Abstract
During its red blood cell stage, the malaria parasite Plasmodium falciparum can switch its variant surface proteins (P. falciparum erythrocyte membrane protein 1) to evade the host immune response. The var gene family encodes P. falciparum erythrocyte membrane protein 1, different versions of which have unique binding specificities to various human endothelial surface molecules. Individual parasites each contain ≈60 var genes at various locations within their chromosomes; however, parasite isolates contain different complements of var genes, thus, the gene family is enormous with a virtually unlimited number of members. A single var gene is expressed by each parasite in a mutually exclusive manner. We report that control of var gene transcription and antigenic variation is associated with a chromatin memory that includes methylation of histone H3 at lysine K9 as an epigenetic mark. We also discuss how gene transcription memory may affect the mechanism of pathogenesis and immune evasion.
Keywords: antigenic variation, chromatin, monoallelic expression, histone modification
Red blood cells infected with Plasmodium falciparum can sequester in deep tissues, including the brain and placenta. Sequestration is important for parasite survival and malaria pathogenesis and is facilitated by a parasite-encoded protein presented on the red-blood cell surface called P. falciparum erythrocyte membrane protein 1 (PfEMP1). Different forms of PfEMP1 are encoded by members of the polymorphic var gene family. Various PfEMP1 proteins have distinct binding affinities to different host molecules present on the surface of endothelial cells and blood cells, and they also bind with chondroitin sulfate A, a glycosaminoglycan found on the lining of the placental intervillous space. The high degree of polymorphism displayed by this family is likely to be the result of positive selection by the host–immune system. Importantly, PfEMP1 can undergo switches to different antigenic forms and evade the host immune response against infected erythrocytes.
Switches in PfEMP1 expression are the result of changes in the transcription of individual var genes (1). As the parasite progresses into the early trophozoite stage, expression is limited to a single gene that encodes the PfEMP1 form that is featured on the surface of the infected cell. The remaining members of the var gene family are not transcribed and are said to be silenced (1–4). All var genes have a two-exon structure (5). The long exon 1 encodes the polymorphic extracellular part of the protein, whereas the short exon 2 encodes a more conserved intracellular part. The silencing mechanism is at least in part the result of a cooperative interaction between the promoter and intron of each individual var gene, both of which are necessary for complete silencing (4, 6–8). The var intron has bidirectional promoter activity (9), and this promoter activity appears to be required for gene silencing. Interestingly, “sterile” transcripts driven by the intron's promoter are observed in late-stage parasites (5).
Several groups have tried to estimate the switching rate and temporal order of var gene expression (10–12), however, the data are somewhat discordant, perhaps reflecting dissimilar experimental systems and methods used to monitor var expression. In this study, we took advantage of a previously undescribed system by using a blasticidin drug-resistance marker gene to select for the expression of one particular var gene. Our analysis of the transcribed and silenced var genes reveals histone modifications reminiscent of those associated with gene-transcription memory found in the homeotic genes of Drosophila melanogaster. We identify a specific epigenetic mark associated with silenced var genes. The persistence of this epigenetic mark in silenced var genes contributes to transcriptional memory that can provide advantages to the parasites in pathogenesis and immune evasion.
Results and Discussion
Transcription Memory at var Genes.
In a previous study, a transgenic parasite line was created in which a gene encoding blasticidin-S-deaminase (BSD) was placed under the control of the regulatory elements of a chromosomal var gene. Analysis of this parasite line confirmed that the recombinant gene, called varBSD, demonstrated activation and silencing typical of var genes (13). In this line, when varBSD was expressed, the parasites were resistant to blasticidin exposure, and all other var genes within the genome became silenced. Thus, the growth of parasites under blasticidin pressure allowed the selection of a homogeneous population exclusively expressing varBSD.
