Abstract
Aging is a universal but poorly understood biological process. Free radicals accumulate with age and have been proposed to be a major cause of aging. We measured genome-wide changes in transcript levels as a function of age in Drosophila melanogaster and compared these changes with those caused by paraquat, a free-radical generator. A number of genes exhibited changes in transcript levels with both age and paraquat treatment. We also found genes whose transcript levels changed with age but not with paraquat treatment. This study suggests that free radicals play an important role in regulating transcript levels in aging but that they are not the only factors. This genome-wide survey also identifies candidates for molecular markers of aging.
Aging is a biological process universal to eukaryotic organisms, and its underlying mechanisms are under intensive study. Genetic analyses of yeast, nematode, fly, and mouse have uncovered a number of genes, whether mutated or misexpressed, that would increase the lifespans of these organisms (1). These genes include superoxide dismutase, a free-radical scavenger; methuselah, a potential G protein-coupled receptor, in Drosophila melanogaster; and p66shc, an oxidative stress-response gene, in mice (2–5). Lifespan-related genes in Caenorhabditis elegans include clk-1, a gene involved in ubiquinone biosynthesis, and a group of genes involved in an insulin receptor-like signaling pathway: daf-2, age-1, and daf-16 (6, 7). Many mutations that increase lifespan also confer resistance to oxidative stress (1). This finding supports the free-radical hypothesis of aging, which suggests that reactive oxygen species that accumulate with increasing age cause oxidative damage to macromolecules (including nucleic acids, proteins, and lipids) and are causally linked to aging and death (8, 9). Free radicals have been found to regulate the expression of a number of genes that include antioxidant defense genes involved in repairing oxidative damage, as well as genes involved in inducing apoptosis (10, 11). The aging process is also accompanied by changes in the expression patterns of a number of genes (12–14). How the regulation of gene expression in aging correlates with that in response to oxidative stress, however, is understood poorly.
Materials and Methods
Construction of Microarray.
We constructed a microarray containing 7,829 expressed sequence tags (ESTs). This set includes 221 ESTs provided by our lab and 7,608 ESTs generously supplied by K. White (Stanford University, Stanford, CA) and K. Burtis (University of California, Davis). The ESTs were amplified (as described by White et al., ref. 15), and the DNA was mechanically spotted onto polylysine-coated slides (as described by DeRisi et al., ref. 16).
Calculation of Survival Rate and Preparation of Fly Tissues.
Male flies (w1118) raised in standard cornmeal agar medium, were collected within 24 h after eclosion (17). Approximately 200 flies were maintained in constant darkness in each food bottle at 26–27°C and 60–70% humidity and were transferred to fresh bottles every 3–4 days. To calculate the lifespan, the number of dead flies was counted at the time of transfer. For microarray experiments, different batches of flies were used to extract RNA from the thorax and abdomen. For the aging experiments, flies were collected at 3, 10, 15, 25, 30, 40, and 50 days of age. We had to pool flies that eclosed over a period of 2–4 days, for each of these time points, because only a small percent of flies survived longer than 30 days. The actual ages of flies at 30-, 40-, and 50-day time points were therefore 30–32, 40–42, and 47–50 days old, respectively. For each of the other time points, males that eclosed on the same day were used. For the paraquat experiments, 3-day-old males were fed either 5% (weight/vol) sucrose or 5% (weight/vol) sucrose with 15 mM paraquat for 3, 12, 25, or 34 h after being starved for 6 h.
Microarray Analysis.
Total RNA was extracted by using the reagent Trizol (GIBCO/BRL). Poly(A) RNA was isolated by using oligo(dT)linked Oligotex resin (Qiagen, Chatsworth, CA). Equal amounts (2–4 μg) of poly(A) RNA from both reference and experimental samples were used as templates to synthesize fluorescently labeled cDNA probes for hybridization to the microarrays. The RNA was reverse transcribed in the presence of 5-(3-aminoallyl)-2′-deoxyuridine 5′ triphosphate (18). The reference sample was coupled to fluorescent dye (Cy)3, and the experimental sample was coupled to Cy5. Hybridization was performed in a humidified chamber for 8–12 h at 63°C. Cy5 and Cy3 fluorescence intensities were measured by using GenePix (Axon Instruments, Foster City, CA). A detailed protocol including reagents can be found at http://derisilab.ucsf.edu/index.html. For aging experiments, cDNA from 3-day-old flies labeled with Cy3 was cohybridized with cDNA from 3-, 10-, 15-, 25-, 40-, or 50-day-old flies labeled with Cy5. For paraquat experiments, cDNA from sucrose-fed flies was labeled with Cy3 and cohybridized with cDNA from paraquat-fed flies labeled with Cy5. The differences in transcript levels for an EST were indicated by the ratios of Cy5/Cy3.
