Abstract
The 5′-untranslated region (5′-UTR) of retroviral genomes contains elements required for genome packaging during virus assembly. For many retroviruses, the packaging elements reside in non-contiguous segments that span most or all of the 5′-UTR. The Rous sarcoma virus (RSV) is an exception in that its genome can be efficiently packaged by a relatively short, 82-nucleotide segment of the 5′-UTR called μΨ. The RSV 5′-UTR also contains three translational start codons (AUG-1, -2 and –3) that have been controvertibly implicated in translation initiation and genome packaging, one of which (AUG-3) resides within the μΨ sequence. We recently demonstrated that μΨ is capable of binding to the cognate RSV nucleocapsid protein (NC) with high affinity (dissociation constant Kd ~2 nM), and that residues of AUG-3 are essential for tight binding. We now report the solution structure of the NC:μΨ complex, determined using NMR data obtained for samples containing 13C,15N-labeled NC and 2H-enriched, nucleotide-specifically-protonated RNAs. Upon NC binding, μΨ adopts a stable secondary structure that consists of three stem loops (SL-A, SL-B and SL-C) and an 8-base pair stem (O3). Binding is mediated by NC’s two zinc knuckle domains. The N-terminal knuckle interacts with a conserved U(217)GCG tetraloop (a member of the UNCG family; N = A,U,G or C), and the C-terminal zinc knuckle binds to residues that flank SL-A, including residues of AUG-3. Mutations of critical nucleotides in these sequences compromise or abolish viral infectivity. Our studies reveal novel structural features important for NC:RNA binding, and support the hypothesis that AUG-3 is conserved for genome packaging rather than translational control.
Keywords: Rous sarcoma virus, ribonucleic acid (RNA), psi-site (μΨ), nucleocapsid (NC) protein, UNCG tetraloop, nuclear magnetic resonance (NMR)
Abbreviations used: A, adenosine; C, cytidine; G, guanosine; GST, glutathione-S-transferase; HIV-1, human immunodeficiency virus type-1; HMQC, heteronuclear multiple quantum coherence; HSQC, heteronuclear single quantumn coherence; ITC, isothermal titration calorimetry; MLV, Moloney Murine Leukaemia Virus; NC, nucleocapsid protein; NOE, nuclear Overhauser effect; NOESY, NOE spectroscopy; ORF, open reading frame; PBS, primer binding site; RMSD, root-mean-square deviation; ROESY, rotating frame Overhauser effect spectroscopy; RSV, Rous sarcoma virus; SD, splice-donor site; U, uridine; UTR, unstranslated region
Introduction
Retroviral genomes are selected for packaging during viral assembly with exquisite efficiency.1 Two copies of the full-length, unspliced genome are selected from a cellular pool that contains a ~100-fold excess of cellular mRNAs and spliced viral RNAs.1 Genome selection is mediated by interactions between the nucleocapsid (NC) domains of the retroviral Gag polyproteins and a packaging signal located within the 5′-untranslated region (5′-UTR) of the viral genome called the Ψ-site.1–4 Nucleotides of the Ψ-site generally overlap with, or reside downstream of, the 5′ splice donor site (SD), providing a potential mechanism for discriminating between full-length and spliced viral transcripts.5 Ψ-site residues also usually overlap with elements that promote RNA dimerization,6–9 suggesting that genome dimerization and packaging may be intimately coupled.1,2,10 Dimerization-dependent conformational changes that expose NC-binding residues in the Ψ-site of the Moloney murine leukemia virus (MLV) have been proposed to regulate diploid genome packaging,11 and a similar mechanism may be utilized by the human immunodeficiency viruses type-1 and –2 (HIV-112 and HIV-213).
Current understanding of the molecular mechanism of retroviral genome selection has been obtained mainly from studies of MLV and HIV-1.14 Mutagenesis studies have identified regions of the HIV and MLV 5′-UTRs that are essential for genome packaging. Deletion or mutation of residues near dimer-promoting elements can severely attenuate genome selection, although for both retroviruses, efficient packaging of heterologous RNAs requires most (MLV) or all (HIV) of the the 5′-UTR.14 The Rous sarcoma virus (RSV), an alpharetrovirus that induces connective tissue tumors in chickens via the integration of a viral src gene of cellular origin,15 is unusual in that efficient RNA packaging can be directed by fragments of the 5′-UTR comprising as few as 82 nucleotides.16 Deletions between the primer binding site (PBS) and SD can significantly attenuate viral infectivity and genome packaging,17,18 whereas deletions upstream19,20 and downstream17,21 of this region have little or no effect on packaging. In addition, segments of the 5′-UTR between the PBS and SD were shown to efficiently package heterologous RNAs into virus like particles.16,22–25 A 160-nucleotide fragment of the RSV 5′-UTR, termed MΨ (Figure 1), is capable of directing heterologous RNA packaging with efficiency that is only 2.6-fold less than that of the native, intact genome.24,25 Although the L3 stem loop is required for infectivity of the virus, the actual sequence of the L3 stem of MΨ appears not to be essential for genome packaging,26 and an even smaller 82-nucleotide segment (μΨ) was more recently shown to direct RNA packaging with efficiency equal to that of MΨ, Figure 1.16
Figure 1.
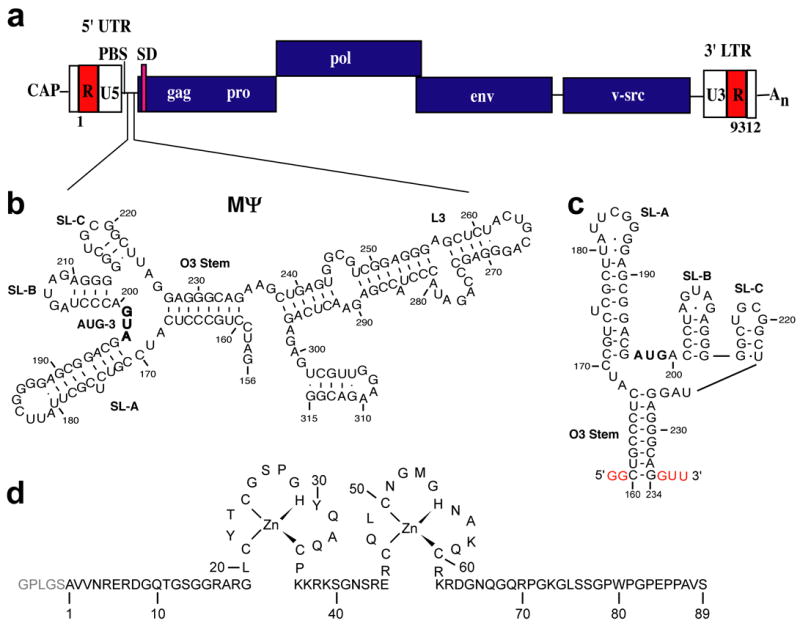
(a) Representation of the Rous sarcoma virus (RSV) genome showing relative locations of the primer binding site (PBS), splice donor (SD) site and MΨ RNA packaging signal. (b) Predicted secondary structure of the MΨ packaging signal (Prague C strain). Nucleotides are numbered using the first residue after the 5′ cap as position +1. (c) Predicted secondary structure of μΨ RNA used in this study. Non-native nucleotides that were added to facilitate in vitro transcription are shown in red. (d) Amino acid sequence and zinc binding mode of the RSV NC protein. Non-native residues derived from the PreScission protease cleavage site (used to cleave GST during purification) are shown in light grey.
The 5′-UTRs of the alpharetroviruses are also unusual in that they contain three translational start codons (AUG-1, -2 and –3), two of which (AUG-1 and AUG-3) are critical for viral replication and gene expression. AUG-3 resides within MΨ, Figure 1, and although some studies suggested that RNA packaging efficiency may be coupled with translation efficiency of the corresponding short open reading frames (ORFs),27–29 other studies indicated that these process are not coupled,30 and that these AUG sequences may instead function by promoting binding to Gag or inhibiting binding to the ribosome.30 Phylogenetic variability in the region between AUG-3 and the gag open reading frame suggested that this region is structurally heterogeneous and might adopt alternate conformations that differentially promote packaging, splicing, or other RNA-dependent processes.31 Consistent with this hypothesis, we recently demonstrated that RSV NC binds μΨ RNA with high affinity, and that mutation of AUG-3 substantially inhibits NC binding.32 We now report the solution structure of the NC:μΨ complex, determined using a combination of isotope-edited NMR experiments. Our studies show that AUG-3 and a conserved UNCG tetraloop participate directly in binding to the NC protein, and that these binding sites are critical for viral fitness. The N- and C-terminal zinc knuckle domains of the NC protein interact with these elements in a manner different from that observed in previously characterized NC:RNA complexes, indicating that the conserved CCHC-type zinc knuckles can direct genome packaging via diverse structural mechanisms.