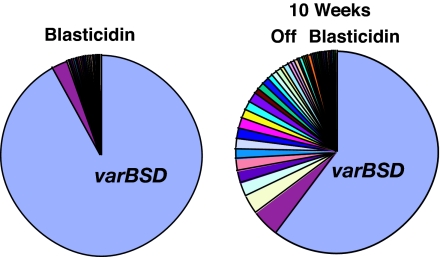
Parasites were able to switch var transcription away from varBSD when drug selection pressure was removed (13), resulting in the expression of other var copies over time. After several weeks of growth in the absence of blasticidin, the presence of parasites expressing other var genes was observed within the parasite population; however, the majority of parasites continued to express varBSD even when the selection pressure was absent for >10 weeks [Fig. 1 and supporting information (SI) Table 1]. After invasion of the red blood cell, a parasite requires ≈48 h to complete its cycle of growth and release 8–24 new invasive forms (14). Therefore, over 10 weeks, each parasite completed 35 cycles and multiplied to a population of 280–840 parasites; nonetheless, most of the parasites had not switched to express other var genes, and varBSD remained the dominant var transcript within the population.
Fig. 1.
Gene transcription memory at var loci. The pie charts present relative levels of transcription of each var gene from the transgenic P. falciparum clone C7G12 as discussed in ref. 13. Under blasticidin pressure, only varBSD is highly expressed. After 10 weeks without the drug, subpopulations of parasites expressing other var genes have expanded, but varBSD remains the dominant gene detected. The analyzed data for var genes and expression values from ref. 13 are presented in SI Table 1.
This type of molecular memory for gene expression has been recognized in many multicellular eukaryotic organisms and is especially well documented among genes that determine the identity of the body segments in D. melanogaster (15). Triggering signals determine cell fate early in the larval stages and commit these cells to form segment-specific organs. Despite the loss of the signals during later developmental stages, the cells retain the memory of their fate and the corresponding gene expression patterns. However, a similar role for chromatin memory in single-celled eukaryotes has not been widely anticipated because of their lack of complex developmental processes.
Epigenetic Mark at var Loci.
We decided to take advantage of the varBSD system to identify possible epigenetic memory factors involved in the regulation of P. falciparum var genes. In preliminary experiments using chromatin immunoprecipitation (ChIP), we noticed that trimethylated histone H3 at lysine-9 (H3K9me3) is significantly enriched at silent var genes (data shown below). H3K9me3 was described as a mark for silenced chromatin in fission yeast and several metazoans (16). We therefore monitored the enrichment of histone modifications at varBSD both in its transcriptionally active and in its silent state.
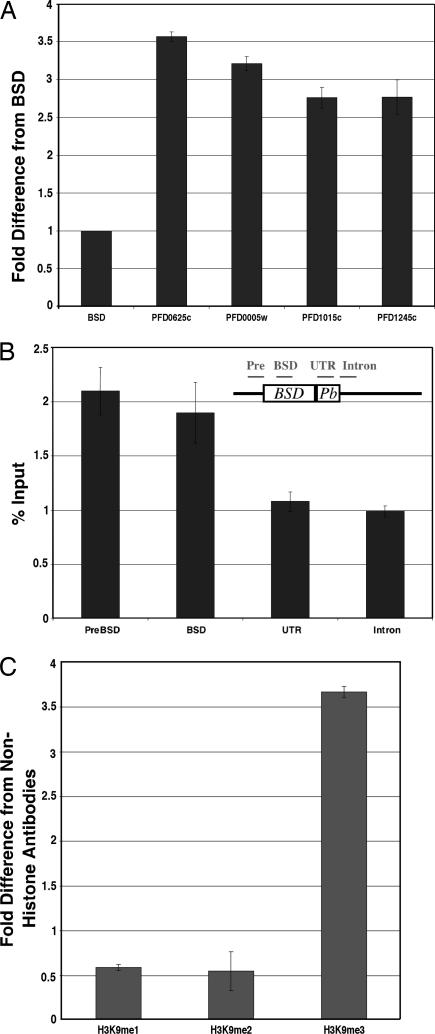
When the varBSD gene was selected to be transcriptionally active, H3K9me3 was undetectable in the varBSD coding region, and there was no detectable difference in H3K9me3 enrichment when compared with the signal obtained from control antibodies (1.19 ± 0.06-fold enrichment compared with the signal from nonhistone antibodies). We then compared the value at the active varBSD gene to other transcriptionally silent var genes on chromosome IV, which contains multiple var genes in subtelomeric regions as well as internal clusters (Fig. 2A). The enrichment of H3K9me3 at the silent var genes was significantly greater than the signal at the active varBSD gene, and this strong enrichment was observed for all types of var genes on chromosome IV, both subtelomeric and internal. The enrichment of H3K9me3 was tested by two distinct antibodies (see Methods) and gave consistent results by ChIP (SI Fig. 4).
