Abstract
The gonadotropin follicle-stimulating hormone (FSH) is required for initiation and maintenance of normal gametogenesis and acts through a specific, cell-surface receptor (Fshr) present only on Sertoli and granulosa cells in the gonads. Despite extensive examination of the transcriptional mechanisms regulating Fshr, the sequences directing its expression to these cells remain unidentified. To establish the minimal region necessary for Fshr expression, we generated transgenic mice carrying a yeast artificial chromosome (YAC) that contained 413 kilobases (kb) of the rat Fshr locus (YAC60). Transgene expression, as determined by RT-PCR, was absent from immature testis and Sertoli cells, limited to germ cells of the adult testis, and never observed in the ovary. While the data is limited to only one transgenic line, it suggests that the 413kb region does not specify the normal spatiotemporal expression pattern of Fshr. Comparative genomics was used to identify potential distal regulatory elements, revealing seven regions of high evolutionary conservation (≥80% identity over 100bp or more), six of which were absent from the transgene. Functional examination of the evolutionary conserved regions (ECRs) by transient transfection revealed that all of the ECRs had modest transcriptional activity in Sertoli or myoid cells with two, ECR4 and ECR5, showing differential effects in expressing and non-expressing cells. These data reveal that distal regulatory regions (outside the 413kb in YAC60) are required for appropriate temporal and spatial Fshr expression and implicate the identified ECRs in transcriptional regulation of Fshr.
Keywords: Fshr, Sertoli cell, yeast artificial chromosome, comparative genomics, transgenics
Introduction
Mammalian gonad function is regulated by a hierarchical axis of hormones that modulate gametogenesis and steroidogenesis. Pulsatile secretion of gonadotropin releasing hormone by a population of neurons in the hypothalamus dominates control of the reproductive endocrine axis. This hormone stimulates anterior pituitary gonadotropes to produce the gonadotropin hormones, LH (lutenizing hormone) and FSH, which, in turn, stimulate somatic cells in the gonads via their specific G-protein coupled receptors. Both FSH and LH, along with their receptors, are requisite for normal gonad function and fertility in males and females (reviewed in Themmen and Huhtaniemi, 2000). The receptor for FSH, Fshr, is found only on testicular Sertoli cells and ovarian granulosa cells, and therefore restricts FSH action to these two cell types and defines the specificity of the hormone (Camp et al., 1991; Tilly et al., 1992; Rannikki et al., 1995; Tisdall et al., 1995). In the rodent testis and ovary, Fshr is expressed late in embryonic development and remains restricted to Sertoli and granulosa cells throughout postnatal life (reviewed in Simoni et al., 1997). Thus, transcriptional activation of the gene encoding Fshr is exquisitely controlled to limit its expression to only two cell types, providing an excellent model for examining cell-specific gene regulation in granulosa and Sertoli cells and the underlying control of FSH signaling.
The transcriptional mechanisms regulating Fshr expression have been examined in a number of species, including human, mouse, rat, and sheep, with the majority of these studies utilizing transient transfection and protein-DNA binding experiments to identify regulatory elements within the promoter region (Huhtaniemi et al., 1992; Gromoll et al., 1994; Sairam and Subbarayan, 1997; Heckert et al., 1998; Heckert and Griswold, 2002). Importantly, these studies highlighted several conserved features of the Fshr promoter. In particular, the minimal region required for maximal promoter activity generally resides within the first few hundred base pairs of 5’ flanking sequence (Gromoll et al., 1994; Linder et al., 1994; Heckert et al., 1998; Xing and Sairam, 2001). This promoter region contains a conserved E-box (5’-CACRTG-3’) in all four species, AP-1 (5’-TTARTCA-3’) and inverted GATA elements in the rodent promoters, and an E2F site (5’-TTTTCGCTGC-3’) in the mouse, rat and human promoters (reviewed in Heckert, 2005). Principle among these elements is the E-box, which is responsible for the majority of Fshr promoter activity and is required for binding and transcriptional induction of Fshr by the ubiquitous basic helix-loop-helix transcription factors Usf1 and Usf2 (Goetz et al., 1996; Heckert et al., 1998; Heckert et al., 2000). Additionally, the orphan nuclear receptor steroidogenic factor 1 (SF-1) activates the Fshr promoters from mouse, rat, and sheep (Heckert, 2001; Levallet et al., 2001; Xing et al., 2002). Since Sertoli and granulosa cells are a subset of SF-1 expressing cells, SF-1 may participate in activating cell-specific Fshr expression (Val et al., 2003). In the rat, Fshr activation by SF-1 requires active Usf1 and Usf2 proteins as well as an intact E-box, suggesting that the E-box coordinates transcriptional regulation of the gene (Heckert, 2001). While informative, data from the above studies were largely generated by in vitro and cell culture analyses, and thus, a significant limitation of these studies was their inability to accurately evaluate transcriptional features associated with cell-specific gene expression. Therefore, to examine the promoter’s ability to direct transcription to Sertoli and granulosa cells, animal models were employed.
To evaluate Fshr promoter function, in vivo, studies employed transgenic mice using promoter sequences to drive expression of marker genes (Linder et al., 1994; Heckert et al., 2000; Nordhoff etal., 2003). Two of these studies concluded that sequences outside the promoter region were needed to properly direct cell-specific expression (Heckert et al., 2000; Nordhoff et al., 2003). One examined 16 lines of transgenic mice that contained transgenes with either 5000bp or 198bp of promoter sequence, and found that cell-specific expression is not controlled through these promoter regions (Heckert et al., 2000). While each promoter fragment directed some transgene expression in the testis and ovary, there was also extensive expression in Fshr-negative tissues, demonstrating inappropriate or ectopic activation of the transgenes (Heckert et al., 2000). Most importantly, transgene expression in the testis originated primarily from the germ cell population, suggesting that the promoters did not recapitulate Fshr expression. Similar findings with respect to inaccurate temporal and germ cell expression were observed in a study employing a transgene with 1500bp of the human FSHR promoter (Nordhoff et al., 2003). Thus, the evidence indicates that regulatory elements located outside of the promoter region (5000bp and 1500bp of 5’flanking sequence in the rat and human genes, respectively) must play a key role in establishing proper cell-specific and temporal regulation of Fshr.
In support of the above conclusion, studies evaluating sequence conservation and DNase I hypersensitivity, two hallmarks of transcriptional regulatory regions, also revealed that important elements for Fshr expression are located outside its promoter region (Hermann and Heckert, 2005). Numerous highly conserved, non-coding sequences, which serve as candidate regulatory elements, were shown to extend across Fshr and some of these colocalized with DNase I hypersensitive sites (Hermann and Heckert, 2005). However, given the large number of these conserved sites and the potential for them to act at large distances from the gene’s coding region, it is clear that additional criteria are needed to help select those sites most likely to be functional. In the current study, we examined expression of a 413kb yeast artificial chromosome in one line of transgenic mice in order to help define the sequences required for appropriate Fshr expression, and thus, restrict analyses of potential regulatory elements to those residing within a transcriptionally competent region. We also employed more advanced comparative sequence analysis to refine our search to only the most highly conserved sequences and thus ones most likely to be functional. The findings indicate that regulatory elements required for proper Fshr expression are located at a significant distance from the coding region and point to seven highly conserved regions as important factors in Fshr cell-specificity.
Materials and Methods
Yeast strain and propagation.
Original YACs purchased from Invitrogen Life Technologies, Inc (Carlsbad, CA) were propagated in the host Saccharomyces cerevisiae strain J57D (MATa, ψ+, ura3-52 trp1 ade2-101 his3-2,15 can1-100 leu2-3, 112) (Zhong et al., 1998) and grown in complete medium lacking uracil and tryptophan. Modified YACs containing the rat Fshr gene were grown in complete media lacking at least one selective nutrient required for YAC maintenance. All media were prepared as described elsewhere and all yeast cell cultures were carried out overnight at 30°C under vigorous agitation (Green et al., 1997; Riethman et al., 1997). Transformation of spheroplasted yeast cells was performed as described except yeast cells were treated with lyticase for approximately 15 min at 37°C (Riethman et al., 1997; Karpova et al., 2005).