Results and Discussion
NMR signal assignments for the NC:μΨ complex
1H NMR spectra obtained for μΨ in the absence of NC exhibited very broad lines, consistent with exchange on the millisecond timescale between two or more RNA conformations. We were unable to obtain assignable NMR spectra for the free μΨ RNA under various conditions of salt ([NaCl = 5 to 100 mM, [MgCl2] = 0 to 5 mM), and temperature (15 to 35 °C). However, high quality spectra were obtained upon addition of one equivalent of NC, which enabled analysis of the NMR spectra.
NMR spectral assignments were made for complexes containing nucleotide-specifically protonated, 2H-labeled μΨ RNA. Nucleotide-specific protonation significantly simplifies the spectra, allowing for the identification of sequential nucleotides of a given type.33 In the case of the RSV μΨ, there are relatively few examples of such sequentially repeating residues: GGGGG (1), GGGG (1), GGG (1), GG (5), CCC (2), CC (1) and UU (4). Thus, a stretch of five sequentially correlated guanosine residues (G211-G215) can be uniquely identified in the NOESY data obtained for the NC:μΨ complex containing fully protonated guanosines and perdeuterated U, C and A nucleotides (GH-μΨ; Supplementary Figure S1). Signals for the remaining guanosines were sequentially assigned by comparisons with spectra obtained for samples containing selectively protonated adenosines (AH-μΨ), uridines (UH-μΨ), and cytidines (CH-μΨ). Representative 2D NMR data obtained for the NC:AH-μΨ complex showing aromatic H8 and H2 to H1′ NOE connectivities are shown in Figure 2a. Partial deuteration (~85%) of the purine bases gave rise to weak but observable internucleotide (but not intranucleotide) NOE cross-peaks with the adenosine and guanosine H8 protons, which facilitated or confirmed assignments for sequential N-to-G/A (N = any nucleotide) and G/A-to-U/C stretches, Figure 2b. Representative data showing sequential NOE connectivities for residues G187–G193 are shown in Figure 2b. Finally, 2D 1H NMR NOESY spectra were obtained for fragments of μΨ corresponding to the predicted stem loops SL-A, SL-B and a hairpin containing the stem residues of O3 tethered by a GAGA tetraloop (Figure 3), for comparison with spectra obtained for the intact μΨ RNA. Except for the terminal nucleotides, NMR chemical shifts and NOE cross-peak patterns observed for the fragments matched the corresponding sub-spectral elements observed for μΨ. With this approach, 100% of the aromatic C-H protons, 100% of the H1′, 73% of the H2′, 70% of the H3′, 61% of the H4′ and 36% of the H5′, H5″ protons were assigned.
Figure 2.
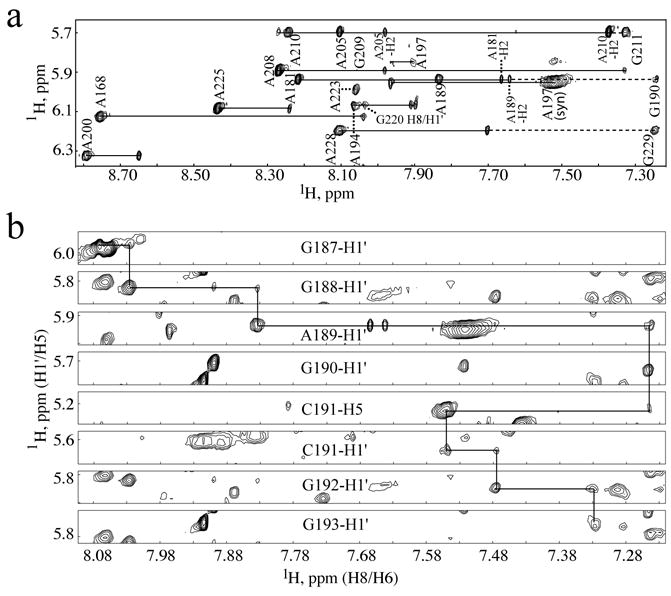
(a) Portion of the 2D NOESY spectrum obtained for A-protonated μΨ (AH-μΨ) bound to NC showing the NOE cross-peaks associated with the fully protonated adenosines and partially protonated guanosine residues. The H1′/H8 (more intense) and H1′/H2 (less intense) cross-peaks from one adenosine residue appear as pairs and are labeled by residue numbers. The sequential H1′/H8 cross-peak from an adenosine residue to the following partially protonated guanosine residue is connected by a dashed line. (b) Representative portions of 2D NOESY data obtained for selectively-protonated (from top to bottom are G, G, A, G, C, C, G and G, respectively, protonated) μΨ RNA bound to NC showing the sequential H1′ to H8 (G187 through G190, C191 through G193) and H8 to H5 (from G190 to C191) NOEs.
Figure 3.
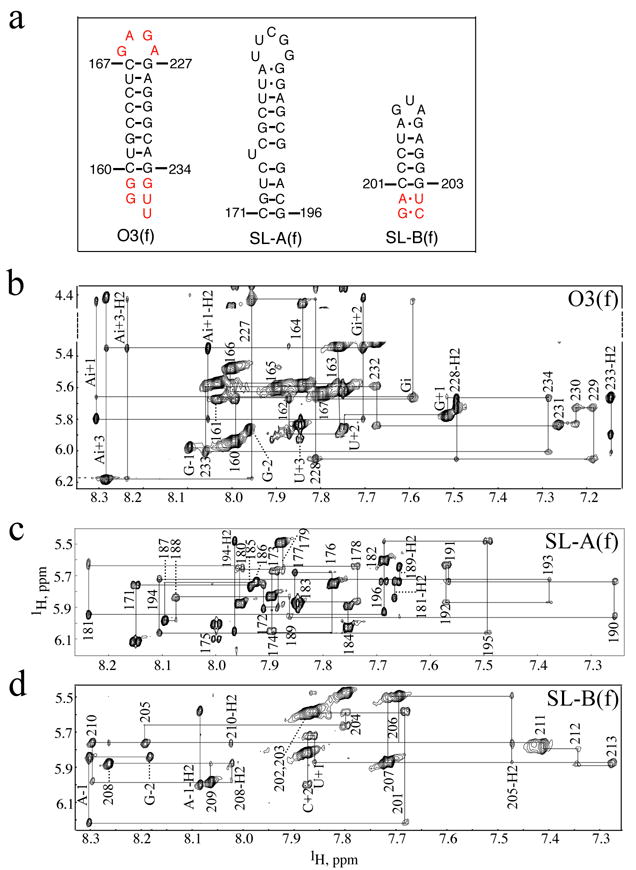
(a) Secondary structures of the μΨ RNA fragments used to facilitate the assignment of the intact μΨ. The fragments corresponding to the O3 stem, SL-A and SL-B are named as O3(f), SL-A(f) and SL-B(f), respectively. Non-native residues are shown in red. In O3(f), non-native residues in the termini corresponding to those in the intact μΨ (see Figure 1) were added as well as a GAGA tetraloop in order to form a stable hairpin. In SL-B(f), two additional GC and AU base pairs were added to stabilize the stem. (b)–(d) Portion of the 2D NOESY spectrum obtained for O3(f), SL-A(f) and SL-B(f), respectively, showing the sequential H1′-to-aromatic proton NOE cross-peaks. In O3(f), non-native residues at the 5′ terminus are labeled as G-2 and G-1 and residues at the 3′ terminus are labeled as G+1, U+2 and U+3, respectively. The GAGA tetraloop residues are labeled as Gi, Ai+1, Gi+2 and Ai+3, respectively. In SL-B(f), the non-native residues are labeled as G-2, A-1 and U+1 and C+2, respectively.
Two sets of signals were observed for residues A197 and C201, indicating that these residues exist in two different conformations. In both cases, one subset of signals was substantially more intense, and NOEs associated with these signals were used for the structure calculations. For A197 (part of AUG-3), the weaker set of signals includes a very intense intraresidue A197-H8 to H1′ NOE (Figure 2a, which is indicative of a syn glycosidic torsion angle) but no direct intermolecular NOEs with the protein. The more intense set of signals includes a very weak intramolecular H8 to H1′ NOE and intense NOEs with the protein, including A197-H8 to His55-Hδ, His55-Hɛ, Cys60-Hα, Arg61-Hα (and sidechain protons) and Lys62-Hα (and sidechain protons), and A197-H1′ to His55-Hɛ. Structures were calculated using distance restraints derived from the latter set of NOEs. Only a single set of signals were observed for U198 and G199 of AUG-3. An intense intraresidue G220-H8 to H1′ NOE and an unusually downfield-shifted G220-H3′ signal (5.77 ppm; Supplementary Figure S2) are consistent with results obtained previously for UNCG tetraloop RNA structures.34–37 The combined NOE data are consistent with the secondary structure of μΨ shown in Figure 1.