Fig. 2.
Histone H3K9 trimethylation at the var loci. (A) Enrichment of H3K9me3 at var genes on chromosome IV. The amount of H3K9me3 was measured by ChIP and presented as the fold difference from the value at the active varBSD. The OFF var genes shown here are located both within subtelomeric regions and in an internal cluster; these particular genes were chosen for study because their amplification by real-time PCR gave specific amplicons. (B) Location of the H3K9me3 enrichment at the OFF varBSD gene. The data are presented as percent input, which is the percent ratio of precipitated DNA compared with the total amount of DNA added into the reaction. The coding region of varBSD is shown as a rectangle. The location of each region is depicted in the diagram. The BSD/Plasmodium berghei hsp86 (Pb) 3′UTR cassette was introduced between the endogenous var promoter and the var intron, replacing exon 1 (13). The ability to study sequences further upstream of the gene was compromised because of sequence homology with numerous other var genes across the upstream region. (C) Nature of H3K9 methylation at the OFF PFD0625w var gene. The data are presented as the fold difference from the signal obtained from nonhistone antibodies (PFI1780w). Both H3K9me1 and H3K9me2 antibodies have low enrichment even when compared with nonhistone antibodies.
To confirm our findings, we checked the H3K9me3 signal at the varBSD gene when it was transcriptionally silent. This was done by obtaining a line of parasites that had been grown in the absence of drug selection and then passed through limiting dilution to generate individual subclones that had each switched to the expression of a different var gene. We found that H3K9me3 becomes highly enriched at the varBSD gene when it is transcriptionally silent (shown below). We checked the location of H3K9me3 at the silent varBSD gene and found the signal upstream of the gene (Fig. 2B, preBSD) and within the coding regions (Fig. 2B, varBSD) to be similar. However, the signal significantly decreased at the 3′ UTR and intron (Fig. 2B) (for discussions on the biological significance of each part, see ref. 17). The enrichment of H3K9 methylation is limited to trimethylation, with no significant enrichment observed for mono- or dimethylation (Fig. 2C). We also could not detect any significant H3K27me3 (trimethylated lysine-27) enrichment at varBSD.
The signal from H3K4 (lysine-4) methylation also was detected to be highly enriched at var genes. We obtained signal greater than background from the mono-, di-, and trimethylation states of H3K4. Because of the relatively high signal from H3K4me2 and H3K4me3 within the same region, as well as variation among several var genes, the significance of the H3K4 pattern remains obscure. The ratio of the H3K4me3 signal between active and silent varBSD genes seems to be consistent with the amount of core histone, and, hence, the H3K4 pattern might simply reflect the enrichment of histones in general at a particular region rather than of a specific histone modification (18). Future studies will be needed to determine what, if any, role H3K4 modification plays in gene activity.
Memory Mark by H3K9 Trimethylation.