Rat Fshr YAC isolation and manipulation.
DNA pools of a rat YAC library (constructed by the Whitehead Institute for Biomedical Research and MIT Center for Genome Research; average insert size of 830 kb) were purchased from Invitrogen Life Technologies and screened for rat Fshr using the polymerase chain reaction (PCR) and oligodeoxynucleotide primers directed against exon 1 (5’Fshr, Table 1) and exon 10 (5’Fshr, Table 1). Six Fshr-containing YACs, 124A9, 135C2, 197B11, 341C7, 368E4 and 392G6 were identified and purchased from Invitrogen. Rat Fshr YAC clone 135C2 was used for all further manipulation. High-molecular weight DNA was analyzed and mapped by pulsed-field gel electrophoresis (PFGE) and Southern blot analysis as described (Riethman et al., 1997; Karpova et al., 2005). Hybridization probes for Southern blot analysis were as follows: 5’Fshr (Fshr exon 1), 3’Fshr (Fshr exon 10), HIS3 (to detect the acentric arm of YAC rFshr-HIS3), TRP1 (to detect the centric pRML1 vector arm), and URA3 (to detect the acentric pRML2 vector arm of the original YACs). DNA for Southern blot probes were PCR-amplified from yeast chromosomal or rat genomic DNA using the primers noted in Table 1 and radiolabeled with a random primer labeling system according to the vendor’s recommendations (New England Nuclear, Boston, MA).
Table 1.
Oligodeoxynucleotide primers used to generate DNA used as probes for Southern blot analysis of YAC60, for PCR screening of YAC60 in line 92, and for RT-PCR analysis of line 92.
| Primer 1 | Primer 1 | Use* | |
|---|---|---|---|
| 5’Fshr | 5’- CTCTTTTCTGTCATTTTGGG-3’ | 5’-GAAAGACCCTATTAGAGCAATG-3’ | Southern blot PCR screening |
| 3’Fshr | 5’- TTTGCCATTTCTGCCTCCCT-3’ | 5’- ATTGGTGACTCTGGGAGCTG-3’ | Southern blot PCR screening |
| URA3 | 5’-GTACCACCAAGGAATTACTGG-3’ | 5’-CGGGTGTATACAGAATAGCAG-3’ | Southern blot |
| TRP1 | 5’-AAATAGTTCAGGCACTCCG-3’ | 5’-TCTGTGAAGCTGCACTGAG-3’ | Southern blot |
| HIS3 | 5’-TTCCCGTTTTAAGAGCTTGG-3’ | 5’-TCCTGATGCGGTATTTTCTC-3’ | Southern blot |
| Cre | 5’-CTGGTCGAAATCAGTGCGTTC-3’ | 5’- TTACCGGTCGATGCAACGAGT -3’ | PCR screening |
| ECR1 3’A | 5’-AACTGTAAAGAACTCACCCTGT-3’ | 5’-ATGAGTTTTCAATGTCACCTTT-3’ | Southern blot |
| site139 | 5’-GCAAGTCATCAAGATCAGGCAAG-3’ | 5’-GCTAGTTGCTGGGAAATTCTCTTG-3’ | Southern blot |
| i1A | 5’-GCTAGTTGCTGGGAAATTCTCTTG-3’ | 5’-GTCATCTGGAATCCTGCCTGTAGTAG-3’ | PCR screening |
| i1B | 5’-GTCATCTGGAATCCTGCCTGTAGTAG-3’ | 5’-CAATGTTGTTCTTTTCTCTTCGGAATC-3’ | PCR screening |
| i1C | 5’-CAATGTTGTTCTTTTCTCTTCGGAATC-3’ | 5’-CTGCGGCATAGGATAGAGGTTGTC-3’ | PCR screening |
| Site141 | 5’-GAGACCCAAATGGAGGAACAAAC-3’ | 5’-TATTGTCCATTCTTCTCCCCAGG-3’ | Southern blot |
| Site146 | 5’-CACAGGGTGTCACTTCTGCCAAAC-3’ | 5’-GGATCTGTGCCCTGTGCTTCAG-3’ | Southern blot |
| Site148 | 5’-CACAAAGCATATCAGAGAAATGAACC-3’ | 5’-GTGTTTTTGGCTTGTGTTATGTCC-3’ | Southern blot |
| Site153 | 5’-CATCCCGCCACAGCCCAC-3’ | 5’-CCTGACTTGAAGGACGGAAACC-3’ | Southern blot |
| Site154 | 5’-CCTTGTGAGTCTTGACCCTGAGC-3′ | 5’-GGACTGTGGGTACACTGGTAATGG-3′ | Southern blot |
| 11b12b | 5’-GTTTTGTATCCGATGAACTGGTC-3’ | 5’-GTCTTGGTCTCACATGACAACTATAC-3’ | Southern blot |
| rat Fshr-Cre | 5’-GGATTTCCGTCTCTGGTGTAG-3’ | 5’-GGCACAGTTAGTTTGTATTGGC-3’ | RT-PCR |
| mouse Fshr | 5’-AATCCGTGGAGGTTTTCGC-3’ | 5’-GGCACAGTTAGTTTGTATTGGC-3’ | RT-PCR |
| Sox9 | 5’-CGTGGACATCGGTGAACTGAGC-3’ | 5’-CTGCTGCGGGGGTGCTTG-3’ | RT-PCR |
| Androgen receptor | 5’-AATGGGACCTTGGATGGAGAAC-3’ | 5’-TCCCTGCTTCATAACATTTCCG-3’ | RT-PCR |
Template DNA for each is noted in the Materials and Methods section.
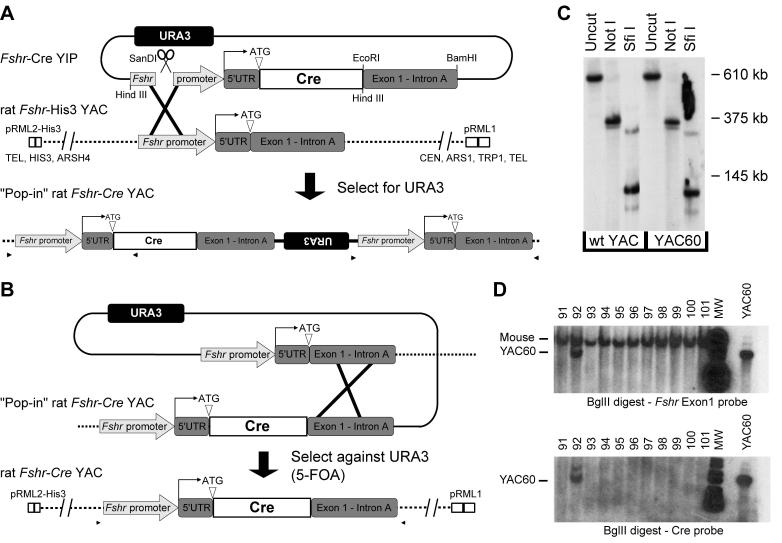
YAC rFshr-HIS3 was generated by retrofitting 135C2 with a pRML2-HIS3 vector, resulting in the replacement of the URA3 marker with a HIS3 marker. pRML2-HIS3 was generated by removing the URA3 cassette from pRML2 by Hind III restriction digestion, blunting the overhangs with Klenow fragment (Roche Diagnostics Corporation, Indianapolis, IN), and inserting a 1.2kb blunt-end HIS3 cassette generated by PCR (Table 1). All restriction enzymes were purchased from either Roche Diagnostics Corporation, New England Biolabs (Ipswich, MA) or Invitrogen Life Technologies. Yeast containing the 135C2 YAC were transformed with a 4.5-kb Sca I fragment of pRLM2-HIS3 and selection for growth on minimal medium lacking histidine and tryptophan. Positive colonies were screened by PFGE and Southern blot analysis utilizing a HIS3 probe (described above). Insertion of the bacteriophage P1 Cre recombinase gene was performed by two-step replacement using a yeast integration plasmid (YIP), pRS406-Fshr-Cre, which was generated by insertion of a 0.9kb Bam HI-EcoRI fragment from a Fshr-Cre transgene (Heckert et al., 2000) into the BamHI-EcoRI sites of the phagemid YIP pRS406, followed by insertion of a 2kb Hind III fragment from the Fshr-Cre transgene into the pRS406 HindIII site (Fig. 1A). The pRS406-Fshr-Cre YIP was linearized by restriction digestion with San DI and transformed into yeast containing the YAC rat Fshr-His3. YIP integration by homologous recombination was screened by selection for growth on minimal medium lacking histidine, uracil and tryptophan (Fig. 1A). Positive “pop-in” clones were screened for correct YIP integration by PCR using primers spanning the insertion site (details available from authors upon request) and with PFGE and Southern blot analysis (URA3 probe). Excision of the vector and wildtype Fshr sequence from “pop-in” YACs was selected for by growth on media lacking histidine and tryptophan and containing both 5-fluororotic acid (Sigma-Aldrich Corp., St. Louis, MO) and uracil (Fig. 1B). Positive clones were screened for correct excision using primers spanning the entire Fshr- Cre mutation (5’-GTCTGGGCAGGTAATAAGTTG-3’ and 5’-GAACAATCAAAGAGCCCATG-3’), PCR products were sequenced, and integrity of positive YACs was confirmed by PFGE and southern blot using the 5’Fshr probe. One clone was obtained that contained Cre recombinase correctly inserted at the ATG of rat Fshr in the YAC (YAC60). Limited restriction endonuclease mapping of YAC60 and the parent clone 135C2 with pulsed field gel electrophoresis (PFGE) and Southern blot analysis demonstrated that no gross rearrangements or deletions occurred during the retrofitting and two-step replacement modifications (Fig. 1C).