1H, 15N and 13C NMR signal assignments were made for all backbone and side-chain atoms of residues Ala17 – Lys37 and Cys47 – Arg63 (which correspond to the N- and C-terminal zinc knuckles, respectively) of the free RSV NC protein (15N,13C-labeled) from 3D 13C-edited NOESY-HMQC and 15N-edited HSQC-NOESY NMR data. 1H and 13C signals were assigned for these residues in the NC:μΨ complex, except for Gly25 and Gly28 of the N-terminal zinc knuckle and Met53 and Gly54 of the C-terminal zinc knuckle, due to the broader linewidths of NMR signals in the complex and severe overlap of the glycine Hα signals. Overlap also precluded unambiguous assignment of Gln59. Most of the remaining residues of the N- and C-terminal tails, as well as several residues in the linker that connects the zinc knuckles, could not be unambiguously assigned for the free or RNA-bound NC protein due to rapid exchange of backbone NH and water protons and apparent conformational flexibility. Large chemical shift changes were observed for several of the NC protein NMR signals upon binding to μΨ, including the aromatic protons of Tyr22, Tyr30 and His55, and these residues gave rise to intermolecular NOEs. However, the intra-zinc knuckle NOE cross-peak patterns and intensities were similar in the free and μΨ-bound NC protein, indicating that binding to μΨ does not affect the folding of zinc knuckles. Portions of the 2D and 3D NOESY data obtained for the NC complex with GH-μΨ and UH-μΨ are shown in Figure 4. No inter-knuckle NOEs were observed for NC protein in the absence or presence of μΨ, indicating that the two knuckles form independently folded domains and bind to separate regions of the μΨ RNA.
Figure 4.
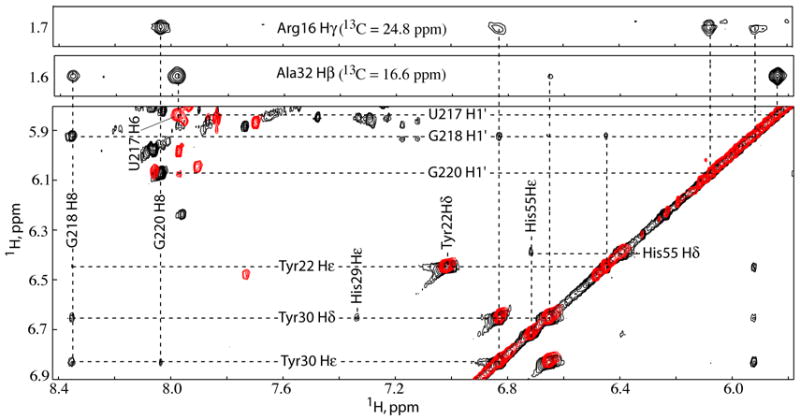
Top: Representative strips from the 3D 13C-edited NOESY-HMQC obtained for 13C, 15N-labeled NC bound to unlabeled μΨ showing the intermolecular NOEs associated with NC N-terminal zinc knuckle residues Arg16 and Ala32, respectively. Bottom: Overlay of the 2D NOESY spectra obtained for GH-μΨ (black) and UH-μΨ (red) bound to NC showing the intermolecular NOEs from SL-C tetraloop residues U217, G218 and G220 to NC N-terminal zinc knuckle aromatic residues Tyr22 and Tyr30.
Structure of the NC:μΨ complex
NMR chemical shifts and NOE cross-peak patterns of the SL-A and SL-B stem loops and O3 stem in the NC:μΨ complex were very similar to those observed for the isolated RNA fragments, indicating that these elements form similar three-dimensional A-helical structures. A total of 20 refined structures with target functions of 4.59±0.35 Å2 were calculated with CYANA using a total of 680 1H-1H distance, 239 RNA torsion angle, 170 inter-phosphate, and 608 H-bond restraints (4 restraints per H-bond), for an average of 28.5 restraints per refined residue. Lower-limit inter-phosphate distance restraints were employed for longer A-helical segments (SL-A and O3), which significantly increases the rate of production of structures with low target functions and affords structures normal helical parameters and major groove widths.33 Additional lower-limit restraints were employed to ensure that the closest approach of non-neighboring phosphate atoms (Pi to Pi+n; n > 4) was greater than 6.0 Å. The O3 helix, SL-C, and the stems of SL-A and SL-B are structurally well defined in the NC:μΨ complex. Best-fit superpositions of the heavy atoms of the stems of O3, SL-A, SL-B and stem loop SL-C afforded pairwise RMS deviations of 0.44 ± 0.13, 0.78 ± 0.20, 0.49 ± 0.18 and 0.70 ± 0.11 Å, respectively (Table 1 and Figure 5). Stem loop SL-A contains two mismatched base pairs (U180•G188, A181•G187) and SL-B contains one mismatched base pair (A205•G209). Residue U175 on the 5′ side of the SL-A stem forms a bulge, with the adjacent C174 and C176 co-stacked. The loop residues of SL-A and SL-B were not restrained during structural calculations, and these regions therefore appear disordered. Standard sequential NOEs were observed between G213 and G214, indicating that SL-B and SL-C are co-stacked. No other inter-stem loop NOEs were observed, and as such, the relative positions and orientations of the remaining stem loops were established loosely by stereochemical and van der Waals restraints and intermolecular restraints to the NC protein. As such, the relative positions and orientations of SL-B, SL-C, O3, and the NC zinc knuckles were reasonably well defined, whereas the position and orientation of SL-A was poorly defined, Figure 6.
Table 1.
Statistics for 20 calculated RSV NC:μΨ structures
| NMR-derived restraints | ||
| 1H-1H distance restraints | ||
| Intraresidue | 326 | |
| Sequential (|i-j|=1) | 182 | |
| Long range (|i-j|>1) | 141 | |
| Intermolecular | 31 | |
| Hydrogen-bond restraints | 608 | |
| Torsion angle restraints | 239 | |
| Inter-phosphate restraints | 170 | |
| Average restraints/refined residue | 28.5 | |
| Target function (Å2) | ||
| Mean ± SD | 4.59±0.35 | |
| Minimum | 4.02 | |
| Maximum | 5.39 | |
| Restraint violations | ||
| Av. upper distance viol. (Å) | 0.028 ± 0.004 | |
| Av. max. upper distance viol. (Å) | 0.38 ± 0.03 | |
| Av. lower distance viol. (Å) | 0.008 ± 0.002 | |
| Av. max. lower distance viol. (Å) | 0.09 ± 0.03 | |
| Av. sum of VDW viol. (Å) | 14.0 ± 1.3 | |
| Av. max. VDW viol. (Å) | 0.72 ± 0.07 | |
| Av. torsion angle viol. (deg.) | 0.043 ± 0.062 | |
| Av. max. torsion angle viol. (deg.) | 0.57 ± 0.85 | |
| Structure convergence (Å)2 | All heavy atom | Main chain atoms |
| μΨ (C160-G234) | 5.69±1.86 | |
| O3 (C160-C167,G227-G234) | 0.44±0.13 | |
| SL-A (C171-C174,C176-A181,G187-G196) | 0.78±0.20 | |
| SL-B (C201-A205,G209-G213) | 0.49±0.18 | |
| SL-C (G214-C223) | 0.70±0.11 | |
| NC-F1 (Arg16-Lys37) | 1.04±0.14 | 0.41±0.05 |
| NC-F2 (Arg46-Lys62) | 1.16±0.22 | 0.36±0.15 |
Statistics for the 39 NC residues (Arg16-Lys37, Arg46-Lys62) restrained during the structure calculations.
Reported as average pairwise RMS deviations for the 20 refined structures.
Cartoon of a representative NMR structure of the Rous sarcoma virus nucleocapsid protein (slate) bound to the cognate μΨ RNA packaging element. Cysteine and histidine residues that bind zinc (gray spheres) are shown in yellow and blue, respectively, and independently structured regions of the RNA are displayed in brown, green, red and purple colors.
Figure 5.
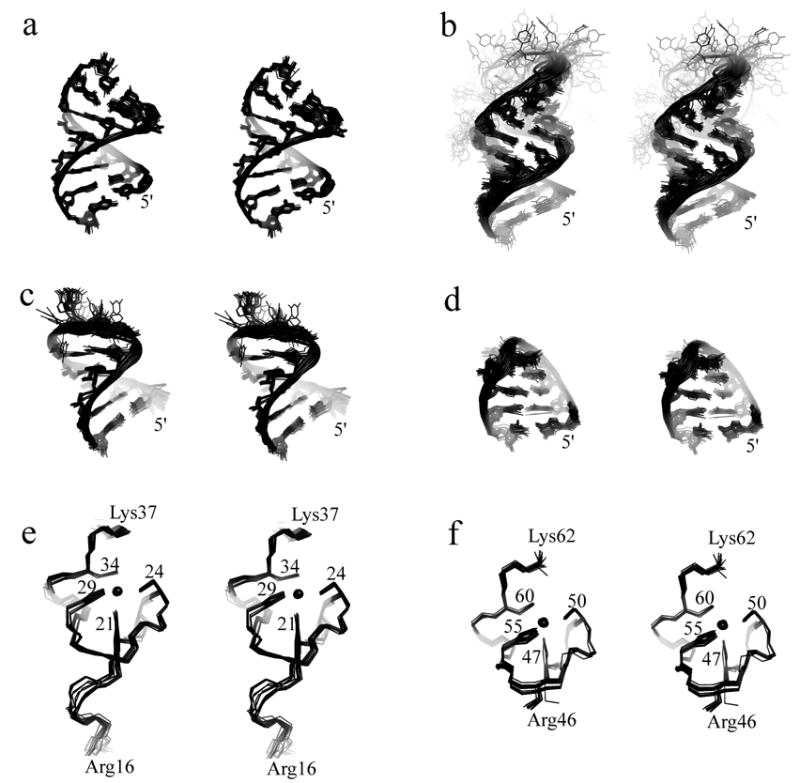
(a)–(d) Stereoviews of the best-fit superpositions of the stems of O3, SL-A, SL-B and stem-loop SL-C (including the UGCG tetraloop), respectively, from the ensemble of 20 refined NC:μΨ structures. Disordered loop nucleotides U182-G186 (SL-A), G206-A208 (SL-B) and the bulge residue U175 (SL-A) are also shown (b and c). (e),(f) Stereoviews of the best-fit superpositions of the main chain atoms of the N- and C-terminal zinc knuckle domains, respectively, from the ensemble of 20 refined NC: μΨ structures.