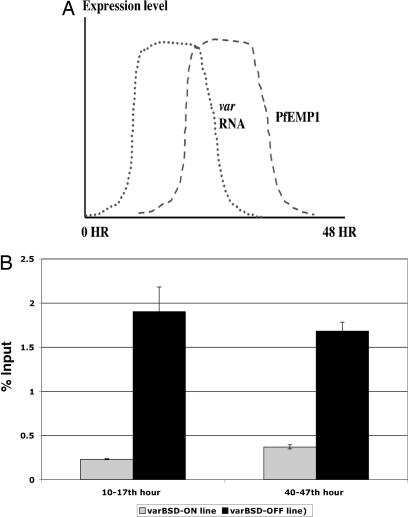
For a molecular modification to serve as an epigenetic memory factor through several rounds of cell division, it needs to be associated with the gene even after transcriptional activation or repression has occurred within each individual cell cycle. We took advantage of the fact that an active var gene is highly transcribed only during the early part of the 48-h parasite asexual cell cycle (10–17 h) (Fig. 3A). All var genes are uniformly not transcribed during the late stages (Fig. 3A), regardless of their transcriptional state in the early part of the cycle. At 40–47 h, we found that the H3K9me3 signal at the transcriptionally active varBSD gene is still negligible (Fig. 3B). Interestingly, the signal at the silent varBSD gene for H3K9me3 is consistently high and similar to what was observed at 10–17 h (Fig. 3B). Thus, the H3K9me3 pattern is consistent with what would be required for a chromatin memory mark.
Fig. 3.
Temporal pattern of the transcription memory mark H3K9me3 at a var gene. (A) Diagram presenting the transcription kinetics for var RNA (dotted line) and PfEMP1 (dashed line) over the course of the P. falciparum cell cycle (modified from ref. 25). The transcription peak for var genes is ≈10–17 h, after which the signal drops significantly. (B) H3K9me3 signal at different states of transcriptional activity. The H3K9me3 signal is shown as the percent input from P. falciparum lines with ON and OFF varBSD transcription. Synchronized parasites were analyzed at 10–17 h (expressing a single var gene) and at 40–47 h (no var expression).
Even though epigenetic memory is well established in metazoans, its possible role in single-celled organisms has not been widely considered. Although the majority of genes in P. falciparum follow a strictly programmed cascade of activation and repression over the course of each cell cycle, the var gene family represents a unique example of genes that are reversibly activated or silenced, each with its transcriptional state maintained for numerous subsequent cell generations. The ability to stabilize var transcription patterns for many cell generations is demonstrated in the relatively slow switching rate that parasites display. This slow rate of switching may aid the parasite in maintaining a persistent infection. Random, rapid switching of var gene expression would result in premature expenditure of the parasite's antigenic repertoire, whereas a complete lack of switching would result in clearance of the infection by the host's immune response. The evolution of a mechanism maintaining cellular memory allows a sort of arms race between the antigenic repertoire of the parasite and the host immune response, which results in a finely balanced equilibrium reflected in the waves of parasitemia displayed in P. falciparum malaria (19). Another implication of gene-transcription memory is that it would allow the progeny of sequestered parasites to maintain the binding affinity of the parent.
The molecular details underlying cellular memory in malaria parasites remain largely a mystery. For instance, P. falciparum lacks a recognized ortholog of heterochromatin protein HP1, which recognizes H3K9me3 and usually is conserved (16). It will be of great interest to learn which proteins or mechanisms have evolved to replace the function of HP1, as well as the other molecular players that control antigenic variation (20, 21).
Materials and Methods
Chromatin Preparation.
The ChIP protocol was modified from refs. 22 and 23. A 5% culture of the P. falciparum clone B12E3 (13) was used for the study (100- to 200-ml culture for 10–15 reactions). Reagents were from Sigma (St. Louis, MO) unless stated otherwise. Each culture was treated with 0.1% saponin until cell lysis was observed and then cross-linked with 0.5% formaldehyde at room temperature for 10 min with occasional swirling (prolonged incubation will limit the sonication efficiency). The cross-linking reaction was stopped by the addition of 125 mM glycine. The culture was centrifuged, and the pellet was washed twice with ice-cold Tris-buffered saline. The extent of cross-linked DNA-protein was checked by ethidium bromide staining. The pellet was mixed in 2 ml of IP buffer [50 mM Hepes pH 7.5/140 mM NaCl/1 mM EDTA/1% Triton X-100/0.1% Na-deoxycholate/5 mM dithiotreitol (Invitrogen, Carlsbad, CA)] with protease inhibitor (Roche, Indianapolis, IN) and the protease inhibitor PMSF. The solution then was stroked by a homogenizer 200 times on ice. Sonication was carried out at 4°C and on ice with a Digital Sonifer Model 450 (Branson, Danbury, CT) at 50%, six times for 20 s each, with an interval between each sonication of 2 min except for the third, which was 5 min to avoid overheating. The average size of sonicated DNA was 300–400 bp. The long exposure to formaldehyde resulted in a smear ranging from 500 to 1,000 bp.