Figure 1.
YAC60 was generated by a two-step replacement via homologous recombination to insert Cre-recombinase at the Fshr translational start site in an Fshr-containing rat YAC. (A) Step one: a yeast integrating plasmid (YIP) with Fshr sequence flanking Cre recombinase was transformed into the YAC-containing yeast and selected for gain of the URA3 marker. Restriction endonuclease sites used to generate the pRS406-Fshr-Cre YIP are noted. (B) Step two: spontaneous revertants with excised YIP vector sequence (i.e.: retaining either the wildtype Fshr or Fshr-Cre sequence in the YAC) were identified by growth in the presence of 5-FOA, which selects against YACs retaining URA3. (C) Limited restriction endonuclease mapping of YAC60. YAC DNA was digested with the noted restriction endonuclease, resolved by PFGE, and analyzed by Southern blot analysis by hybridization with a probe to exon 10. Molecular weight marker migration is shown to the right of the autoradiogram. (D) Southern blot analysis of tail DNA from potential transgenic founder mice was performed using Bgl II restriction digestion and hybridization with probes against exon1 and Cre. Positive transgenic mice were identified by the presence of a faster migrating band (noted in line 92).
YAC purification and screening of transgenic mice.
For the generation of transgenic mice, YAC60 DNA was purified as reported, with minor modifications noted below (Peterson et al., 1995; Karpova et al., 2005). Initial fractionation by PFGE was performed using the following conditions; 0.5X TBE (44.5 mM Tris, 44.5 mM boric acid, 1 mM EDTA), 6 V/cm, 40 sec switch time, 24 hours, at 14°C. Excess carbohydrates were removed from the β-Agarase I treated YAC DNA by spot dialysis against injection buffer for 2hr at room temperature on a 0.025 μm MF disc mixed cellulose esters hydrophilic membrane filter (Millipore, Billerica, MA).
The genotypes of all founder transgenic mice and their offspring were determined by PCR and Southern blot of mouse tail biopsy DNA, which was prepared as described (Karpova et al., 2005). Transgene amplification was performed using a pair of primers specific for Cre recombinase (Table 1). Tail DNA was also digested with Bgl II and analyzed by Southern blot analysis using the 5’Fshr probe and the Pin AI - Cla I restriction fragment of Cre recombinase (Cre probe). One positive founder animal was obtained (Line 92) and used for all subsequent analysis. Animals were bred to the C57BL/6J mouse strain and manipulations were conducted according to the National Academy of Sciences Guide for Care and Use of Experimental Animals.
Integrated transgene mapping in Line 92.
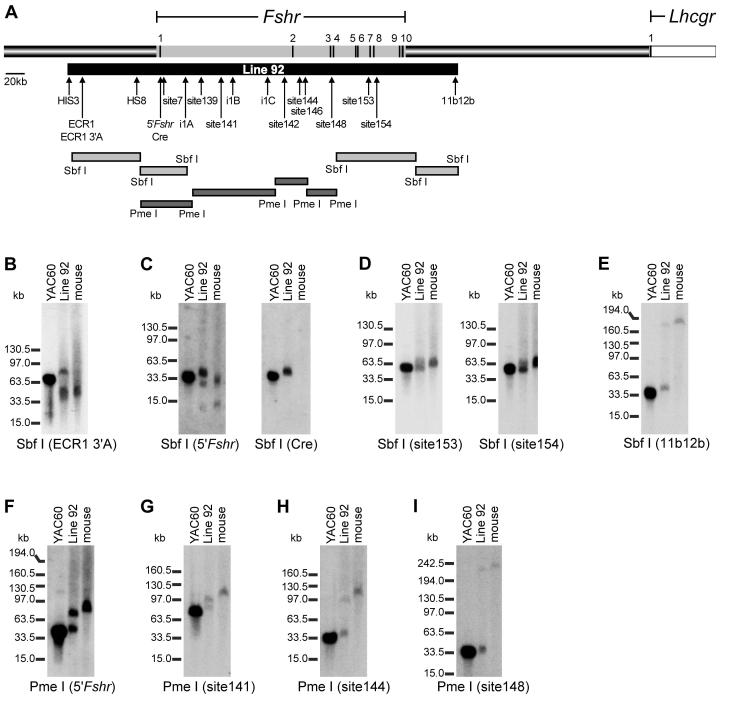
Adult transgenic and control mice (C57BL/6J) were used to generate high molecular weight genomic DNA as described (Navas et al., 2001; Karpova et al., 2005). Restriction endonuclease digestion was performed by first equilibrating slices of agarose plugs in the appropriate endonuclease buffer, followed by overnight incubation in the presence of the enzyme (0.375U/μl) at 4°C. Plugs were then digested at the optimal digestion temperature for each restriction endonuclease for 8 hours. Additional enzyme (0.25U/μl) was added, incubated at 4°C overnight, and digested for 24 hours at the optimal temperature. The digested DNA was fractionated by PFGE using the following conditions; 0.5X TBE, 6V/cm, initial switching time 1 sec ramped to the final switching time of 35.5 sec, 20 hours, at 14°C. DNA fragments of approximately 15kb to 400kb were resolved under these conditions. Southern blot analysis was performed using various hybridization probes spanning YAC60 (Fig. 2A). Probe DNA was PCR-amplified using rat genomic DNA and Bio-X-Act DNA polymerase (Bioline, Randolph, MA) with primers shown in Table 1 and radiolabeled as described above.
Figure 2.