Figure 6.
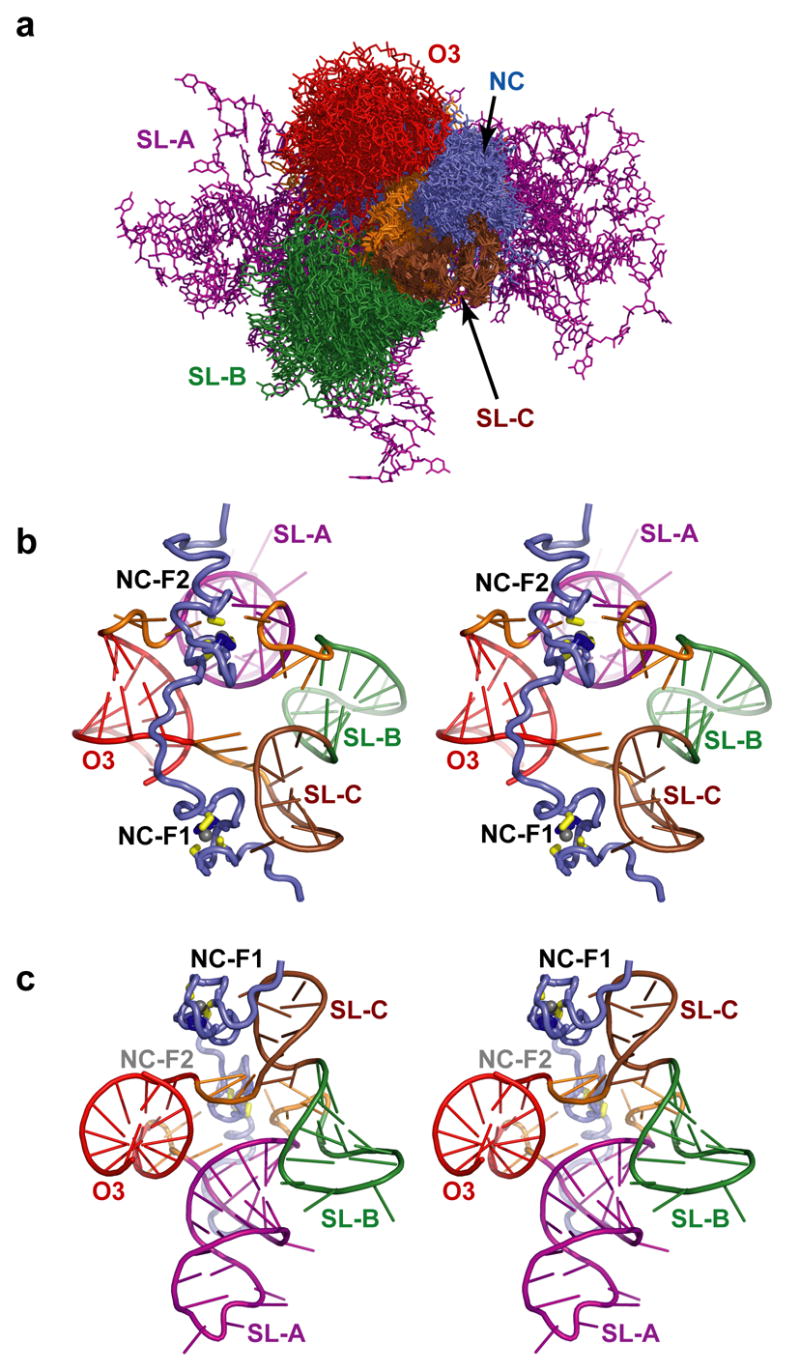
(a) Superposition of the 20 NC:μΨ structures showing the relative convergence of the secondary structure elements. The figure was generated by superpositioning the carbon atoms of the SL-C stem. The relative positions of SL-B (green), SL-C (brown) O3 (red), the linkers (orange), and the NC zinc knuckles (slate) are fairly well defined by the NMR data, but the position of SL-A (purple) is poorly defined. (b,c) Stereo views (differing by a 90 degree rotation) of a representative structure (with lowest target function), colored as in (a), showing the relative positions of the NC and μΨ secondary structures.
The U217GCG tetraloop of SL-C in NC:μΨ complex adopts a canonical UNCG tetraloop structure similar to those observed in previous NMR34–37 and X-ray crystal38 structures. U217 and G220 form a UG base pair that is stabilized by hydrogen bonding between the 2′-hydroxyl proton of U217 and the O6 oxygen of G220, and between the the O2 oxygen of U217 and imino and amino protons of G220. In addition, the amino group of C219 is poised to form a hydrogen bond with the phosphate group of G218. G220 adopts a syn glycosidic torsion angle and is stacked on top of G221, while U217 is stacked upon the C216•G221 base pair and C219 is stacked upon U217•G220 base pair. The base of G218 does not interact with the other nucleotides of SL-C, but instead makes extensive contacts with the NC protein (see below). Residues U224-A225-G226 of the linker that connects SL-C with the O3 stem fold back into the minor groove of the SL-C stem, with A225-H2 in close proximity to C216-H1′ and Ala32 of the N-terminal zinc knuckle (see below), Figure 7.
Figure 7.
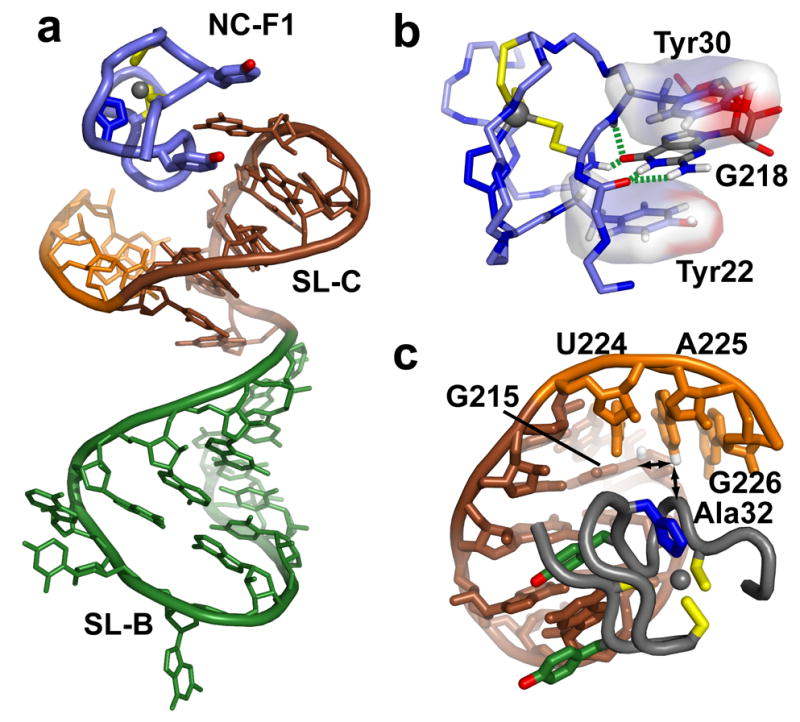
(a) The N-terminal NC zinc knuckle (NC-F1) binds to the UGCG tetraloop of SL-C (brown), which itself co-stacks with stem loop SL-B (green). (b) Binding is mediated, in part, by the insertion of G218 between the aromatic side chains of Tyr22 and Tyr30, and by hydrogen bonds with backbone atoms of the zinc knuckle (dashed green lines). (c) Linker residues U224, A225 and G226 fold back and pack in the minor groove of SL-C. Long-range intra- and inter-molecular NOEs involving the A225-H2 proton are shown with arrows.
