proteins should allow selective modification of the activities of individual alleles. The generation and use of new peptide aptamer derivatives should facilitate high-resolution study of regulatory pathways and could possibly inspire new therapeutic strategies.
Acknowledgments
We are grateful to Sandrine Mouradian for the flow cytometry experiments, Ron Geyer and Alejandro Colman-Lerner for anti-Ste5 aptamers, Fred Winston for RSP5, John McCoy for rabbit anti-TrxA antiserum, Rosine Haguenauer-Tsapis for Yep105, Stan Tabor for advice about TrxA mutants, and Mark Stahl for advice with the evanescent wave experiments. We thank Ron Geyer, Alejandro Colman-Lerner, and Jeffrey C. Way for comments on the manuscript. These experiments were supported by a grant from the National Institute of General Medical Sciences (to R.B.) and by grants from the Association pour la Recherche sur le Cancer, the Ligue Nationale contre le Cancer, and the Fondation pour la Recherche Médicale (to P.C.).
Abbreviations
- NLS
nuclear localization sequence
- GFP
green fluorescent protein D
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Colas P, Cohen B, Jessen T, Grishina I, McCoy J, Brent R. Nature (London) 1996;380:548–550. doi: 10.1038/380548a0. [DOI] [PubMed] [Google Scholar]
- 2.Xu C W, Mendelsohn A, Brent R. Proc Natl Acad Sci USA. 1997;94:12473–12478. doi: 10.1073/pnas.94.23.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen B A, Colas P, Brent R. Proc Natl Acad Sci USA. 1998;95:14272–14277. doi: 10.1073/pnas.95.24.14272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fabrizzio E, Le Cam J, Polanowska J, Kakzorek M, Lamb N, Brent R, Sardet C. Oncogene. 1999;18:4357–4363. doi: 10.1038/sj.onc.1202825. [DOI] [PubMed] [Google Scholar]
- 5.Butz K, Denk C, Ullmann A, Scheffner M, Hoppe-Seyler F. Proc Natl Acad Sci USA. 2000;97:6693–6697. doi: 10.1073/pnas.110538897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolonin M G, Finley R L., Jr Proc Natl Acad Sci USA. 1998;95:14266–14271. doi: 10.1073/pnas.95.24.14266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geyer C R, Colman-Lerner A, Brent R. Proc Natl Acad Sci USA. 1999;96:8567–8572. doi: 10.1073/pnas.96.15.8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Norman T C, Smith D L, Sorger P K, Drees B L, O'Rourke S M, Hughes T R, Roberts C J, Friend S H, Fields S, Murray A W. Science. 1999;285:591–595. doi: 10.1126/science.285.5427.591. [DOI] [PubMed] [Google Scholar]
- 9.Blum J H, Dove S L, Hochschild A, Mekalanos J J. Proc Natl Acad Sci USA. 2000;97:2241–2246. doi: 10.1073/pnas.040573397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colas P. Curr Opin Chem Biol. 2000;4:54–59. doi: 10.1016/s1367-5931(99)00051-4. [DOI] [PubMed] [Google Scholar]
- 11.Cadwell R C, Joyce G F. PCR Methods Appl. 1992;2:28–33. doi: 10.1101/gr.2.1.28. [DOI] [PubMed] [Google Scholar]
- 12.Gietz D, St. Jean A, Woods R A, Schiestl R H. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Estojak J, Brent R, Golemis E A. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finley R L, Jr, Brent R. In: DNA Cloning, Expression Systems: A Practical Approach. Hames B D, Glover D M, editors. Oxford: Oxford Univ. Press; 1995. pp. pp.169–203. [Google Scholar]
- 15.Ellison M J, Hochstrasser M. J Biol Chem. 1991;266:21150–21157. [PubMed] [Google Scholar]
- 16.Finley R L, Jr, Brent R. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Golemis E A, Brent R. Mol Cell Biol. 1992;12:3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ausubel F M, Brent R, Kingston R E, Moore D, Seidman J G, Struhl K. Current Protocols in Molecular Biology. New York: Greene and Wiley-Interscience; 1987–1997. [Google Scholar]
- 19.Brent R, Ptashne M. Nature (London) 1984;312:612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- 20.LaVallie E R, DiBlasio E A, Kovacic S, Grant K L, Schendel P F, McCoy J M. Bio/Technology. 1993;11:187–193. doi: 10.1038/nbt0293-187. [DOI] [PubMed] [Google Scholar]
- 21.Ferrigno P, Silver P A. Methods Cell Biol. 1999;58:107–122. doi: 10.1016/s0091-679x(08)61951-2. [DOI] [PubMed] [Google Scholar]
- 22.Scheffner M, Nuber U, Huibregtse J M. Nature (London) 1995;373:81–83. doi: 10.1038/373081a0. [DOI] [PubMed] [Google Scholar]
- 23.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jennissen H P. Eur J Biochem. 1995;231:1–30. [PubMed] [Google Scholar]
- 25.Katti S K, LeMaster D M, Eklund H. J Mol Biol. 1990;212:167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- 26.Silver P A, Brent R, Ptashne M. Mol Cell Biol. 1986;6:4763–4766. doi: 10.1128/mcb.6.12.4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahanty S K, Wang Y, Farley F W, Elion E A. Cell. 1999;98:501–512. doi: 10.1016/s0092-8674(00)81978-9. [DOI] [PubMed] [Google Scholar]
- 28.Brent R, Ptashne M. Cell. 1985;43:729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- 29.Oertel-Buchheit P, Lamerichs R M, Schnarr M, Granger-Schnarr M. Mol Gen Genet. 1990;1:40–48. doi: 10.1007/BF00315795. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Yang J, Huibregtse J M. Mol Cell Biol. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gosink M M, Vierstra R D. Proc Natl Acad Sci USA. 1995;92:9117–9121. doi: 10.1073/pnas.92.20.9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hershko A. Curr Opin Cell Biol. 1997;9:788–799. doi: 10.1016/s0955-0674(97)80079-8. [DOI] [PubMed] [Google Scholar]
- 33.Mahaffey D T, Yoo Y, Rechsteiner M. FEBS Lett. 1995;370:109–112. doi: 10.1016/0014-5793(95)00799-f. [DOI] [PubMed] [Google Scholar]
- 34.Terrell J, Shih S, Dunn R, Hicke L. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 35.Schneider S, Buchert M, Georgiev O, Catimel B, Halford M, Stacker S A, Baechi T, Moelling K, Hovens C M. Nat Biotechnol. 1999;17:170–175. doi: 10.1038/6172. [DOI] [PubMed] [Google Scholar]
- 36.Pelham H R. Cell Struct Funct. 1996;21:413–419. doi: 10.1247/csf.21.413. [DOI] [PubMed] [Google Scholar]
- 37.Haucke V, Lithgow T. J Bioenerg Biomembr. 1997;29:11–17. doi: 10.1023/a:1022451520203. [DOI] [PubMed] [Google Scholar]
- 38.Boutin J A. Cell Signalling. 1997;9:15–35. doi: 10.1016/s0898-6568(96)00100-3. [DOI] [PubMed] [Google Scholar]
- 39.Vita C. Curr Opin Biotechnol. 1997;8:429–434. doi: 10.1016/s0958-1669(97)80064-x. [DOI] [PubMed] [Google Scholar]
- 40.Gilbert W, de Souza S J, Long M. Proc Natl Acad Sci USA. 1997;94:7698–7703. doi: 10.1073/pnas.94.15.7698. [DOI] [PMC free article] [PubMed] [Google Scholar]