Mapping of YAC60 in line 92. (A) Schematic representation of 750kb of rat genomic sequence that spans the Fshr coding sequence (grey shading on top bar), including the neighboring gene, LH receptor (Lhcgr, white). Arrows denote the positions of markers (HIS3, ECR1, HS8, Cre, site7, site139, i1A, i1B, i1C, and 11b12b) used for PCR-based mapping of YAC60 in line 92 as well as probes used for restriction endonuclease mapping in Southern blot hybridization analysis (ECR1 3’A, 5’Fshr, Cre, site141, site142, site144, site146, site148, site153, site154, 11b12b). The position of each Fshr exon is noted as a dark line on the grey bar and its number indicated above the bar (note: exon sizes are not to scale). The region of the locus identified within Line 92 is noted by the black bar below, beginning 98kb 5’ to the first transcriptional start site and extending 80kb 3’ of exon 10. Shaded bars indicate restriction fragments used for mapping. Mapping of the YAC60 transgene in line 92 was performed by digestion of high molecular weight DNA with Sbf I or Pme I followed by Southern blot analysis using the indicated probes. In each experiment, digestion of YAC60, line 92, and wildtype mouse DNA were compared. Sbf I digested DNA was analyzed with the following probes: (B) ECR1 3’A probe, (C) 5’Fshr and Cre, (D) site153 and site154, and (E) 11b12b. Pme I digested DNA was analyzed with the probes (F) 5’Fshr, (G) site141, (H) site144, and (I) site148. Restriction endonucleases and hybridization probes are noted below each autoradiogram. Migration of molecular weight standards is noted to the left of each autoradiogram.
RT-PCR Analysis of Transgene Expression.
Briefly, adult and 16 day transgenic and control littermates (C57BL/6J) were sacrificed and total RNA was isolated from a panel of 21 tissues or from isolated Sertoli and germ cells, using Trizol reagent according to manufacturer’s recommendations (Invitrogen). Complementary DNA (cDNA) was synthesized as described in the presence and absence of reverse transcriptase (Heckert et al., 2000). Oligodeoxynucleotide primers against either Cre recombinase or the mouse Fshr 5’UTR were used together with a primer against mouse and rat Fshr exon 10 to amplify the cDNA for either rat Fshr-Cre or endogenous mouse Fshr, respectively (Table 1). The identity of PCR-amplified transgenic or endogenous Fshr was confirmed by sequencing. Amplification of ribosomal protein L7 cDNA was used to control for cDNA synthesis as described (Heckert et al., 2000). Mouse Sox9 and Androgen receptor cDNAs were detected by PCR using the oligodeoxynucleotide primers shown in Table 1. PCR reactions were resolved by agarose gel electrophoresis.
Cell Culture and Transient Transfection.
Enriched populations of germ cells and Sertoli cells were prepared from adult transgenic mouse testes as described and used to prepare RNA (Heckert et al., 2000). Preparation and transient transfection of day 15 primary rat Sertoli cells and peritubular myoid cells as well as firefly and Renilla luciferase reporter assays were performed as described elsewhere (Hermann and Heckert, 2005). Data are presented as the firefly/Renilla luciferase activity ratio of chimeric Fshr constructs relative to the firefly/Renilla luciferase activity ratio of the Fshr (-220/+123) Luc minimal promoter. Statistical significance was determined using a two-tailed heteroscedastic student’s T-Test comparing the relative data.
Evolutionary conservation analysis of the Fshr locus.
Pre-computed whole genome alignments of the human, rat and chicken assemblies available through ECR Browser(http://ecrbrowser.dcode.org/) were used to visualize the windowed-average sequence identity score for alignment of the Fshr locus between Lhcgr and Nrxn1 using 80% identity over 100bp as minimal conservation parameters (Ovcharenko et al., 2004). The sequence of the rat Fshr gene was extracted from the NCBI database as described (Hermann and Heckert, 2005). Sequences for each rat ECR were identified through the ECR browser and annotated on the rat Fshr gene using Discovery Studio Gene v1.5 software (Accelrys, Inc, San Diego, CA).
Reporter clone construction.
Fragments corresponding to ECRs 1-7 were PCR-amplified (primers shown in Table 2) using rat genomic DNA and Bio-X-Act DNA polymerase (Bioline). Each amplified ECR was digested with the restriction endonuclease EcoRV. The Fshr(-220/+123)Luc (Heckert et al., 1998) reporter was linearized by digestion with Kpn I, located 5’ to the promoter, and the overhangs blunted with Klenow fragment (Roche Diagnostics Corp) and dephosphorylated by treatment with Calf Intestinal Phosphatase (New England Biolabs) according to each manufacturers’ recommendations. Fshr/ECR clones were generated by ligating the ECR PCR products into the Kpn I-blunted reporter using Quick Stick Ligase (Bioline).
Table 2.
Evolutionarily conserved regions (ECRs) in the human, rat and chicken Fshr loci.
| Previous designation* | % Identity# | Position (kb)§ | Size (bp) | Cloning primers† | |
|---|---|---|---|---|---|
| ECR1 | site110 | 89.4% | -81 | 501 | 5’-GTACACACCTCTTTCTGCCTGGG-3’ 5’-GGTGGGGGCTCAGTCTGCTG-3’ |
| ECR2 | site160 | 85.6% | +401 | 543 | 5’-GGTCACCGAACACAGAATAATG-3’ 5’-GGAGGTGCCCATATTTTTGA-3’ |
| ECR3 | site161 | 84.4% | +429 | 284 | 5’-CTAATACATCCACAGCAGCCC-3’ 5’-CAATCCACAGATACTTTCGCAC-3’ |
| ECR4 | site73 | 83.3% | -265 | 202 | 5’-GGTGAACACTGGTCTTATTTGTC-3’ 5’-GTTTACGGTTGTGAAGGAGAGG-3’ |
| ECR5 | site63 | 83.3% | -334 | 153 | 5’-CTAAAAATGTTCCACGAGGTAGG-3’ 5’-CTTTTAAGTCCTGTTTTAAATCTTC -3’ |
| ECR6 | site38 | 81.8% | -474 | 294 | 5’-CACCCACAGATGCTTTCCTAC-3’ 5’-GTAAAACTAACTAGCACATAGCC-3’ |
| ECR7 | site34 | 83.0% | -489 | 316 | 5’-CTTTGCCTATAAATAAGATGTCAAC-3’ 5’-GGTGATTTCTAATCTCATTATACCC -3’ |
As identified in (Hermann and Heckert, 2005) at 70% sequence identity over 100bp or more between the rat and human loci.
Windowed-average sequence identity score between human, rat, and chicken Fshr(≥80%;≥100bp).
Relative to the Fshr transcriptional start sites.
The recognition sequence for EcoRV (5’-GCGCGATATC-3’) was present on the 5’end of each oligodeoxynucleotide (not shown).
Results
Characterization of a rat Fshr-Cre yeast artificial chromosome in transgenic mice.
Previous studies demonstrated that 5000bp of sequence 5’ to the Fshr transcriptional start site was insufficient to direct appropriate expression of the gene, presumably due to a lack of key regulatory elements located outside this region (Heckert et al., 2000). In order to understand the mechanisms controlling Fshr expression, the requisite sequences must first be defined, requiring the use of an in vivo model, such as transgenic mice, that can accurately evaluate cell-specific gene regulation. Transgenic analysis of larger genomic regions in mice was employed to delineate the extent of sequence required for Fshr expression and serve as a reference to effectively identify cis-acting regulatory elements. Several features of Fshr, including its significant size and its shared characteristics with a class of genes that reside in gene deserts, suggested that the amount of DNA required for appropriate Fshr expression would be large (Nobrega et al., 2003; Hermann and Heckert, 2005; Ovcharenko et al., 2005). Furthermore, the presence of numerous highly conserved, non-coding sequences associated with Fshr, but located at large distances from the promoter, implicated distal cis-acting elements in its regulation (Hermann and Heckert, 2005). In an attempt to accommodate all required regulatory sequences, we generated transgenic mice with a yeast artificial chromosome (YAC) that contained 413kb of the rat Fshr locus modified to contain bacteriophage P1 Cre recombinase gene inserted at the initiation ATG of Fshr, which served as a reporter for transgene expression (YAC60; see Materials and Methods for a detailed description of transgene generation; Figs 1A, B and C).
