Abstract
The asymmetric localization of messenger RNA (mRNA) and protein determinants plays an important role in the establishment of complex body plans. In Drosophila oocytes, the anterior localization of bicoid mRNA and the posterior localization of oskar mRNA are key events in establishing the anterior-posterior axis. Although the mechanisms that drive bicoid and oskar localization have been elusive, oocyte microtubules are known to be essential. Here we report that the plus end–directed microtubule motor kinesin I is required for the posterior localization of oskar mRNA and an associated protein, Staufen, but not for the anterior-posterior localization of other asymmetric factors. Thus, a complex containing oskar mRNA and Staufen may be transported along microtubules to the posterior pole by kinesin I.
The Drosophila oocyte provides an excellent model system for studies of how the localization of mRNAs helps to establish complex body plans (1, 2). During the late stages of oogenesis, bicoid (bcd) and oskar (osk) mRNAs are localized to the anterior and posterior poles of the oocyte, respectively. Subsequent localized expression of Bicoid protein is necessary for anterior patterning, whereas localized expression of Oskar is necessary for posterior patterning and germ cell development. Genes required for the localization of bcd mRNA (e.g., exuperentia and swallow) and osk mRNA (e.g., staufen and orb) have been identified (1, 2), but the mechanisms of localization are not well understood.
Before stage 7 of oogenesis, the oocyte nucleus and a microtubule organizing center (MTOC) are positioned at the posterior pole, generating a high concentration of microtubule minus ends near the posterior cortex (3-6). During stages 7 and 8, MTOC components switch to the anterior end, resulting in the reorganization of microtubules to place plus ends toward the posterior cortex (3-6). The oocyte nucleus then migrates from the posterior to the anterior end in a microtubule-dependent manner (1, 2, 7).
The localization of bcd and osk mRNAs between stages 8 and 12 is sensitive to microtubule-depolymerizing drugs, consistent with active transport of the mRNAs along microtubules by motor proteins (5, 8, 9). According to this model, bcd is moved to the anterior cortex by minus end–directed motors, such as dyneins, and osk is moved to the posterior cortex by plus end–directed motors, such as kinesins. Indeed, a dynein light chain can bind directly to Swallow, a putative RNA-binding protein necessary for anterior bcd localization (10).
If osk is transported to the posterior pole along microtubules, one or more of the more than 20 Drosophila kinesin superfamily members (11) might drive the movement. We focused our attention on Kinesin heavy chain (Khc) and its encoded protein KHC. KHC is the force-producing subunit of the tetrameric adenosine triphosphatase kinesin I (conventional kinesin), which in Drosophila and other metazoan animals serves as a motor for plus end–directed fast axonal transport of membranous organelles (12).
To determine if kinesin I is involved in oocyte patterning, we used mitotic recombination to generate mosaic female flies that contained clones of homozygous Khc null germ line stem cells (13-16). Western blots confirmed the depletion of KHC in embryos produced from such germ line clones (Fig. 1) (17). The production of eggs and embryos by the mosaic females suggests that germ line stem cells can proliferate and proceed through oogenesis without kinesin I. However, embryogenesis failed, despite fertilization by wild-type males. Most embryos arrested before blastoderm formation, but a few proceeded into early gastrulation stages. This maternal lethal effect was completely rescued by a wild-type Khc transgene (14, 18). Thus, germ line expression of KHC was required for normal embryogenesis, contrary to previous results from studies of a hypomorphic temperature-sensitive Khc allele (13).
Fig. 1.

Depletion of KHC in embryos produced from germ line stem cell clones. Embryos from wild-type female flies (left lane) or females that carried Khc27/Khc27 germ line clones (right lane) were homogenized and tested for the presence of KHC by Western blotting with anti-KHC (16). Anti-actin staining (Act) showed that the two lanes had equivalent protein loadings.
Examination of embryos that reached the blastoderm stage revealed an absence of pole cells, the germ line precursors. To assay for earlier defects, we examined the distributions of osk and bcd mRNAs in Khc null oocytes (19). The localization of bcd mRNA was normal (Fig. 2, A and B), concentrated at the anterior during stages 8 to 10 (193 observed). In contrast, the localization of osk mRNA was defective (Fig. 2, C to F). It normally accumulates transiently at the anterior pole early in stage 8 and then moves to the posterior pole (20, 21). In Khc null stage 8 to 10 egg chambers, osk mRNA accumulated excessively at the anterior pole and was never concentrated at the posterior pole (72 observed). This localization defect was completely rescued by a wild-type Khc transgene. Thus, although KHC was not required for anterior localization of either bcd or osk, it was required for the posterior localization of osk. Perhaps kinesin I transports osk mRNA along microtubules toward their plus ends and the posterior pole.