ChIP.
The immunoprecipitation was carried out at 4°C for ≈18 h with 1:20 dilution of antibodies against H3 (ab1791; Abcam, Cambridge, MA), H3K4me1 (ab8895; Abcam), H3K4me2 (07-030; Upstate Biotechnology, Charlottesville, VA), H3K4me3 (07-473; Upstate Biotechnology), H3K9me1 (ab9045; Abcam), H3K9me2 (ab7312; Abcam), H3K9me3 [ab8898 (Abcam) and 07-442 (Upstate Biotechnology)], and H3K27me3 (05-581; Upstate Biotechnology). H3K9me3 antibodies from Abcam and Upstate Biotechnology were raised against two kinds of antigens (long-methylated histone H3 peptides and branched short peptide, respectively). The samples first were precleared with salmon sperm DNA-protein A or protein G agarose (Upstate Biotechnology). After the incubation with antibodies, the beads were added as explained in the preclear step and incubated for 3 h at 4°C. The beads were washed on a rotator at room temperature for 5 min each, twice in IP buffer, once in IP buffer/500 mM NaCl, once in LiCl buffer (10 mM Tris·HCl pH 8.0/0.25 M LiCl/0.5% Nonidet P-40/0.5% Na-deoxycholate/1 mM EDTA), and once in TE (10 mM Tris/1 mM EDTA, pH 7.4). The DNA–protein complexes were eluted by incubating the bead with 50 mM Tris·HCl, pH 8.0/10 mM EDTA/1% SDS/5 mM DTT at 65°C for 15 min, then the eluant was collected. The beads were washed again briefly with TE/0.67% SDS/5 mM DTT, and the eluant from both steps was pooled. To reverse the cross-linking reaction, the eluted material was incubated at 65°C for ≈18 h. Samples then were treated with Proteinase K (Invitrogen) before phenol/chloroform extraction.
Quantitative Real-Time PCR.
Real-time PCRs were carried out with QuantiTect SYBR Green PCR (Qiagen, Valencia, CA) in MX3000 (Stratagene, La Jolla, CA). The primer sets are from ref. 24 except PreBSD (5′-CCCATAACATACGCAATACGC-3′ and 5′-GGGTGGATTCTTCTTGAGACA-3′), BSD (5′-AGGTGCTTCTCGATCTGCAT-3′ and 5′-CATAACCAGAGGGCAGCAAT-3′), UTR (5′-CAATGATTCATAAATAGTTGGACTTGA-3′ and 5′-TCGAAATTGAAGGAAAAACCA-3′), and Intron (5′-TCCACAACATCATCGGACTT-3′ and 5′-CCAAACACACCCACACATACA-3′). Each primer pair has its own genomic DNA standard to allow quantitation of its amount and specificity. The PCR conditions were 95°C for 10 min for 1 cycle and 95°C for 1 min, 55°C for 1 min, and 72°C for 30 s for 40 cycles with the melting condition (95°C for 1 min, 55°C for 30 s, and 95°C for 30 s).