Northern Analysis.
Northern analysis was performed to measure transcript levels of three ESTs and the control gene rp49 at four time points: 3 days, 25 days, 40 days, and 50 days. Poly(A) mRNA (1–2 μg) was transferred to Hybond-N nylon membrane and hybridized with 32P-labeled probes (Amersham Pharmacia). mRNA levels were measured by using the Fuji Film Image Reader (Fuji Photo Film). The changes in transcript levels were determined by the ratios of signals for older flies and signals for 3-day-old flies after normalization with rp49 signals.
Data Analysis.
The microarray data were submitted and processed in the AMAD database designed by J. DeRisi (University of California, San Francisco). In each array, the Cy5/Cy3 ratios that indicate changes in transcript levels were normalized with a normalization factor such that the ratio of total Cy5/total Cy3 was equal to 1. For analysis, the data were filtered first to minimize the poorly hybridized spots. An EST spot in a microarray was excluded if it showed pixel-by-pixel regression correlation for the Cy3 and Cy5 measurements of less than 0.75 or if the value of either the Cy3 or the Cy5 fluorescence intensity was less than 150. A total of 7,387 ESTs that had at least one ratio value with fairly good quality of hybridization signals was selected. By using the cluster program, age-regulated and paraquat-regulated ESTs were selected separately. The age-regulated ESTs were those with a greater than 1.8-fold change in at least three of the six 30-, 40-, and 50-day age arrays and with no more than five missing values in the 13 data points for the aging experiments. The paraquat-regulated ESTs were those with a greater than 1.8-fold change in at least three of the six 12-to-34-h arrays. The expression patterns of these ESTs were analyzed with the hierarchical clustering method. The tree view program was used to view the cluster images. All of these programs are available at http://www.microarrays.org. All of the ESTs discussed in this article were confirmed by sequencing.
Results and Discussion
Measuring Changes in Transcript Levels with Age and in Response to Oxidative Stress.
To study aging and its relationship to the oxidative stress response in D. melanogaster, we used the microarray technique to monitor expression patterns of approximately 8,000 ESTs (16). These ESTs represent more than 4,500 unique genes and cover 30–40% of the estimated total number of expressed genes in the Drosophila genome (15). Age-related changes in transcript levels were measured by comparing 3-day-old w1118 males to other males of six different ages: 10, 15, 25, 30, 40, and 50 days old. The median lifespan of w1118 males is approximately 35 days under our assay conditions (Fig. 1). To study the oxidative stress response, we compared 3-day-old males fed paraquat in a sucrose solution to those fed only sucrose. Paraquat is a free-radical generator and has been widely used for testing oxidative stress responses (19). Response to paraquat treatment was examined at 3, 12, 25, and 34 h. The median mortality rate of paraquat-treated w1118 males at these four time points was <0.1%, <0.5%, ≈54%, and ≈64%, respectively. Two microarrays were used for each time point by using samples isolated independently.
Figure 1.
Survival curve of w1118. Two independent experiments, indicated by open or closed squares, were performed by using 172 and 156 males, respectively. The curve was generated by fitting data from both experiments. The median lifespan in these experiments was approximately 35 days.
Validation of Microarray Data.
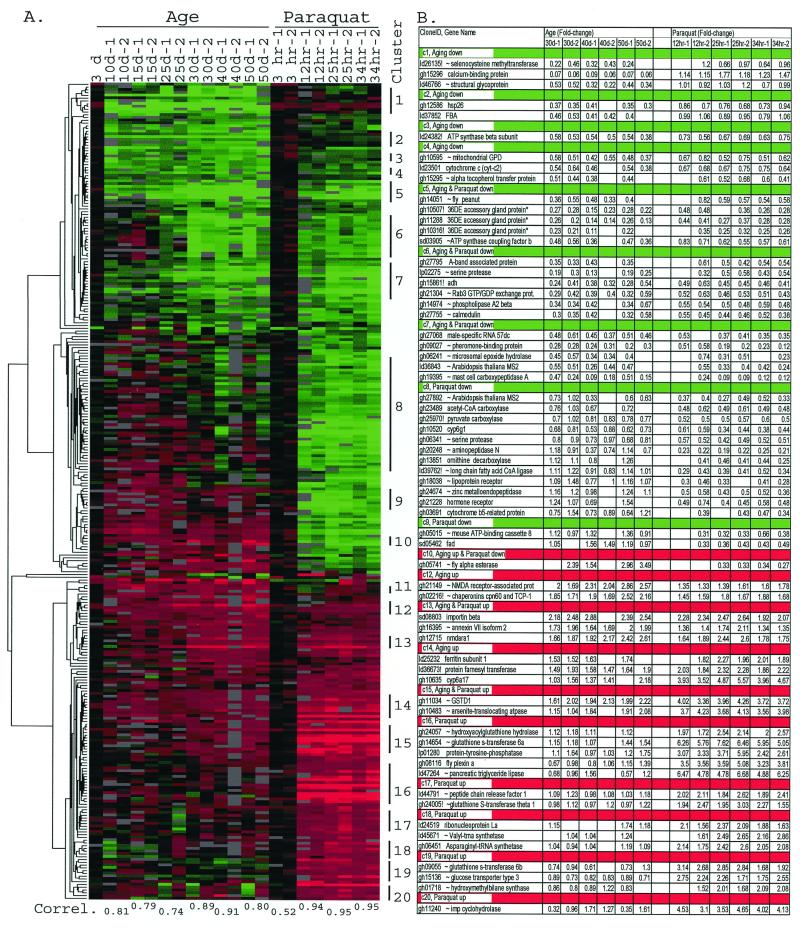