Fig. 2.
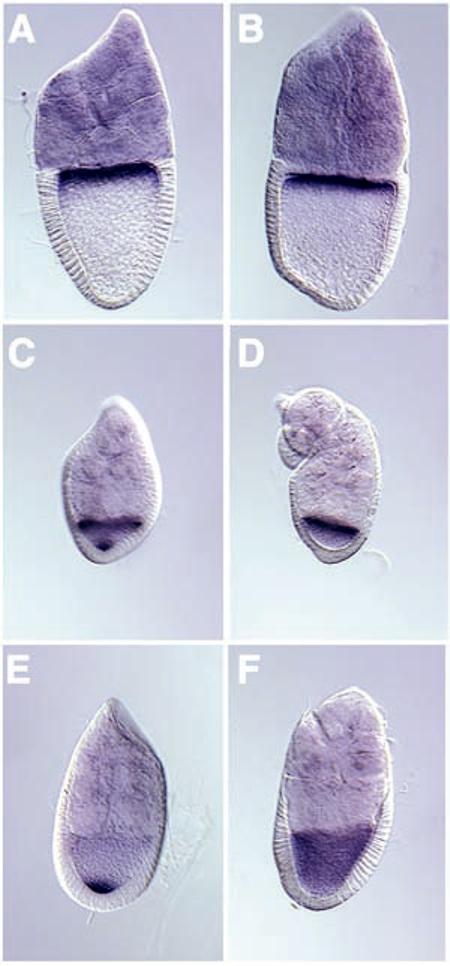
Localization of anterior-posterior mRNA determinants in Khc null oocytes. In situ hybridization was used to test the distribution of bcd and osk mRNAs. bcd is shown in (A) stage 10A wild-type and (B) stage 10B Khc27/Khc27 egg chambers. osk is shown in (C) stage 8 wild-type, (D) stage 8 Khc27/Khc27, (E) stage 9 wild-type, and (F) stage 9 Khc27/Khc27 egg chambers.
Given that microtubule-disrupting drugs prevent the posterior localization of osk mRNA during oogenesis (5, 8), we considered the possibility that the absence of KHC blocks osk localization indirectly by disturbing oocyte microtubules. The shift of the oocyte nucleus from posterior to anterior poles during stage 6, which is microtubule-dependent (7), appeared normal in Khc null oocytes. Furthermore, the anterior localization of the MTOC component Centrosomin was normal (Fig. 3, A and B). As described above, the anterior localization of bcd mRNA, which is microtubule-dependent (9), was not affected by the loss of KHC.
Fig. 3.
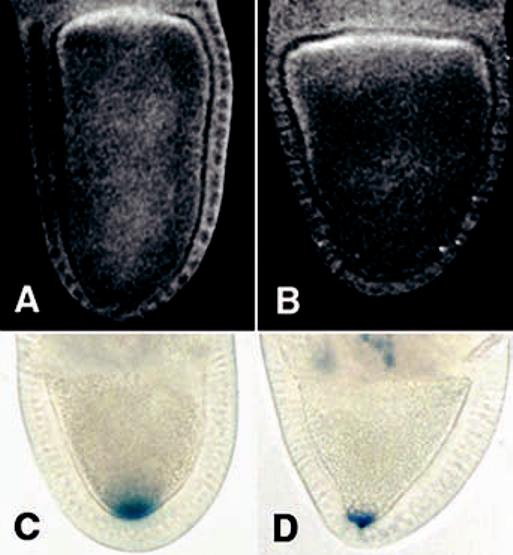
Polarity of the oocyte microtubule cytoskeleton in Khc null oocytes. The localization of MTOC material was tested with antibodies to Centrosomin. (A) Wild-type and (B) Khc27/Khc27 stage 10A oocytes. The orientation of microtubule plus ends was tested by localizing a KHC::β-Gal fusion protein. (C) Wild-type and (D) Khc27/Khc27 late stage 9 egg chambers. The wild-type genotype included two copies of the fusion gene, whereas the mutant included one copy.