The N-terminal NC zinc knuckle binds to the UGCG tetraloop of SL-C
The RSV NC protein contains two zinc knuckle domains that adopt canonical folds observed for all other structurally characterized retroviral zinc knuckle domains.14 Best-fit superposition of the main chain atoms of the N- and C-termainal zinc knuckle domains afforded pairwise RMS deviations of 0.41 ± 0.05 and 0.36 ± 0.15 Å, respectively, Figure 5e,f. The nucleobase of G218 fits deeply into a hydrophobic pocket defined by residues Tyr22, Tyr30, Leu20 and Gln31. Hydrophobic residues at the same positions, which generally include at least one Trp or Phe residue, form similar clefts on other structurally characterized zinc knuckle domains, including those of HIV-1 and MLV NC.14,39 Representations showing the position and orientation of the N-terminal zinc knuckle relative to the co-stacked stem loops SL-B and SL-C and the packing of the G218 nucleobase with the aromatic side chains of Tyr22 and Tyr30 are shown in Figure 7. The importance of these interactions is supported by site-directed mutagenesis studies using a yeast three hybrid system, in which mutation of Tyr22 to Ile decreased the affinity of RSV NC for MΨ by 97%.40 Binding is further stabilized by intermolecular hydrogen bonding with backbone NH and O atoms located at the bottom of the pocket (G218-O6-Tyr22-NH; G218-O6-Gln31-NH; G218-N1H-Leu20-O; G218-NH21-Leu20-O), Figure 7. This mode of guanosine recognition and binding by CCHC-type zinc knuckles has been observed in HIV-1 and MLV NC-RNA and zinc knuckle-DNA complexes.11,41–43 In addition, the nucleobase of G220 packs against the side chains of Tyr30 and Ala17, and the ribose moiety of C216 packs against the side chain of Ala32, Figure 7b.
The C-terminal zinc knuckle binds A197 of AUG-3
The C-terminal zinc knuckle of RSV NC also forms a canonical CCHC zinc knuckle backbone fold, Figure 5f. This domain is unusual in that it does not contain residues with aromatic side chains (other than the zinc-coordinating His55), nor does it contain a well-defined hydrophobic pocket. No NOEs were observed between this zinc knuckle domain and guanosine residues, indicating that the domain functions differently from the the previously characterized zinc knuckle domains. However, a number of intermolecular NOEs were observed between this zinc knuckle and residues in the linkers adjacent to stem loop SL-A (Supplementary Figure S3). The A197 nucleobase packs against His55, Cys60, and Arg61, with the exocyclic amino group and N7 atom poised to form hydrogen bonds with the carbonyl of Gln59 and the amide proton of Arg61, respectively. The side chain of Arg61 is also posed to form a salt bridge with the 5′-phosphate of A197. Mutation of the conserved A197 to G reduced the NC binding affinity of μΨ by ~10,000 times,32 and this could be due in part to the inability of G to form these hydrogen bonds. These findings are also consistent with mutagenesis studies showing that Arg61 is important for the high affinity Gag-Ψ RNA binding in vitro and efficient genome packaging in vivo.40,44,45 The nucleobase of A168 packs against the side chains of Leu49, Cys60 and Lys58, and appears to be stabilized by a hydrogen bond between the A168 phosphate oxygen and Lys58 side chain amino group, Figure 8.
Figure 8.
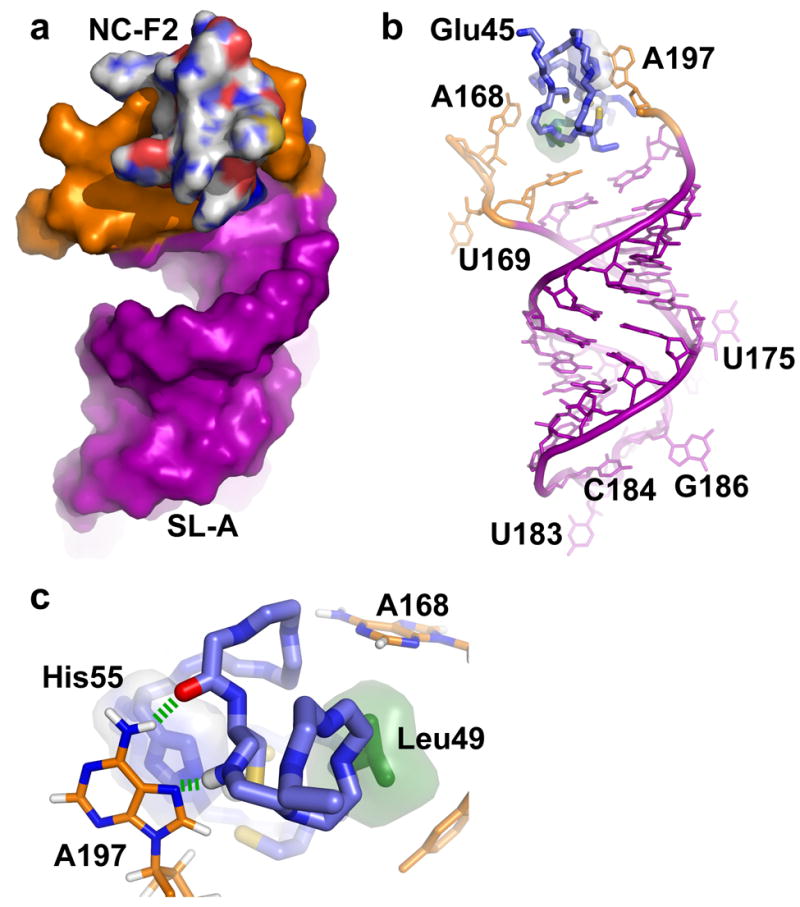
Surface (a) and stick (b) representations of the C-terminal NC zinc knuckle (NC-F2) bound to the linker residues (orange) proximal to stem loop SL-A (purple). (c) Expanded view of the NC-F2 binding site showing the packing of A168 and A197 against the side chains of Leu49 (green) and His55 (blue), respectively. Hydrogen bonds are shown as dashed green lines.
Nucleobases interacting with NC are important for viral infectivity
To assess the biological significance of the RSV NC:μΨ structure, we created mutations in the RNA sequences that appeared critical for NC binding, and then introduced these mutations into an infectious RSV viral DNA. Two days after transfection of the mutant DNA into chicken DF1 cells, virus particles shed into the medium were collected and used to infect new DF1 cells, and the spread of the virus was followed over time by a standard assay for reverse transcriptase in the growth medium.46 Substitution of GAGA for the wild type sequence UGCG217–220 in the tetraloop of SL-C resulted in a large decrease in the rate of viral spread, Figure 9, but did not completely destroy infectivity. Ten days after infection, the sequence of the viral RNA showed no reversion or compensatory mutations in psi or gag (not shown). Substitution of CCU for AUG197–199 or for AUC168–170 (or for both) abrogated viral infectivity, since the mutant viruses were unable to spread at all, Figure 9. These results provide strong support for the relevance of the NC:μΨ structure in vivo.
Figure 9.
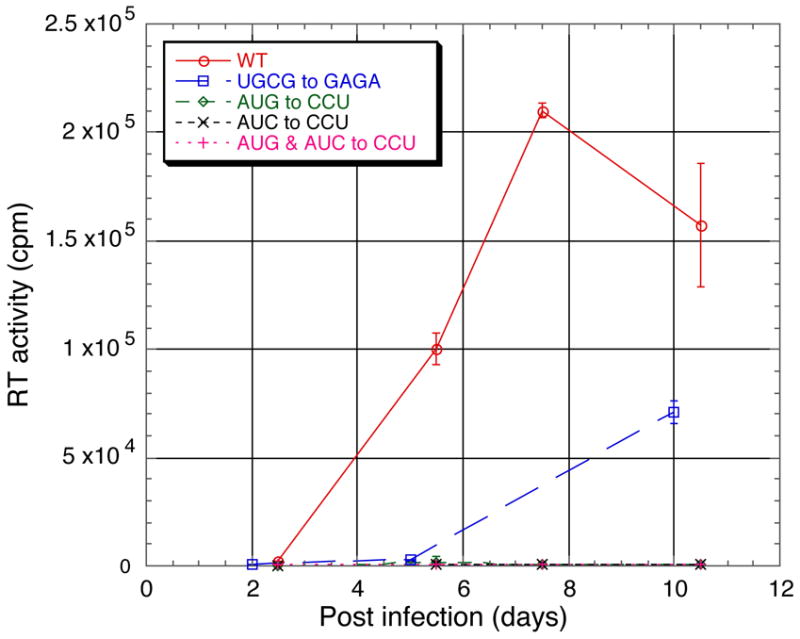
Virus infectivity monitored by RT assays. Compared with wild type virus (circles), mutant virus in which the SL-C tetraloop sequence was changed from UGCG217–220 to GAGA exhibited slower growth kinetics (squares). Mutant viruses in which the sequences AUC168–170 and/or AUG197–199 on either side of SL-A were changed to CCU were not infectious (diamonds, crosses, or plus symbols).