Supplementary Material
Acknowledgments
We thank T. Wellems and the members of the K.W.D. and D.L.H. laboratories for invaluable discussion and support; D. Moazed, M. Motamedi, and members of the Maniatis laboratory for their advice on ChIP; and members of the Allis laboratory for their advice on histone modifications. This work and T.C. were supported by an Ellison Medical Foundation Training Grant and a Merck Genomic Grant. K.W.D., R.D., M.F., and F.L. were supported by National Institutes of Health Grant AI52390. The Department of Microbiology and Immunology at Weill Medical College of Cornell University acknowledges the support of The William Randolph Hearst Foundation. D.L.H. is an Ellison Medical Foundation Senior Scholar in Global Infectious Disease. K.W.D. is a Stavros S. Niarchos Scholar and an Ellison Medical Foundation New Investigator.
Abbreviations
- BSD
blasticidin S deaminase
- H3K9me3
trimethylated histone H3 at lysine-9
- PfEMP1
P. falciparum erythrocyte membrane protein 1.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609084103/DC1.
References
- 1.Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, Gysin J, Lanzer M. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kyes SA, Christodoulou Z, Raza A, Horrocks P, Pinches R, Rowe JA, Newbold CI. Mol Microbiol. 2003;48:1339–1348. doi: 10.1046/j.1365-2958.2003.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Q, Fernandez V, Sundstrom A, Schlichtherle M, Datta S, Hagblom P, Wahlgren M. Nature. 1998;394:392–395. doi: 10.1038/28660. [DOI] [PubMed] [Google Scholar]
- 4.Deitsch KW, Calderwood MS, Wellems TE. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- 5.Su XZ, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, Ravetch JA, Wellems TE. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 6.Voss TS, Healer J, Marty AJ, Duffy MF, Thompson JK, Beeson JG, Reeder JC, Crabb BS, Cowman AF. Nature. 2006;439:926–927. doi: 10.1038/nature04407. [DOI] [PubMed] [Google Scholar]
- 7.Gannoun-Zaki L, Jost A, Mu J, Deitsch KW, Wellems TE. Eukaryot Cell. 2005;4:490–492. doi: 10.1128/EC.4.2.490-492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank M, Dzikowski R, Constantini D, Amulic B, Burdougo E, Deitsch K. J Biol Chem. 2006;281:9942–9952. doi: 10.1074/jbc.M513067200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. J Biol Chem. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- 10.Roberts DJ, Craig AG, Berendt AR, Pinches R, Nash G, Marsh K, Newbold CI. Nature. 1992;357:689–692. doi: 10.1038/357689a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kyes S, Horrocks P, Newbold C. Annu Rev Microbiol. 2001;55:673–707. doi: 10.1146/annurev.micro.55.1.673. [DOI] [PubMed] [Google Scholar]
- 12.Horrocks P, Pinches R, Christodoulou Z, Kyes SA, Newbold CI. Proc Natl Acad Sci USA. 2004;101:11129–11134. doi: 10.1073/pnas.0402347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dzikowski R, Frank M, Deitsch K. PLoS Pathog. 2006;2:e22. doi: 10.1371/journal.ppat.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herbert M, Gilles DAW. Bruce-Chwatt's Essential Malariology. Boston: Butterworth–Heinemann; 1993. [Google Scholar]
- 15.Ringrose L, Paro R. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 16.Grewal SI, Moazed D. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 17.Frank M, Deitsch K. Int J Parasitol. 2006;36:975–985. doi: 10.1016/j.ijpara.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. PLoS Biol. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller LH, Good MF, Milon G. Science. 1994;264:1878–1883. doi: 10.1126/science.8009217. [DOI] [PubMed] [Google Scholar]
- 20.Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- 21.Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- 22.Hecht A, Grunstein M. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- 23.Motamedi MR, Verdel A, Colmenares SU, Gerber SA, Gygi SP, Moazed D. Cell. 2004;119:789–802. doi: 10.1016/j.cell.2004.11.034. [DOI] [PubMed] [Google Scholar]
- 24.Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, Hviid L, Theander TG. Mol Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- 25.Sherman I. Molecular Approches to Malaria. Washington, DC: Am Soc Microbiol; 2005. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.