We tested microtubule organization further by localizing a hybrid protein composed of the motor domain of KHC fused to a reporter enzyme, β-galactosidase (β-Gal). KHC::β-Gal is thought to localize in regions of cells with high concentrations of microtubule plus ends (22). It is important to note that this chimeric protein did not rescue patterning defects in Khc null oocytes. In wild-type stage 9 to 10a oocytes, KHC::β-Gal concentrated at the posterior pole in 79 cases (Fig. 3C) and elsewhere in 2 cases. In Khc null oocytes, KHC::β-Gal concentrated at the posterior pole in 11 instances (Fig. 3C) and elsewhere in 4 instances. This suggests that in 73% of the stage 9 to 10a null oocytes with detectable amounts of KHC::β-Gal, microtubule plus ends were concentrated at the posterior pole. This, and the indications that microtubules in the anterior end are normal, suggests that osk mRNA mislocalization in Khc null oocytes is not due to a disruption of microtubule organization. In Khc null oocytes, KHC::β-Gal staining was often not detected in oocytes, although it was visible in nurse cells. Perhaps efficient transport of the hybrid protein from nurse cells to oocyte requires the presence of native KHC.
Orb protein is concentrated along the posterior and lateral oocyte cortex during stages 7to9(23). It is thought to hold osk mRNA at the posterior pole and help regulate its translation (24, 25). To determine if the osk mislocalization in Khc null oocytes was secondary to a mislocalization of Orb, we stained with antibodies to Orb (anti-Orb) (22). The distribution of Orb in Khc null oocytes was normal (Fig. 4). Thus, the failure of osk localization may be due to a defect in osk transport rather than in posterior retention.
Fig. 4.
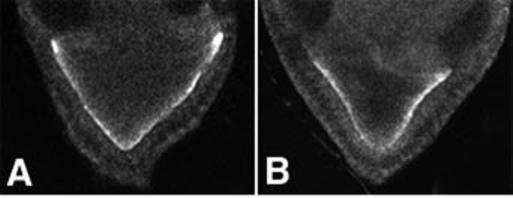
Distribution of Orb, an osk mRNA binding protein that may anchor osk to the posterior cortex. Anti-Orb was used to stain stage 9 egg chambers. (A) Wild-type; (B) Khc27/Khc27.
Posterior transport of osk mRNA is thought to depend on Staufen protein (26). Staufen is transiently localized at the anterior end of the oocyte during stage 8 where it may form a complex with osk mRNA (27). If kinesin I transports such osk-Staufen complexes along microtubules to the posterior pole, then Staufen protein should be mislocalized in Khc null oocytes. Immunostaining with anti-Staufen (28) confirmed this prediction (Fig. 5). In wild-type stage 8 to 10 oocytes, Staufen concentrated at the anterior end early, appeared in granules along the cortex, and then concentrated at the posterior end. Granular Staufen distribution was detected in 61 of the 69 oocytes observed. This is consistent with the hypothesis that Staufen and osk mRNA form complexes at the anterior cortex that are transported to the posterior pole. In Khc mutant oocytes, Staufen protein overaccumulated in the anterior end during stage 8, was not detected in granules, and did not concentrate at the posterior pole (>90 oocytes observed). Normal Staufen distribution patterns were restored in Khc null oocytes by the addition of a wild-type Khc transgene (18).
Fig. 5.
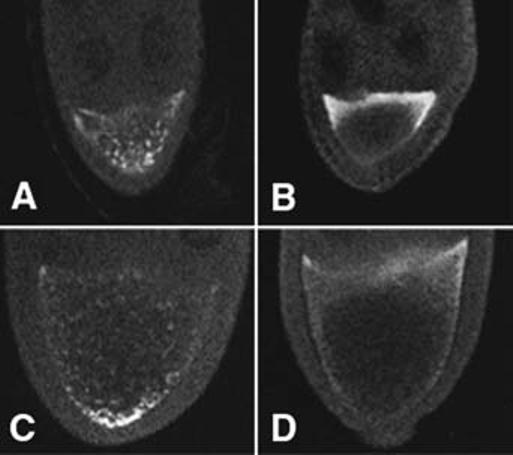
Distribution of Staufen, an osk mRNA binding protein thought to be required for posterior osk transport. Anti-Staufen was used to stain egg chambers. (A) Stage 8 wild type, (B) stage 8 Khc27/Khc27, (C) stage 9 wild type, and (D) stage 9 Khc27/Khc27.