Biological implications
Previous studies have shown that RSV MΨ and μΨ are capable of directing heterologous RNA packaging with efficiencies near that of the full-length, wild-type genome.16,24,25 Studies using yeast three hybrid assays suggest that RSV Gag binds MΨ and μΨ RNAs with similar affinities,47 and the affinity of mutant Gag molecules for the Ψ-site appears to be intrinsically related to the efficiency of packaging.16 Gag constructs that lack the matrix, capsid, or protease domains bind MΨ with similar affinities and package viral RNA into virus-like particles with high efficiency, indicating that interactions between the NC domain of Gag and μΨ serve as the primary determinant of genome selection.44 RSV NC binds μΨ with unusually high affinity (Kd ~2 nM), and truncated μΨ-derived RNAs that lack O3 or any of the stem loops bind NC with substantially reduced affinities.32 These data collectively support the proposal that μΨ functions as the minimal determinant for NC binding and genome packaging.16
Recent NC binding studies showed that the UGCG tetraloop of SL-C is required for high-affinity binding.32 Substitution of the UGCG residues by GAGA (a member of the GNRA family; N=A,U,G or C and R = A or G) resulted in a ~1,500-fold reduction in the affinity of RSV NC for μΨ RNA,32 whereas substitution of the loop residues of SL-A and SL-B by GAGA residues did not affect binding.32 Consistent with these findings, the present studies reveal that the loop residues of SL-A and SL-B do not interact with the protein or make long range RNA-RNA contacts, and that the UGCG tetraloop of SL-C binds directly to the N-terminal zinc knuckle of NC. Furthermore, mutating the tetraloop to GAGA causes a major decrease in viral infectivity. UNCG tetraloops are widely distributed in biology,48 and together with GNRA sequences comprise more than 70 percent of the tetraloops of large and small rRNAs.49,50 These families of tetraloops form unusually stable hairpin structures, and in some cases are known to participate in long-range RNA-RNA interactions.35,51–57 To our knowledge, the present findings provide the first example in which a UGCG tetraloop participates directly in protein-RNA interactions. The syn-conformation of the universally conserved tetraloop guanosine (G220) appears to not only stablize the loop structure, but also allows greater exposure of the G218 nucleobase, which binds to the hydrophobic pocket of NC’s N-terminal zinc knuckle. This binding mode is reminiscent of that observed previously in HIV-1 and MLV NC:RNA complexes, in which exposed guanosine nucleobases are inserted into hydrophobic pockets and form hydrogen bonds with backbone NH and O atoms of the zinc knuckles.11,42,43
While it is conserved in almost all alpharetroviruses and forms part of the NC binding site in vitro in the Prague C (PrC) strain of RSV, the UGCG tetraloop appears not to be strictly required for genome packaging and infectivity. Mutation of this sequence to UUUG did not substantially diminish RNA packaging16, and in randomization studies of a 14 nucleotide stretch including this sequence, infectious viruses were recovered with a variety of sequences different from the canonical UNCG26. Furthermore, when we changed the tetraloop sequence to GAGA, the resulting virus was still infectious, although it was compromised in replication. The same GAGA mutation reduced the affinity of NC for the RNA by a thousand-fold in vitro32, but this reduction is substantially less than that brought about by mutation of the A in AUG-3. Based on all of these results, it seems that the tetraloop is of secondary importance for Gag binding in vivo.
The C-terminal zinc knuckle interacts with the RNA in a manner that has not been observed in other retroviral NC:RNA complexes. This domain does not contain a hydrophobic pocket and does not interact with exposed guanosine residues. Instead, the domain interacts with two adenosine nucleobases, one (A168) associated with the linker that connects O3 and SL-A and the other (A197 of AUG-3) located in the linker that connects SL-A and SL-B. The nucleobase of A168 makes hydrophobic contacts with Leu49, Cys60 and Lys58, and binding appears to be further stabilized by a salt bridge between the 5′-phospate of A168 and the side chain amino group of Lys58. The nucleobase of A197 is poised to form hydrogen bonds with the backbone NH and O atoms of Arg61 and Gln59, respectively, and its 5′-phosphate appears to form a salt bridge with the side chain of Arg61. The nucleobase also appears to partially stack with the aromatic side chain of the zinc-coordinating His55. These findings demonstrate that the retroviral CCHC zinc finger scaffold can be used in significantly different ways to promote RNA binding, and provide a potential explanation for why chimeric viruses containing “swapped” NC domains are sometimes (but not always) capable of competitively packaging the genome of the virus from which NC was derived.58
The present findings support the proposal that the AUG residues of AUG-3 are conserved to promote genome packaging rather than translation initiation.30,59 Mutations and deletions within and/or adjacent to the initiation codon of AUG-3 clearly can cause significant reductions in RNA packaging16,27–29. For example, an RNA containing C195A, G199C and A200U mutations was packaged with efficiency that was 50-fold lower than that of the same RNA containing the native MΨ sequence and comparable to that of a negative control RNA lacking an MΨ sequence16. In addition, mutation of A197 to G reduces the affinity of NC for μΨ by a factor of ~10,000.32 Our findings demonstrate that A197 participates directly in NC binding. The nucleobase of U198 packs against A200 and U224 in a manner that appears to stabilize the fold of the μΨ RNA. The reason for the conservation of G199 is not obvious, as this nucleotide did not exhibit inter-residue or intermolecular NOEs and does not appear to participate in long-range RNA-RNA or RNA-protein interactions. It is conceivable this guanosine is needed to stabilize an alternate conformation that is important for translation, nuclear export, reverse transcription, or some other RNA-dependent function.
The phenotypes of the AUC168–170 and AUG197–199 viral mutants, namely the abolishment of infectivity, support the role of the linker nucleotides on either side of SL-A as being critical for virus replication. In our experience, few mutations in the viral Ψsite have such a drastic effect. Taken together with the previous binding data32, the structure presented here and the in vivo phenotype of the mutations make it likely that the role of AUC168–170 and of AUG197–199 is to create a high-affinity binding site for the NC domain of Gag, and that this binding site is essential for genome packaging.
The O3 stem, which is conserved among the alpharetroviruses,16 is important for both genome packaging and NC binding. Mutations that disrupt O3 stem base pairing lead to dramatic reductions in packaging, which can be restored by co-variational mutations24, and point mutations in the O3 stem can inhibit interactions with RSV Gag.47 Deletion of the O3 stem results in a significant decrease in the affinity of NC for μΨ32. The present studies demonstrate that the O3 stem does not interact directly with NC. Instead, O3 appears to promote NC binding by stabilizing a tertiary structure that is required for high-affinity binding, probably by maintaining the close proximity of the linker residues that are involved in NC binding.
As indicated above, we have been unable to obtain quality NMR spectra for the free μΨ RNA due to conformational heterogeneity and exchange at unfavorable interconversion rates. Mfold calculations indicate that μΨ can adopt two secondary structures with similar free energies. One secondary structure matches that shown in Figure 1, and the other contains the same O3 stem structure but different conformations for stem loops A, B and C. Although we were unable to identify the conformations of the free RNA, it is tempting to speculate that both structures exist in equilibrium in the absence of NC, and that NC binding shifts the equilibrium toward the observed structure shown in Figure 1. This model would be consistent with the proposal that μΨ adopts different structures for translation initiation and genome packaging.30,59 In the absence of NC, the alternate conformation that is responsible for line broadening might play a role in translation initiation. As Gag concentrations increased, the RNA would be shifted by NC binding from the conformation responsible for translation initiation to the observed conformation that directs packaging. Additional studies of the free μΨ and MΨ RNAs, and of RNA constructs with mutations designed to stabilize the putative alternate conformation, should provide insights into the nature and function of the free RNA heterogeneity.
Materials and Methods
Sample preparation
RSV NC was expressed as a GST fusion protein in E. coli BL21 codon plus (DE3)-RIL strain (Invitrogen), and purified by affinity chromatography on Glutathione Sepharose 4B (GE Healthcare) under non-denaturing conditions as previously described.32 Isotopic enrichment with 15N and/or 13C was readily achieved by growing the transformed bacteria in minimal media supplemented with 15NH4Cl and/or 13C glucose (Cambridge Isotopes).
μΨ RNA fragments O3(f), SL-A(f) and SL-B(f) were obtained from Dharmacon Research Inc. with 2′-o-bis (acetoxyethoxy)-methyl (ACE) protection. All these RNAs were de-protected following manufacturer recommendations and ethanol precipitated prior to NMR studies. μΨ RNA was synthesized by in vitro transcription with T7 RNA polymerase and purified by denaturing gel electrophoresis as described.32 Specifically, selectively protonated RNA samples (AH-μΨ, UH-μΨ, GH-μΨ, CH-μΨ) were prepared using a combination of three deuterated NTPs (~85% perdeuteration, Silantes GmbH) and the respective protonated NTP (Sigma-Aldrich) with yields typically in the range of 7–11 mg of purified RNA per 15 mL RNA reaction. Both NC and RNA samples were prepared in Tris-d11-HCl buffer (5 mM at pH 7.0, 5 mM NaCl, 0.1 mM ZnCl2 in 2H2O) for NMR studies. The concentrations of NC and RNA were estimated by the optical absorbance at 280nm and 260nm, respectively. Samples of the 1:1 NC:μΨ complex were prepared by titrating 6.0 mM NC protein into 0.8–1.3 mM μΨ RNA. The completeness of the titrations was confirmed by native polyacrylamide gel electrophoresis. Usually a sample volume of 225–250 μL in a Shigemi tube (Shigemi, Inc.) was used for NMR data collection.