Pronuclear injection of YAC60 produced 101 potential founder pups, from which a single line (line 92) carrying YAC60 was identified by Southern blot analysis (Fig. 1D). PCR-based mapping of 10 regions spanning the transgene (HIS3, ECR1, HS8, site7, i1A, site139, i1B, i1C, 11b12b, TRP1) and extensive restriction endonuclease mapping indicate that the integrated transgene consists of an intact fragment spanning from 98kb upstream of the first transcriptional start site to 80kb downstream of exon 10 (Fig. 2A, black bar below the genomic segment). More specifically, each of the ten regions examined by PCR was present in line 92 and Southern blot analysis of high molecular weight DNA digested with Sbf I revealed similar digestion patterns between YAC60 isolated from yeast and that which integrated into line 92 (Fig. 2A, 2B-E, and data not shown). The slight migration differences were attributed to differences in DNA concentration. Additional restriction endonuclease mapping with Pme I revealed similar digestion patterns for DNA prepared from either yeast or transgenic line 92 (Fig. 2F-I). These data strongly suggested that the transgene is intact in line 92. Importantly, the identity of approximately 195kb of sequence present in YAC60 beginning 80kb downstream of exon 10 (beyond probe 11b12b) is not known and suspected to be chimeric sequence unrelated to Fshr, and thus, could not be mapped in line 92 (Figure 2 and data not shown).
YAC60 is inappropriately expressed in transgenic mice.
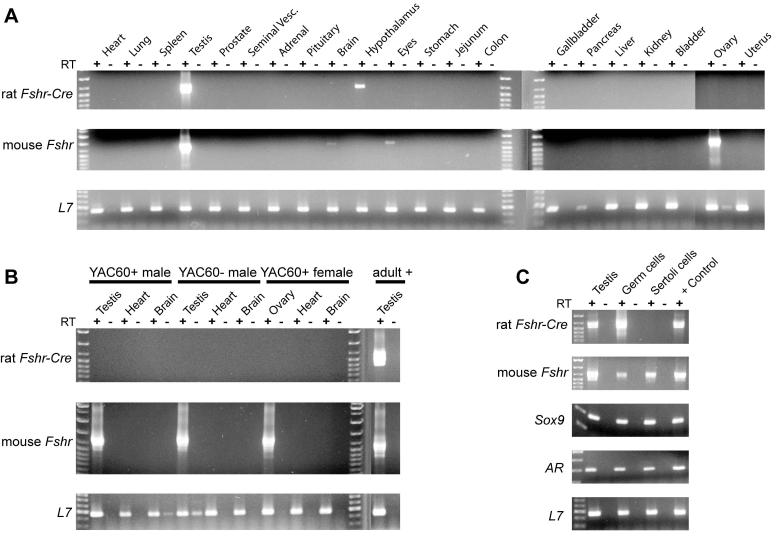
To determine if the transgene sequence in line 92 contained the necessary regulatory information for Fshr expression, we used RT-PCR to evaluate 21 different tissues for expression of both the Fshr transgene and the endogenous gene. Testes from adult transgenic mice strongly expressed transgenic and endogenous Fshr mRNAs, indicating that YAC60 directed testis expression (Fig. 3A). Notably, transgene expression was not observed in the ovary, despite the presence of endogenous Fshr and ribosomal protein L7 transcripts (Fig. 3A). To determine if YAC60 was expressed in a temporally appropriate manner in the testis, we similarly examined transgenic and endogenous Fshr mRNAs in younger animals, when Fshr is expressed and germ cell numbers are minimal. Transgene expression was absent from the testis of 16-day-old animals, while the signal for endogenous Fshr mRNA was significant in the testis and ovary (Fig. 3B). The lack of transgene expression in the gonads of immature mice revealed that YAC60 expression did not follow the normal temporal pattern for Fshr. To determine if expression of the transgene in the adult testis reflected its presence in Sertoli cells, we examined its expression in Sertoli cells and germ cells isolated from testes of adult transgenic mice. Robust transgene expression was observed in adult testis and isolated germ cells, but not in Sertoli cells, while endogenous Fshr mRNA was present in all samples, including germ cells (Fig. 3C). The latter was attributed to contamination of the germ cells with Sertoli cells, which was confirmed by the presence of Sertoli-specific mRNAs for Sox9 and Androgen receptor within the germ cell preparation (Fig. 3C). The lack of transgene expression in Sertoli cells and ovaries of line 92, as well as the ectopic expression in male germ cells, revealed that 413kb of the rat Fshr locus lacks the ability to appropriately regulate the gene, and therefore cis-acting elements required for Fshr expression must reside outside this region. Although modest expression signals were observed in the hypothalamus (transgenic Fshr-Cre) and in brain and eyes (endogenous Fshr), the inconsistency of these findings suggested they were artifactual (Fig. 3A).
Figure 3.
Expression profile of YAC60 transgenic line 92. Tissues from YAC60+ mice were examined for the presence of rat Fshr-Cre, endogenous Fshr, and ribosomal protein L7 mRNAs by RTPCR in (A) adult mice, (B) 16 day old mice, and (C) testes, isolated Sertoli cells and isolated germ cells from adult YAC60+ mice. Sox9 and Androgen receptor (AR) mRNAs were also detected by RT-PCR in C. RT-PCR was performed in the presence or absence of reverse transcriptase (RT) and is noted with + or - below the indicated tissue source of the evaluated RNA, which is indicated above each lane.
Evolutionarily conserved regions implicated in Fshr regulation.
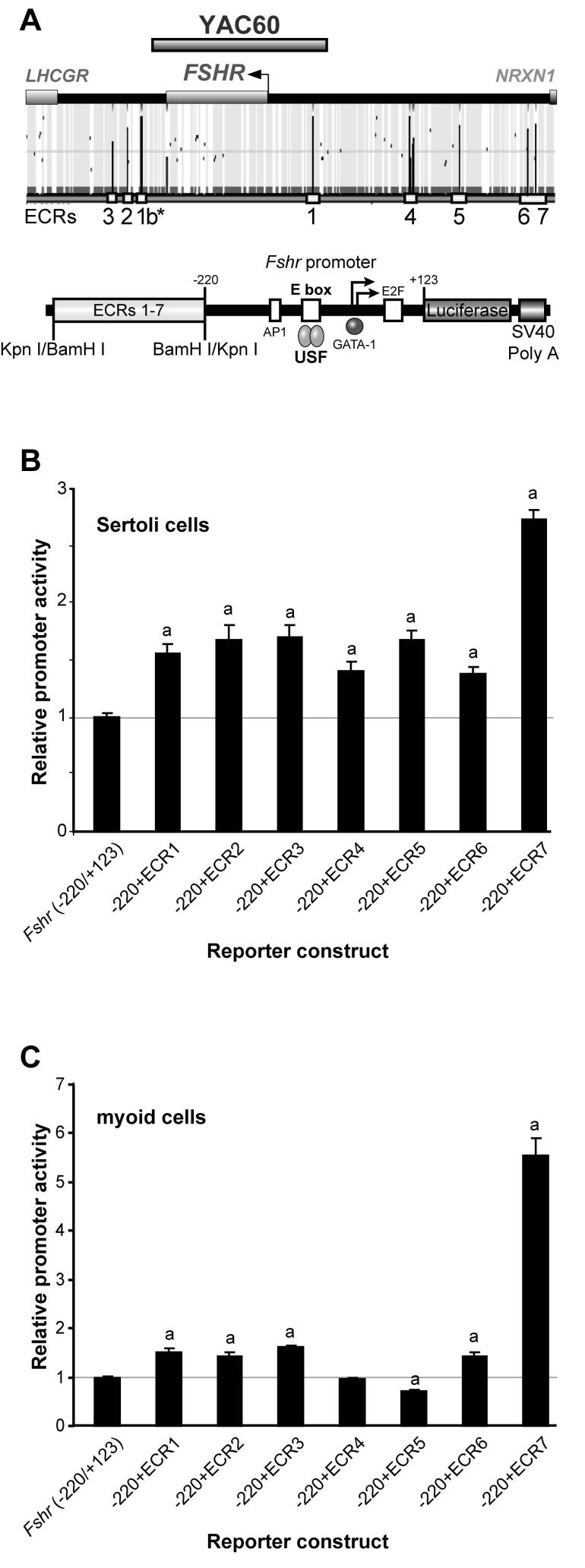
To assist in identifying novel Fshr regulatory sequences, we employed a comparative genomics approach to locate non-coding sequences that are highly conserved throughout evolution, which represent those most likely to be functional and house important cis-acting elements. Evolutionary conserved regions (ECRs) of at least 80% identity over 100bp or more were identified in the human, rat and chicken Fshr genes using an internet-based genome navigation tool (ECR browser; http://ecrbrowser.dcode.org/; (Ovcharenko etal., 2004). Seven ECRs (ECR1-7) were identified in the vicinity of Fshr, and notably only ECR1 was present in YAC60 (Fig. 4A and Table 2). Thus, if one or more of these ECRs have associated transcriptional activity obligatory for normal Fshr transcriptional regulation, their absence from YAC60 likely accounts for the lack of appropriate transgene expression.