Thus, KHC, the force-generating component of the plus end–directed microtubule motor kinesin I, is required for the posterior localization of both osk mRNA and Staufen protein. The participation of kinesin I in this mRNA motility process could be direct. It might attach specifically to osk-Staufen complexes at the anterior pole and transport them toward the posterior pole. However, our initial tests for coimmunoprecipitation of KHC and Staufen from Drosophila ovary cytosol have not revealed any robust association, so perhaps the linkage is less direct. It is generally accepted that kinesin I transports membranous organelles toward microtubule plus ends (12). Thus, osk and Staufen could localize to the posterior pole by virtue of association with mitochondria or other organelles carried by kinesin I. An alternative to these models is derived from the effect of a loss of KHC on the particulate staining pattern of Staufen (Fig. 5). Before stages 7 to 8, while microtubules are still oriented with their plus ends toward the anterior, kinesin I might deliver to the cortex materials necessary for the assembly of transport-competent osk-Staufen complexes. Thus, the lack of visible Staufen particles in Khc null oocytes may indicate that their assembly or persistence depends on kinesin I activity. New studies, using green fluorescent protein tags to follow the localization dynamics of osk mRNA, Staufen, and organelles, may distinguish between these models and provide further insight into the mechanisms that drive the movements of maternal determinants for early developmental patterning.
References and Notes
- 1.Lasko P. FASEB J. 1999;13:421. doi: 10.1096/fasebj.13.3.421. [DOI] [PubMed] [Google Scholar]
- 2.van Eeden F, St Johnston D. Curr. Opin. Genet. Dev. 1999;9:396. doi: 10.1016/S0959-437X(99)80060-4. [DOI] [PubMed] [Google Scholar]
- 3.Theurkauf WE, Smiley S, Wong ML, Alberts BM. Development. 1992;115:923. doi: 10.1242/dev.115.4.923. [DOI] [PubMed] [Google Scholar]
- 4.Clark IE, Jan LY, Jan YN. Development. 1997;124:461. doi: 10.1242/dev.124.2.461. [DOI] [PubMed] [Google Scholar]
- 5.Clark IE, Giniger E, Ruohola-Baker H, Jan LY, Jan YN. Curr. Biol. 1994;4:289. doi: 10.1016/s0960-9822(00)00068-3. [DOI] [PubMed] [Google Scholar]
- 6.Li K, Kaufman TC. Cell. 1996;85:585. doi: 10.1016/s0092-8674(00)81258-1. [DOI] [PubMed] [Google Scholar]
- 7.Koch EA, Spitzer RH. Cell Tissue Res. 1983;228:21. doi: 10.1007/BF00206261. [DOI] [PubMed] [Google Scholar]
- 8.Pokrywka NJ, Stephenson EC. Dev. Biol. 1995;167:363. doi: 10.1006/dbio.1995.1030. [DOI] [PubMed] [Google Scholar]
- 9.Pokrywka NJ, Stephenson EC. Development. 1991;113:55. doi: 10.1242/dev.113.1.55. [DOI] [PubMed] [Google Scholar]
- 10.Schnorrer F, Bohmann K, Nusslein-Volhard C. Nature Cell Biol. 2000;2:185. doi: 10.1038/35008601. [DOI] [PubMed] [Google Scholar]
- 11.Goldstein LSB, Gunawardena S. J. Cell Biol. 2000;150:F63. doi: 10.1083/jcb.150.2.f63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin MAE, Hurd DD, Saxton WM. Cell. Mol. Life Sci. 1999;56:200. doi: 10.1007/s000180050422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saxton WM, Hicks J, Goldstein LSB, Raff EC. Cell. 1991;64:1093. doi: 10.1016/0092-8674(91)90264-y. [DOI] [PubMed] [Google Scholar]
- 14.Brendza RP. Indiana University; 1999. thesis. [Google Scholar]
- 15. We generated germ line clones [T. B. Chou, E. Noll, N. Perrimon, Development 119, 1359 (1993)] by mating y w P{hs-FLP}; P{w+, FRT}42B, P{OvoD1}55D/ CyO males to w; P{w+, FRT}42B c Khc27/ Cyo females. Mitotic recombination was induced in the germ line stem cells of first-instar larvae (60 to 72 hours old) with the genotype y w P{hs-FLP}/ w; P{w+, FRT}42B, P{OvoD1}55D / P{w+, FRT}42B c Khc27. OvoD1 is a dominant female sterile mutation that blocks oogenesis at early stages. Thus, oocytes that developed beyond stage 4 were descendants of Khc27/Khc27 germ cell clones. After development, females producing eggs were collected and mated to wild-type males.