NMR spectroscopy
NMR data were collected with Bruker AVANCE (800 MHz, 1H) and DMX (600 MHz, 1H) spectrometers, processed with NMRPipe/NMRDraw60 and analyzed with NMRView.61 All homonuclear experiments were collected using the States method for quadrature detection in the indirect dimensions.62 Water suppression was accomplished with flip-back pulses,63 pulsed field gradients,64 or presaturation pulses during the relaxation delay.
NMR experiments of the free NC protein, except for the 3D 1H-13C HMQC-NOESY, were collected at 25°C. All other experiments were done at 35°C. Backbone signals of the free NC protein were assigned using standard triple resonance methods,65 and side chain signals were assigned from 3D 1H-15N HSQC-NOESY (τm = 200 ms), 3D 1H-13C HMQC-NOESY (τm = 200 ms) and 4D 15N, 13C- edited NOESY (τm = 200 ms) data.66,67 Signal assignments of NC protein in the NC-μΨ complex were obtained from a 3D 1H-13C HMQC-NOESY (τm = 85 ms) spectrum of a sample containing 13C, 15N-labled NC bound to unlabeled μΨ. μΨ RNA assignments were obtained by analyses of 2D NOESY (τm = 85 ms) spectra for AH-μΨ, UH-μΨ, GH-μΨ, CH-μΨ (containing protonated A, U, G and C, respectively, with the remaining nucleotides perdeuterated) and fully protonated μΨ bound to unlabeled NC, respectively. 2D ROESY data (25 ms mixing time) were obtained to identify chemical exchange cross-peaks and distinguish direct and relayed NOEs.68–70 A mixing time of 200 ms was used in 2D NOESY experiments for μΨ fragments O3(f), SL-A(f) and SL-B(f).
Structure calculations
Upper interproton distance bounds of 2.7, 3.3 and 5.0 Å were employed for NOE cross-peaks of strong, medium and weak intensity, respectively, which were qualitatively assessed following intensity normalization of the different NOE data sets. Structures were calculated in torsion angle space with CYANA71 (http://www.las.jp/index_eg.html) starting from random initial angles (involving both protein and RNA). We were unable to obtain residual dipolar couplings for the NC:μΨ complex (due to severe line broadening in the presence of the alignment media). Therefore, loose inter-phosphate restraints were therefore employed to limit the approach of phosphate atoms across the major grooves of A helices to 8 Å or greater, and to limit the approach of all other long-rang P(i)-to-P(i+4) distance to 6 Å or greater.33 Structure figures were generated with PyMol (DeLano, W.L. The PyMOL Molecular Graphics System (2002): http://pymol.sourceforge.net).
Cell culture and assay for viral infectivity
DF1 cells were maintained in Dulbecco’s modified Eagle’s medium plus 5% fetal calf serum, 5% NuSerum, 1% heat inactivated chick serum, and vitamins and antibiotics. Viral spread was analyzed using a standard reverse transcriptase assay. For the experiment with the GAGA mutation, replicate values represent samples taken from a single infection. The experiment was repeated once with similar results. For the CCU mutations, replicates were taken from three independent infections initiated on the same day. In all cases the virus was collected from 1.5 mL of culture medium by first filtering through a 0.45 μm filter, and then centrifuging the virus through a 20% sucrose cushion. One third of the resuspended virus was submitted to a reverse transcriptase assay as described previously (ref Stewart et al 1990) with poly(A) RNA as a template, oligo(dT) as a primer, and [α-32P] dTTP at a specific activity of 2 x 104 μCi/μmol.
Supplementary Material
Acknowledgments
Supported by NIH grant GM42561 to MFS and USPHS grant CA20081 to VMV. We are grateful to Dr. Victoria D’Souza, Yu Chen, Rob Edwards, and Cindy Finch (HHMI, UMBC) for technical assistance and support.
Footnotes
Depositions: The atomic coordinates are available from the RCSB Protein Data Bank with accession codes rcsb039591 (RCSB) and 2IHX (PDB).
References
- 1.Berkowitz R, Fisher J, Goff SP. RNA packaging. Curr Top Microbiol Immun. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 2.Rein A. Retroviral RNA packaging: A review. Arch Virology. 1994;9:513–522. doi: 10.1007/978-3-7091-9326-6_49. [DOI] [PubMed] [Google Scholar]
- 3.Swanstrom R, Wills JW. Synthesis, assembly and processing of viral proteins. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor Press; Plainview, N.Y.: 1997. pp. 263–334. [PubMed] [Google Scholar]
- 4.Berkhout B. Prog Nucl Acid Res and Mol Biol. Vol. 54. Academic Press, Inc.; 1996. Structure and function of the human immunodeficiency virus leader RNA; pp. 1–34. [DOI] [PubMed] [Google Scholar]
- 5.Mann R, Baltimore D. Varying the position of a retrovirus packaging sequence results in the encapsidation of both unspliced and spliced RNA. J Virol. 1985;54:401–407. doi: 10.1128/jvi.54.2.401-407.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greatorex J, Lever A. Retroviral RNA dimer linkage. J Gen Virol. 1998;79:2877–2882. doi: 10.1099/0022-1317-79-12-2877. [DOI] [PubMed] [Google Scholar]
- 7.Paillart JC, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 8.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: An inseparable pair. Nature Reviews Microbiology. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 9.Greatorex J. The retroviral RNA dimer linkage: different structures may reflect different roles. Retrovirology. 2004;1 doi: 10.1186/1742-4690-1-22. (18 August edition). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prats AC, Roy C, Wang P, Erard M, Housset V, Gabus C, Paoletti C, Darlix JL. Cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. Journal of Virology. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.D’Souza V, Summers MF. Structural basis for packaging the dimeric genome of Moloney Murine Leukaemia Virus. Nature. 2004;431:586–590. doi: 10.1038/nature02944. [DOI] [PubMed] [Google Scholar]
- 12.Berkhout B, Ooms M, Beerens N, Huthoff H, Southern E, Verhoef K. In vitro evidence that the untranslated leader of the HIV-1 genome is an RNA checkpoint that regulates multiple functions through conformational changes. J Biol Chem. 2002;277:19967–19975. doi: 10.1074/jbc.M200950200. [DOI] [PubMed] [Google Scholar]
- 13.Dirac AMG, Huthoff H, Kjems J, Berkhout B. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucl Acids Res. 2002;30:2647–2655. doi: 10.1093/nar/gkf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Souza V, Summers MF. How retroviruses select their genomes. Nature Reviews Microbiology. 2005;3:643–655. doi: 10.1038/nrmicro1210. [DOI] [PubMed] [Google Scholar]
- 15.Brickell PM. The p60c-src family of protein-tyrosine kinases: structure, regulation and function. Crit Rev Oncog. 1992;3:401–446. [PubMed] [Google Scholar]
- 16.Banks JD, Linial ML. Secondary structure analysis of a minimal avian leukosis-sarcoma virus packaging signal. J Virol. 2000;74:456–464. doi: 10.1128/jvi.74.1.456-464.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoker AW, Bissell MJ. Development of avian sarcoma and leukosis virus based vector packaging cell lines. J Virol. 1988;16:1161–1170. doi: 10.1128/jvi.62.3.1008-1015.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katz RA, Terry RW, Skalka AM. A conserved cis-acting sequence in the 5′ leader of avian sarcoma virus RNA is required for packaging. Journal of Virology. 1986;59:163–167. doi: 10.1128/jvi.59.1.163-167.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cobrink D, Soskey L, Leis J. A retroviral RNA secondary structure required for efficient initiation of reverse transcription. J Virol. 1988;62:3622–3630. doi: 10.1128/jvi.62.10.3622-3630.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cobrink D, Aiyar A, Ge Z, Katzman M, Huang H, Leis J. Overlapping retrovirus U5 sequence elements are required for efficient integration and initiation of reverse transcriptiuon. J Virol. 1991;65:3864–3872. doi: 10.1128/jvi.65.7.3864-3872.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Norton PA, Coffin JM. Bacterial b-galactosidase as a marker of Rous sarcoma virus gene expression and replication. Mol Cell Biol. 1985;5:281–290. doi: 10.1128/mcb.5.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arnoff R, Linial M. Specificity of retroviral RNA packaging. J Virol. 1991;65:71–80. doi: 10.1128/jvi.65.1.71-80.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arnoff R, Hajjar AM, Linial ML. Avian retroviral RNA encapsidation: Reexamination of functional 5′ RNA sequences and the role of nucleocapsid Cys-His motifs. J Virol. 1993;67:178–188. doi: 10.1128/jvi.67.1.178-188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Banks JD, Yeo A, Green K, Cepeda F, Linial ML. A minimal avian retroviral packaging sequence has a complex structure. J Virol. 1998;72:6190–6194. doi: 10.1128/jvi.72.7.6190-6194.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Banks JD, Kealoha BO, Linial ML. An MΨ containing heterologous RNA, but not env mRNA, is efficiently packaged into avian retroviral particles. J Virol. 1999;73:8926–8933. doi: 10.1128/jvi.73.11.8926-8933.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doria-Rose NA, Vogt VM. In vivo selection of Rous sarcoma virus mutants with randomized sequences in packaging signals. J Virol. 1998;72:8073–8082. doi: 10.1128/jvi.72.10.8073-8082.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donze O, Spahr PF. Role of the open reading frames of Rous sarcoma virus leader RNA in translation and genome packaging. EMBO J. 1992;11:3747–3757. doi: 10.1002/j.1460-2075.1992.tb05460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moustakas A, Sonstegard TS, Hackett PB. Alterations of the three short open reading frames in the Rous sarcoma virus leader RNA modulate viral replication and gene expression. J Virol. 1993;67:4337–4349. doi: 10.1128/jvi.67.7.4337-4349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donze O, Damay P, Spahr PF. The first and third uORFs in RSV leader RNA are efficiently translated: implications for translational regulation and viral RNA packaging. Nucl Acids Res. 1995;23:861–868. doi: 10.1093/nar/23.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonstegard TS, Hackett PB. Autogenous regulation of RNA translation and packaging by Rous sarcoma virus Pr76gag. J Virol. 1996;70:6642–6652. [PMC free article] [PubMed] [Google Scholar]