To verify the microarray data, Northern analysis was performed to measure transcript levels of three ESTs at 3-, 25-, 40-, and 50-day time points (Table 1). The ratios measured by Northern analysis were similar to those measured by the microarray experiments. The similarity of duplicate microarray experiments at each time point was assessed by calculating the Pearson's correlation coefficient for 329 ESTs whose transcript levels were significantly altered with age and/or in response to oxidative stress as discussed below (20, 21). All duplicate arrays except for those at 15-day, 25-day, and 3-hour time points had a correlation coefficient of 0.8 or greater (Fig. 4A), indicating a fairly good correlation between two sets of data (21). The reliability of our experiments was also evident in the cluster image shown in Fig. 4. The greater the similarity between the expression patterns of two ESTs, the closer these ESTs are located in the cluster image (20). Different ESTs representing the same gene were almost always placed side by side or close to each other in the same cluster (see details at http://www.ucsf.edu/jan). For example, three ESTs representing the same accessory gland protein (Acp) were placed side by side in cluster c5 (see Fig. 4B). These control analyses verify the reliability of the microarray data.
Table 1.
Comparison of ratios measured from Northern and microarray analyses
| EST | Age, days | Northern | Array I | Array II |
|---|---|---|---|---|
| GH21304 | 25 | 0.82 | 0.71 | 1.01 |
| 40 | 0.46 | 0.39 | 0.40 | |
| 50 | 0.30 | 0.32 | 0.59 | |
| GH27755 | 25 | 0.78 | 0.68 | NA |
| 40 | 0.45 | 0.42 | NA | |
| 50 | 0.32 | 0.32 | 0.58 | |
| LD41905 | 25 | 1.65 | 1.09 | 1.07 |
| 40 | 5.90 | 3.74 | NA | |
| 50 | 3.58 | 4.82 | 2.99 |
In Northern analysis, the ratios were calculated by dividing mRNA levels at 25-, 40-, and 50-day time points by those at 3-day time points after normalization with mRNA levels of the control gene rp49. Ratios in microarray analysis are provided from each of the duplicate experiments for comparison.
Figure 4.
CLUSTER image. (A) CLUSTER image of 295 age-regulated and/or paraquat-regulated ESTs. The cluster is subdivided further into 20 clusters, indicated by the number. The symbols and color coding used in this figure are the same as those used in Fig. 3. Correl. represents Pearson's correlation coefficient. (B) Gene list. The ESTs with clone identification codes and sequence information are shown with Cy5/Cy3 ratios at six time points: 30, 40, and 50 days for the aging experiments and 12, 25, and 34 h for the paraquat experiments. Two independent values for each time point are listed side by side. The order of the ESTs in the table is the same as their relative positions in the cluster image shown in A. Cluster identification is indicated by “c” followed by the number, and coded with two colors—green indicates down-regulation, and red indicates up-regulation.
Global Changes in Transcript Levels in Aging.
We defined age-regulated ESTs as those showing a greater than 1.8-fold change in transcript levels in at least three of the six 30-, 40-, and 50-day aging arrays. A total of 132 ESTs, representing 127 genes, met this criterion and was categorized into two major clusters. Of these 132 ESTs, 43 ESTs in one cluster were up-regulated with increasing age, whereas 89 were down-regulated (Fig. 2A; details at http://www.ucsf.edu/jan). Potential functions of previously uncharacterized genes were inferred based on predictions in the D. melanogaster genome database, Gadfly. Most of the 132 ESTs encode proteins with unknown functions or with functions whose roles in aging are unclear. However, some of these genes encode proteins with known or predicted functions that could account for age-related physiological changes. Below, we describe age-dependent regulation of genes involved in reproduction, metabolism, and detoxification.
Figure 2.