Figure 4.
Evolutionary conserved regions in the Fshr locus have associated transcriptional activity. (A) The human, rat and chicken Fshr loci between Nrxn1 and Lhcgr were analyzed for evolutionary conservation using the web-based genome navigation tool ECR browser (http://ecrbrowser.dcode.org/; Ovcharenko et al., 2004). Seven evolutionarily conserved regions (ECRs) were identified and annotated below the sequence (dark peaks marked 1-7). Each ECR was cloned in front of the minimal Fshr (-220/+123) Luc promoter and transfected together with phRL-TK into (B) primary rat Sertoli cells and (C) peritubular myoid cells, a non-expressing cell type. The data are represented as the firefly/Renilla luciferase activity ratio of each ECR-containing construct relative to the firefly/Renilla luciferase activity ratio of Fshr (-220/+123) Luc. Transfections were performed a minimum of three times. Error bars represent the standard errors of the means. Statistically significant differences (p≤0.002) in activity of ECR-containing promoters relative to that of the basal promoter (Fshr - 220bp/+123bp) are noted by “a” above the bars. Statistical significance was determined by a student’s T-Test.
To determine if ECRs 1-7 have associated transcriptional activity, regions containing each were tested for their ability to affect Fshr promoter activity. PCR-amplified fragments containing the ECRs were cloned upstream of the core -220bp Fshr promoter to produce chimeric reporter plasmids that were examined by transient transfection analysis in 15d primary rat Sertoli cells and peritubular myoid cells, a cell type that does not express Fshr (Fig. 4A, Table 2). Clones containing ECRs 1, 2, 3, 6 or 7 were more active than the basal Fshr promoter in both Sertoli cells and myoid cells, suggesting they house elements that activate transcription (Figs. 4B and 4C). Interestingly, ECR5 repressed Fshr promoter activity to 70% of wildtype in myoid cells but activated the promoter in Sertoli cells (1.7-fold), suggesting it functions to both activate and silence Fshr in order to help restrict expression to the appropriate cells (Figs. 4B and 4C). Similarly, ECR4 activated the Fshr promoter in Sertoli cells, but did not alter promoter activity in myoid cells, suggesting its activity is limited to specific cell types, including Sertoli cells (Figs. 4B and 4C).
Discussion
To date, little attention has been paid to regulatory elements located outside the promoter region in transcriptional control of Fshr. Numerous protein-DNA binding and transient transfection experiments have examined the promoter and elucidated the basal transcriptional mechanisms for Fshr promoter activity, but failed to identify a mechanism controlling the gene’s spatiotemporal expression (Heckert and Griswold, 2002). Earlier transgenic analyses suggest that this results from a lack of critical sequence information within the promoter, as neither 5000bp of rat Fshr nor 1500bp of human FSHR 5’ flanking sequence correctly directed cell-specific expression in transgenic mice (Heckert et al., 2000; Nordhoff et al., 2003). The current study provides additional and more compelling evidence that long-range transcriptional regulation is crucial to establishing Fshr expression in Sertoli and granulosa cells, while prohibiting ectopic expression in germ cells.
In an effort to uncover novel regulatory elements, we sought to define a region that would express Fshr appropriately in transgenic mice and employed a large genomic segment, which included the entire Fshr coding sequence plus substantial flanking sequence, as a transgene. In the transgene design, the 413kb YAC was engineered to include the bacteriophage P1 Cre recombinase gene inserted at the ATG of Fshr to facilitate evaluation of cellular transgene expression and for use in future experiments to evaluate the genetics of Sertoli cell and granulosa cell function. Reporter mouse lines such as GtRosa26tm1sor and Z/EG robustly express β-galactosidase or green fluorescent protein in cells expressing Cre recombinase, thus, when crossed to Cre-transgenic lines, cells expressing Cre are robustly marked, even if Cre expression is low, as with weakly expressing genes such as Fshr (Soriano, 1999; Novak et al., 2000). Further, the addition of Cre recombinase to a transgene that mimics Fshr expression would provide a valuable research tool that allows specific gene ablations in Sertoli and granulosa cells through Cre-lox recombination (Sauer and Henderson, 1988; Sauer, 1998). The Fshr-Cre YAC construct was used to generate transgenic mice and analysis of its expression revealed incorrect tissue and cellular profiles, suggesting critical elements were absent from the transgene. However, in considering such a conclusion, one must weigh an important caveat of the study, in that only a single transgene was analyzed. Thus, despite evidence that large transgenes, such as the Fshr-Cre YAC, are known to minimize position effects, they cannot be eliminated from the potential mechanisms directing transgene expression (Giraldo and Montoliu, 2001). Furthermore, while the most likely explanation holds that important regulatory elements are missing from the transgene, until additional experiments confirm this, consideration must be given to the possibility that neighboring chromatin provides transcriptional cues that both redirect Fshr expression to germ cells and extinguish it within granulosa and Sertoli cells. Importantly, the results indicated that inappropriate transgene expression was not due to truncation of the transgene, as extensive PCR and restriction mapping of the integrated transgene in line 92 demonstrated it was intact (Fig. 2). This was also supported by our ability to amplify a transcript between Cre recombinase (inserted in exon 1) and Fshr exon 10 (Fig. 3). Notably, we were unable to identify the sequence of a 195kb segment of YAC60 (beginning 80kb downstream of exon 10) based on PCR-mapping to amplify known genomic sequences between the Fshr and Lhcgr coding sequences (data not shown). YAC60 may therefore be chimeric (i.e. contain unrelated sequence) at the far 3’ end. And while we have no direct evidence to support this conclusion, nor does it provide a likely explanation for the results, it is important to note that chimeric sequence may have influenced transgene expression. However, our favored hypothesis, and the one best supported by the data and published scientific studies, is one in which sequences outside the region defined by the transgene are needed for Fshr expression.
The absence of transgene expression in either Sertoli or granulosa cells, incorrect temporal regulation, and ectopic expression in germ cells of the testis was similar to previously reported transgenic studies (Heckert et al., 2000; Nordhoff et al., 2003). In those cases, 5000bp and 198bp of the rat Fshr promoter and 1500bp of the human FSHR promoter were also insufficient for appropriate Fshr expression (Heckert et al., 2000; Nordhoff et al., 2003). Unlike the promoter transgenic studies, however, ectopic expression of the YAC60 transgene was limited to male germ cells, indicating YAC60 largely prevented ectopic expression and contained sufficient sequence to shield it from integration site effects (Heckert et al., 2000). The inclusion of a recently identified OCT-1/GATA silencing element in the YAC60 transgene, that was absent from previous transgenes, is one factor that may account for the diminished ectopic expression (Hermann and Heckert, 2005). On the surface, these transgenic studies appear to contrast with a similar study that observed restricted expression of a β-galactosidase reporter in both testis and ovary using the 5000bp rat Fshr promoter (Linder et al., 1994). However, the limited number of tissues examined (6) and lack of cellular expression data obscured evaluation of the promoter’s true ability to drive cell-specificity (Linder et al., 1994). Thus, the majority of evidence indicates that distal regulatory elements are essential for correct expression of Fshr.