- 31.Hackett P, Dalton MW, Johnson DP, Peterson RB. Phylogenetic and physical analysis of the 5′ leader RNA sequences of avian retroviruses. Nucl Acids Res. 1991;19:6929–6934. doi: 10.1093/nar/19.24.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou J, McAllen K, Tailor Y, Summers MF. High affinity nucleocapsid protein binding to the μΨ RNA packaging signal of Rous sarcoma virus. J Mol Biol. 2005;349:976–988. doi: 10.1016/j.jmb.2005.04.046. [DOI] [PubMed] [Google Scholar]
- 33.D’Souza V, Dey A, Habib D, Summers MF. NMR structure of the 101 nucleotide core encapsidation signal of the Moloney Murine Leukemia Virus. J Mol Biol. 2004;337:427–442. doi: 10.1016/j.jmb.2004.01.037. [DOI] [PubMed] [Google Scholar]
- 34.Cheong C, Varani G, Tinoco I., Jr Solution structure of an unusually stable RNA hairpin 5′GGAC(UUCG)GUCC. Nature. 1990;346:680–682. doi: 10.1038/346680a0. [DOI] [PubMed] [Google Scholar]
- 35.Varani G, Cheong C, Tinoco I., Jr Structure of an Unusually stable RNA hairpin. Biochemistry. 1991;30:3280–3289. doi: 10.1021/bi00227a016. [DOI] [PubMed] [Google Scholar]
- 36.Allain FHT, Varani G. Structure of the P1 helix from Group I self-splicing introns. J Mol Biol. 1995;250:333–353. doi: 10.1006/jmbi.1995.0381. [DOI] [PubMed] [Google Scholar]
- 37.Comolli LR, Ulyanov NB, Soto AM, Marky LA, James TL, Gmeiner WH. NMR structure of the 3′ stem-loop from human U4 snRNA. Nucl Acids Res. 2002;30:4371–4379. doi: 10.1093/nar/gkf560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ennifar e, Nikulin A, Tishchenko S, Serganov A, Nevskaya N, Garbver M, Ehresmann B, Ehresmann C, Nikonov S, Dumas P. The crystal structure of UUCG tetraloop. J Mol Biol. 2000;304:35–42. doi: 10.1006/jmbi.2000.4204. [DOI] [PubMed] [Google Scholar]
- 39.Summers MF, South TL, Kim B, Hare DR. High-resolution structure of an HIV zinc fingerlike domain via a new NMR-based distance geometry approach. Biochemistry. 1990;29:329–340. doi: 10.1021/bi00454a005. [DOI] [PubMed] [Google Scholar]
- 40.Lee EG, Alidina A, May C, Linial ML. importance of basic residues in binding of Rous sarcoma virus nucleocapsid to the RNA packaging signal. J Virol. 2003;77:2010–2020. doi: 10.1128/JVI.77.3.2010-2020.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.South TL, Summers MF. Zinc- and sequence-dependent binding to nucleic acids by the N-terminal zinc finger of the HIV-1 nucleocapsid protein: NMR structure of the complex with the Psi-site analog, dACGCC. Protein Science. 1993;2:3–19. doi: 10.1002/pro.5560020102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 43.Amarasinghe GK, De Guzman RN, Turner RB, Chancellor K, Wu ZR, Summers MF. NMR structure of the HIV-1 nucleocapsid protein bound to stem-loop SL2 of the Ψ-RNA packaging signal. J Mol Biol. 2000;301:491–511. doi: 10.1006/jmbi.2000.3979. [DOI] [PubMed] [Google Scholar]
- 44.Lee EG, Yeo A, Kraemer B, Wickens M, Linial ML. The Gag domains required for avian retroviral RNA encapsidation determined by using two independent assays. J Virol. 1999;73:6282–6292. doi: 10.1128/jvi.73.8.6282-6292.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee EG, Linial ML. Basic residues of the retroviral nucleocapsid play different roles in Gag-Gag and Gag-Y RNA interactions. J Virol. 2004;78:8486–8495. doi: 10.1128/JVI.78.16.8486-8495.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stewart L, Schartz G, Vogt VM. Properties of avian retrovirus particles defective in viral protease. J Virol. 1990;64:5076–5079. doi: 10.1128/jvi.64.10.5076-5092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee EG, Linial ML. Yeast three-hybrid screening of Rous sarcoma virus mutants with randomly mutagenized minimal packaging signals reveals regions important for Gag interactions. J Virol. 2000;74:9167–9174. doi: 10.1128/jvi.74.19.9167-9174.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlenbeck OC. Tetraloops and RNA folding. Nature. 1990;346:613–614. doi: 10.1038/346613a0. [DOI] [PubMed] [Google Scholar]
- 49.Woese CR, Gutell R, Gupta R, Noller HF. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983;47:621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guttel rR, Fox GE. A compilation of large subunit RNA sequences presented in a structural format. Nucl Acids Res. 1988;16 (Suppl):r175–r269. doi: 10.1093/nar/16.suppl.r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abramovitz DL, Pyle AM. Remarkable morphological variability of a common RNA folding motif: the GNRA tetraloop-receptor interaction. J Mol Biol. 1997;266:493–506. doi: 10.1006/jmbi.1996.0810. [DOI] [PubMed] [Google Scholar]
- 52.Michel F, Westhof E. Modeling of the three-dimensional architecture of Group I catralytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 53.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: Principles of RNA packaging. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 54.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 56.Murphy FL, Cech TR. GAAA tetraloop and conserved bulge stabilize tertiary structure of a group-I intron domain. J Mol Biol. 1994;236:49–63. doi: 10.1006/jmbi.1994.1117. [DOI] [PubMed] [Google Scholar]
- 57.Pley HM, Flaherty KM, McKay DB. Model for an RNA tertiary interaction from the structure of an intermolecular complex between a GAAA tetraloop and an RNA helix. Nature. 1994;372:111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]
- 58.Dupraz P, Spahr PF. Specificity of Rous sarcoma virus nucleocapsid protein in genomic RNA packaging. Journal of Virology. 1992;66:4662–4670. doi: 10.1128/jvi.66.8.4662-4670.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deffaud C, Darlix JL. Rous sarcoma virus translation revisited: Characterization of an internal ribosome entry segment in the 5′ leader of the genomic RNA. J Virol. 2000;74:11581–11588. doi: 10.1128/jvi.74.24.11581-11588.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 61.Johnson BA, Blevins RA. NMRview: a Computer Program for the Visualization and Analysis of NMR Data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 62.States DJ, Haberkorn RA, Ruben DJ. A two-dimensional nuclear Overhauser experiment with pure absorption phase in four quadrants. J Magn Reson. 1982;48:286–292. [Google Scholar]
- 63.Grzesiek S, Bax A. The Importance of Not Saturating H2O in Protein NMR. Application to Sensitivity Enhancement and NOE Measurement. J Am Chem Soc. 1993;115:12593–12593. [Google Scholar]
- 64.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J Biomol NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 65.Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. Journal of Magnetic Resonance. 1992;96:432–440. [Google Scholar]
- 66.Kay EL, Ikura M, Bax A. The design and optimization of complex NMR experiments. Application to a triple-resonance pulse scheme correlating ha, NH,15N chemical shifts in 15N-13C-labeled proteins. Journal of Magnetic Resonance. 1991;91:84–92. [Google Scholar]
- 67.Grzesiek S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114:6291–6293. [Google Scholar]
- 68.Bax A, Sklenar V, Summers MF. Direct identification of relayed nuclear Overhauser effects. J Magn Reson. 1986;70:327–331. [Google Scholar]
- 69.Bothner-by A, Stephens RL, Lee J-m, Warren CD, Jeanloz RW. Structure determination of a tetrasaccharide: Transient nuclear Overhauser effects in the rotating frame. Journal of the American Chemical Society. 1984;106:811–813. [Google Scholar]
- 70.Bax A, Davis DG. Separation of chemical exchange and cross relaxation effects in two-dimensional NMR spectroscopy. J Magn Reson. 1985;63:201–213. [Google Scholar]
- 71.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for protein structure calculations with a new program, DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