- 16.Brendza RP, Sheehan KB, Turner FR, Saxton WM. Mol. Biol. Cell. 2000;11:1329. doi: 10.1091/mbc.11.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Total protein was extracted from embryos (0 to 2 hours old), and Western blotting was performed on serial dilutions of each extract (13). Flyk-2 antibody (1:200 dilution) was used to localize KHC (13). Anti-actin (Amersham) was used at 1:2000 dilution to identify lanes that had equivalent protein loadings. Blots were developed with the use of an ECL kit (Amersham)
- 18. All defects detected in Khc null oocytes and embryos were rescued by a transgenic copy of the wild-type Khc gene (13). This was determined by generating germ line clones in females carrying the genotype y w P{hs-FLP}/ w; P{w+, FRT}42B, P{OvoD1}55D / P{w+, FRT}42B c Khc27; P{w+, Khc+}/ +
- 19.Gonzalez-Reyes A, Elliott H, St Johnston D. Nature. 1995;375:654. doi: 10.1038/375654a0. [DOI] [PubMed] [Google Scholar]
- 20.Ephrussi A, Dickinson LK, Lehmann R. Cell. 1991;66:37. doi: 10.1016/0092-8674(91)90137-n. [DOI] [PubMed] [Google Scholar]
- 21.Kim-Ha J, Smith JL, Macdonald PM. Cell. 1991;66:23. doi: 10.1016/0092-8674(91)90136-m. [DOI] [PubMed] [Google Scholar]
- 22. Khc null clones were induced in females as described (15) with the addition of a third chromosome carrying P[Khc::LacZ] (5). Localization of KHC::β-Gal activity in oocytes was performed according to Ghiglione [Cell 96, 847 (1999)]. Centrosomin was localized as described by Roth et al. [S. Roth, F. S. Neuman-Silberberg, G. Barcelo, T. Schupbach, Cell 81, 967 (1995)] using anti-CNN at 1: 800 dilution (6). Orb was localized with undiluted mouse anti-Orb as described (23, 25)
- 23.Lantz V, Chang JS, Horabin JI, Bopp D, Schedl P. Genes Dev. 1994;8:598. doi: 10.1101/gad.8.5.598. [DOI] [PubMed] [Google Scholar]
- 24.Chang JS, Tan L, Schedl P. Dev. Biol. 1999;215:91. doi: 10.1006/dbio.1999.9444. [DOI] [PubMed] [Google Scholar]
- 25.Christerson LB, McKearin DM. Genes Dev. 1994;8:614. doi: 10.1101/gad.8.5.614. [DOI] [PubMed] [Google Scholar]
- 26.St Johnston D, Beuchle D, Nusslein-Volhard C. Cell. 1991;66:51. doi: 10.1016/0092-8674(91)90138-o. [DOI] [PubMed] [Google Scholar]
- 27.Micklem DR, Adams J, Grunert S, St Johnston D. EMBO. J. 2000;19:1366. doi: 10.1093/emboj/19.6.1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ovarioles were fixed in 200 μl of 2% paraformaldehyde, 0.5% NP-40 in phosphate-buffered saline (PBS) mixed with 600 μl of heptane (20 min), rinsed in PBS–0.2% Tween-20, and incubated in rabbit anti-Staufen (25) at 1:1600 dilution (overnight at 4°C). After extensive rinsing, ovarioles were stained with fluorescein-conjugated goat anti-rabbit immunoglobulin G (Jackson Laboratory, Bar Harbor, ME) at 1:500. All fluorescence images were generated with a Bio-Rad MRC600 scanning confocal fluorescence microscope.
- 29. Supported by NIH grant R01GM46295 (W.M.S.)