These findings implicate a transcriptional mechanism for Fshr that places it among a growing class of genes that employ distal elements in their transcriptional control. In some cases, such as with Sox9 and Sonic hedgehog, distal regulatory elements were shown to act over genomic distances as great as 1Mb, even extending into neighboring genes (Bishop et al., 2000; Gottgens et al., 2000; Loots et al., 2000; Lettice et al., 2002; Qin et al., 2004; Zheng et al., 2004; Sagai et al., 2005). Knowing this, it is clear that criteria are needed to help select functional sequences from the massive amount of non-functional DNA. Comparative genomics is a powerful tool used to identify functional sequences by virtue of their evolutionary conservation, and now, with the availability of extensive genome sequence data can be applied to the identification of elements located at significant distances from a gene’s promoter region (reviewed in Frazer et al., 2003). Recent application of comparative genomics across the COUP-TFII and Apolipoprotein E loci identified novel tissue-specific enhancers located 66kb and 46kb distal to their genes’ promoters, respectively, demonstrating the potential of this approach (Zheng et al., 2004; Baroukh et al., 2005). A similar approach was applied to Fshr and identified seven highly conserved sequences (ECRs1-7) between the human, rat, and chicken Fshr loci, which had associated transcriptional activity, suggesting they house distal cis-acting regulatory sequences (Table 2, Figure 4). Notably, each ECR was defined in a previous, less stringent, comparative study of the rat and human genes that revealed over 100 conserved sites (Table 2; Hermann and Heckert, 2005). That these seven ECRs all had transcriptional activity, and all but ECR1 were absent from the YAC60 transgene, suggested they participate individually or in different combinations to influence Fshr transcription. A combinatorial function may explain the weak transcriptional activity associated with each ECR alone and testing their combined activities may reveal such a mechanism. Alternatively, weak ECR activity may reflect limitations of the cell culture system, which may not provide the appropriate cellular environment for their function. Additional studies are underway to evaluate ECR function, in vivo, to more clearly resolve their participation in Fshr regulation. It is also possible that integration of ECR regulatory activity with promoter transcription may require additional promoter sequences absent from the -220bp/+123bp construct used in these experiments, such as the SF-1 binding sites located between -743bp and -2700bp (Heckert, 2001). Previous studies postulated that SF-1 plays a combinatorial role in directing cell-specific Fshr expression by interacting with other transcription factors that have overlapping limited expression profiles (Heckert, 2001; Levallet et al., 2001; Xing et al., 2002). Thus, ECR regulation of Fshr transcription may only be revealed in the presence of SF-1 and its responsive region, and experiments are underway to explore the role of SF-1 in ECR transcriptional activity.
In summary, the vast majority of studies examining Fshr transcription have focused on the promoter region, which we now know is incapable of directing accurate expression of the gene. This information provides an important starting point for determining the precise cis- and trans-acting components of Fshr cell-specific expression and a conceptual model for Fshr transcription that, in Sertoli and granulosa cells, includes the combined actions of proteins bound to distal elements with those already known to work through the gene’s promoter. Together, this array of cis-acting DNA elements and trans-acting proteins form a highly specialized transcriptional program that specifies gene expression in Sertoli and granulosa cells. Future experiments designed to identify the distal regulatory elements and define their functional contribution is critical to elucidating the mechanism that directs Fshr transcription, and evaluation of the ECRs represents an excellent starting point.
Acknowledgments
The authors wish to thank Drs. Kenneth Peterson and Susanna Harju for significant technical assistance with YACs, Daren Rice for general assistance and advice, Drs. Barbara Sotolongo and Ning Lei for assistance with transgenic mice, Lovella Tejada for assistance with cell culture, the Transgenic and Gene Targeting Institutional Facility for the generation of transgenic mice and the University of Kansas Medical Center and the Kansas IDeA Network of Biomedical Research Excellence (K-INBRE) for access to DS Gene.
Footnotes
This work was supported by the National Institutes of Child Health & Human Development (HD35217 to LLH), a University of Kansas Medical Center Biomedical Training fellowship to BPH and the University of Kansas Medical Center’s Center for Reproductive Sciences (NICHD SCCPRR; U54-HD33994).
References
- Baroukh N, Ahituv N, Chang J, Shoukry M, Afzal V, Rubin EM, Pennacchio LA. Comparative genomic analysis reveals a distant liver enhancer upstream of the COUP-TFII gene. Mamm. Genome. 2005;16:91–95. doi: 10.1007/s00335-004-2442-9. [DOI] [PubMed] [Google Scholar]
- Bishop CE, Whitworth DJ, Qin Y, Agoulnik AI, Agoulnik IU, Harrison WR, Behringer RR, Overbeek PA. A transgenic insertion upstream of sox9 is associated with dominant XX sex reversal in the mouse. Nat.Genet. 2000;26:490–494. doi: 10.1038/82652. [DOI] [PubMed] [Google Scholar]
- Camp TA, Rahal JO, Mayo KE. Cellular localization and hormonal regulation of follicle-stimulating hormone and luteinizaing hormone receptor messenger RNAs in the rat ovary. Mol. Endocrinol. 1991;5:1405–1417. doi: 10.1210/mend-5-10-1405. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Elnitski L, Church DM, Dubchak I, Hardison RC. Cross-species sequence comparisons: a review of methods and available resources. Genome Res. 2003;13:1–12. doi: 10.1101/gr.222003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo P, Montoliu L. Size matters: use of YACs, BACs and PACs in transgenic animals. Transgenic.Res. 2001;10:83–103. doi: 10.1023/a:1008918913249. [DOI] [PubMed] [Google Scholar]
- Goetz TL, Lloyd TL, Griswold MD. Role of E box and initiatory region in the expression of the rat Follicle-stimulating hormone receptor. J.Biol.Chem. 1996;271:33317–33324. doi: 10.1074/jbc.271.52.33317. [DOI] [PubMed] [Google Scholar]
- Gottgens B, others Analysis of vertebrate SCL loci identifies conserved enhancers. Nat.Biotechnol. 2000;18:181–186. doi: 10.1038/72635. [DOI] [PubMed] [Google Scholar]
- Green ED, Hieter P, Spencer FA. Yeast Artificial Chromosomes. In: Birren B, et al., editors. Genome Analysis; A laboratory Manual. Cold Spring Harbor Laboratory Press; Plainview: 1997. pp. 297–565. [Google Scholar]
- Gromoll J, Dankbar B, Gudermann T. Characterization of the 5’ flanking region of the human Follicle-stimulating hormone receptor gene. Mol Cell Endocrinol. 1994;102:93–102. doi: 10.1016/0303-7207(94)90102-3. [DOI] [PubMed] [Google Scholar]
- Heckert LL. Structure and regulation of the FSH Receptor gene. In: Skinner MK, Griswold MD, editors. Sertoli Cell Biology. Elsevier Academic Press; San Diego, CA: 2005. pp. 281–302. [Google Scholar]
- Heckert LL. Activation of the rat Follicle-stimulating hormone receptor promoter by steroidogenic factor 1 is blocked by protein kinase a and requires upstream stimulatory factor binding to a proximal E-box element. Mol.Endocrinol. 2001;15:704–715. doi: 10.1210/mend.15.5.0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckert LL, Daggett MA, Chen J. Multiple promoter elements contribute to activity of the Follicle-stimulating hormone receptor (FSHR) gene in testicular Sertoli cells. Mol Endocrinol. 1998;12:1499–1512. doi: 10.1210/mend.12.10.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckert LL, Griswold MD. The expression of the follicle-stimulating hormone receptor in spermatogenesis. Recent.Prog.Horm.Res. 2002;57:129–148. doi: 10.1210/rp.57.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heckert LL, Sawadogo M, Daggett MA, Chen J. The USF proteins regulate transcription of the Follicle-stimulating hormone receptor but are insufficient for cell-specific expression. Mol.Endocrinol. 2000;14:1836–1848. doi: 10.1210/mend.14.11.0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Heckert LL. Silencing of Fshr occurs through a conserved, hypersensitive site in the first intron. Mol.Endocrinol. 2005;19:2112–2131. doi: 10.1210/me.2004-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huhtaniemi I, Eskola V, Pakarinen P, Matikainen T, Sprengel R. The murine Luteinizing hormone and Follicle-stimulating hormone receptor genes: transcripional initiation sites, putative promoter sequences, and promoter activity. Mol.Cell.Endocrinol. 1992;88:55–66. doi: 10.1016/0303-7207(92)90009-u. [DOI] [PubMed] [Google Scholar]
- Karpova T, Presley J, Manimaran RR, Scherrer SP, Tejada L, Peterson KR, Heckert LL. A Ftz-F1-containing yeast artificial chromosome recapitulates expression of steroidogenic factor 1 in vivo. Mol.Endocrinol. 2005 doi: 10.1210/me.2004-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lettice LA, others Disruption of a long-range cis-acting regulator for Shh causes preaxial polydactyly. Proc.Natl.Acad.Sci.U.S.A. 2002;99:7548–7553. doi: 10.1073/pnas.112212199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levallet J, Koskimies P, Rahman N, Huhtaniemi I. The promoter of murine Follicle-stimulating hormone receptor: functional characterization and regulation by transcription factor steroidogenic factor 1. Mol Endocrinol. 2001;15:80–92. doi: 10.1210/mend.15.1.0583. [DOI] [PubMed] [Google Scholar]
- Linder CC, Heckert LL, Goetz TL, Griswold MD. Follicle-stimulating hormone receptor gene promoter activity. Endocrine. 1994;2:957–966. [Google Scholar]
- Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- Navas PA, Peterson KR, Li Q, McArthur M, Stamatoyannopoulos G. The 5’HS4 core element of the human beta-globin locus control region is required for high-level globin gene expression in definitive but not in primitive erythropoiesis. J.Mol.Biol. 2001;312:17–26. doi: 10.1006/jmbi.2001.4939. [DOI] [PubMed] [Google Scholar]
- Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Scanning human gene deserts for long-range enhancers. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- Nordhoff V, Gromoll J, Foppiani L, Luetjens CM, Schlatt S, Kostova E, Huhtaniemi I, Nieschlag E, Simoni M. Targeted expression of human FSH receptor Asp567Gly mutant mRNA in testis of transgenic mice: role of human FSH receptor promoter. Asian.J.Androl. 2003;5:267–275. [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Ovcharenko I, Loots GG, Nobrega MA, Hardison RC, Miller W, Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovcharenko I, Nobrega MA, Loots GG, Stubbs L. ECR Browser: a tool for visualizing and accessing data from comparisons of multiple vertebrate genomes. Nucleic.Acids.Res. 2004;32:W280–W286. doi: 10.1093/nar/gkh355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KR, Li QL, Clegg CH, Furukawa T, Navas PA, Norton EJ, Kimbrough TG, Stamatoyannopoulos G. Use of yeast artificial chromosomes (YACs) in studies of mammalian development: production of beta-globin locus YAC mice carrying human globin developmental mutants. Proc.Natl.Acad.Sci.U.S.A. 1995;92:5655–5659. doi: 10.1073/pnas.92.12.5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Kong LK, Poirier C, Truong C, Overbeek PA, Bishop CE. Long-range activation of Sox9 in Odd Sex (Ods) mice. Hum.Mol Genet. 2004;13:1213–1218. doi: 10.1093/hmg/ddh141. [DOI] [PubMed] [Google Scholar]
- Rannikki AS, Zhang F-P, Huhtaniemi IT. Ontogeny of Follicle-stimulating hormone receptor gene expression in the rat testis and ovary. Mol.Cell.Endocrinol. 1995;107:199–210. doi: 10.1016/0303-7207(94)03444-x. [DOI] [PubMed] [Google Scholar]
- Riethman H, Birren B, Gnirke A. Preparation, manipulation, and mapping of HMW DNA. In: Birren B, et al., editors. Genome Analysis; A laboratory Manual. Cold Spring Harbor Laboratory Press; Plainview: 1997. pp. 83–248. [Google Scholar]
- Sagai T, Hosoya M, Mizushina Y, Tamura M, Shiroishi T. Elimination of a long-range cis-regulatory module causes complete loss of limb-specific Shh expression and truncation of the mouse limb. Development. 2005;132:797–803. doi: 10.1242/dev.01613. [DOI] [PubMed] [Google Scholar]
- Sairam MR, Subbarayan VSR. Characterization of the 5’ flanking region and potential control elements of the ovine Follitropin receptor gene. Molecular Reproduction and Development. 1997;48:480–487. doi: 10.1002/(SICI)1098-2795(199712)48:4<480::AID-MRD8>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Sauer B. Inducible gene targeting in mice using the Cre/lox system. Methods. 1998;14:381–392. doi: 10.1006/meth.1998.0593. [DOI] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoni M, Gromoll J, Nieschlag E. The Follicle-stimulating hormone receptor: biochemistry, molecular biology, physiology, and pathophysiology. Endocr.Rev. 1997;18:739–773. doi: 10.1210/edrv.18.6.0320. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat.Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Themmen APN, Huhtaniemi IT. Mutations of gonadotropins and gonadotropin receptors: elucidating the physiology and pathophysiology of pituitary-gonadal function. Endocr.Rev. 2000;21:551–583. doi: 10.1210/edrv.21.5.0409. [DOI] [PubMed] [Google Scholar]
- Tilly JL, LaPolt PS, Hsueh AJ. Hormonal regulation of Follicle-stimulating hormone receptor messenger ribonucleic acid levels in cultured rat granulosa cells. Endocrinology. 1992;130:1296–1302. doi: 10.1210/endo.130.3.1311235. [DOI] [PubMed] [Google Scholar]
- Tisdall DJ, Watanabe K, Hudson NL, Smith P, McNatty KP. FSH receptor gene expression during ovarian follicle development in sheep. J.Mol.Endocrinol. 1995;15:273–281. doi: 10.1677/jme.0.0150273. [DOI] [PubMed] [Google Scholar]
- Val P, Lefrancois-Martinez AM, Veyssiere G, Martinez A. SF-1 a key player in the development and differentiation of steroidogenic tissues. Nucl.Recept. 2003;1:1–8. doi: 10.1186/1478-1336-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing W, Danilovich N, Sairam MR. Orphan receptor chicken ovalbumin upstream promoter transcription factors inhibit steroid factor-1, upstream stimulatory factor, and activator protein-1 activation of ovine Follicle-stimulating hormone receptor expression via composite cis-elements. Biol Reprod. 2002;66:1656–1666. doi: 10.1095/biolreprod66.6.1656. [DOI] [PubMed] [Google Scholar]
- Xing W, Sairam MR. Characterization of regulatory elements of ovine Follicle-stimulating hormone (FSH) receptor gene: the role of E-box in the regulation of ovine FSH receptor expression. Biol.Reprod. 2001;64:579–589. doi: 10.1095/biolreprod64.2.579. [DOI] [PubMed] [Google Scholar]
- Zheng P, Pennacchio LA, Le Goff W, Rubin EM, Smith JD. Identification of a novel enhancer of brain expression near the apoE gene cluster by comparative genomics. Biochim.Biophys.Acta. 2004;1676:41–50. doi: 10.1016/j.bbaexp.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Zhong TP, Kaphingst K, Akella U, Haldi M, Lander ES, Fishman MC. Zebrafish genomic library in yeast artificial chromosomes. Genomics. 1998;48:136–138. [PubMed] [Google Scholar]