CLUSTER image of 132 age-regulated ESTs. Each column represents one of seven time points: 3, 10, 15, 25, 30, 40, and 50 days. Except for 3 days (the control), each time point has two sets of data, which are shown side by side in columns. Each row represents the expression pattern of a single EST. Red indicates up-regulation in the experimental sample, and green represents down-regulation. Gray indicates missing data. Identities of some age-regulated ESTs are provided on the right side of the CLUSTER image, placed in the same order as their relative position in the cluster image. Each EST is labeled with its clone identification code followed by the gene name or the name of its homolog. ESTs with sequence error in this study are indicated by the symbol !. ESTs encoding an uncharacterized protein with homology to a known protein are marked by ˜. The ESTs marked by three asterisks (***) represent those coregulated with age and oxidative stress.
Age-Regulated Genes Involved in Reproductive Capacity.
Decline in reproductive capacity is an age-related phenotype, and the reproductive system seems to play an important role in longevity (22). For example, signals from germ cells can affect lifespan in C. elegans (23). In our study, we observed decreased RNA levels for several genes involved in reproduction (Fig. 3). These include two genes that encode members of the Acp family. The Acp from male flies stimulates female egg-laying and facilitates storage of sperm in the female genital tract (24). In addition, two ESTs showing age-regulated decrease of transcript levels represent different genes with homology to Arabidopsis MALE STERILITY 2 (MS2; ref. 25), a gene involved in gametogenesis. Furthermore, an EST with homology to peanut, a member of the septin family (26), is down-regulated in older flies. This down-regulation may reflect a decrease in spermatogenesis.
Figure 3.
Transcript profiles of four classes of genes whose transcript levels changed with age or oxidative stress. These genes were selected from the 329 ESTs whose transcript levels are significantly altered with aging, oxidative stress, or both. Each column represents 1 of 11 time points: 3, 10, 15, 25, 30, 40, and 50 days for the aging experiments and 3, 12, 25, and 34 h for paraquat experiments. Except for 3 days (the control), each time point has two sets of data, which are shown side by side in columns. The ESTs with clone identification codes and gene names are listed in the same order as their relative positions in the CLUSTER image. The symbols and color coding used in this figure are the same as those used in Fig. 2. The ESTs with the same names marked by a single asterisk (*) have the same sequences.
Age-Regulated Genes Involved in Metabolism.
Metabolism is a physiological process that changes with age. We found age-regulated reduction in transcript levels of genes involved in various steps of several metabolic pathways (Fig. 3). These include genes that encode fructose-1,6-bisphosphate aldolase (FBA), a homolog of aldose-1-epimerase, and a homolog of mitochondrial glycerol-3-phosphate dehydrogenase (GPD), all of which function in the metabolism of glucose and other sugars in glycolysis (27). Interestingly, transcript levels of FBA and GPD have been shown to be up-regulated in calorically restricted mice (12), which have up to a 2-fold longer lifespan than controls (28). After glycolysis, a central step in metabolism is the tricarboxylic acid cycle in mitochondria (27). We identified a gene encoding a homolog of aconitase, whose RNA levels decreased with increasing age (Fig. 3). Age-regulated decrease of aconitase activity because of protein oxidation has been demonstrated in houseflies (29). Our study indicates that down-regulation of transcript levels may reduce aconitase activities further in older flies. Another important step in metabolism in mitochondria is oxidative phosphorylation, which generates most of the energy in aerobically respirating cells. We observed that transcript levels of cytochrome c, a homolog of ubiquinol-cytochrome c reductase ubiquinone binding protein, ATP synthase β-subunit, and ADP/ATP translocase all decreased with age (Fig. 4). Enzymes encoded by these genes are components of the electron transfer chain in mitochondria (27).
Age-Regulated Genes Functioning as Detoxification Agents and Chaperones.
Another feature of aging is a gradual decrease in resistance to various stresses such as heat shock and oxidative stress. In our study, we found age-related down-regulation of genes functioning as chaperones or detoxification agents (Fig. 4). These include a gene encoding the small heat-shock protein Hsp26, alcohol dehydrogenase (Adh), α-tocopherol transfer-related protein, and a homolog of microsomal epoxide hydrolase (mEH). Small heat-shock proteins serve as molecular chaperones to protect proteins from various stressors (30). mEH functions in detoxification by metabolizing reactive epoxide intermediates (31). Adh is involved in alcohol detoxification (32). α-Tocopherol, one form of vitamin E, is a major antioxidant in higher eukaryotes (9).
Not all age-regulated genes encoding proteins with functions as chaperones or detoxification agents are down-regulated. Some of these genes are actually up-regulated, including a gene encoding glutathione S-transferase D1, a gene encoding a homolog of arsenite translocating ATPase, and a gene encoding a homolog of chaperone 60. Arsenite translocating ATPase functions in eliminating toxic arsenite (33). Chaperone 60 is a major chaperone involved in proper protein folding (34). Glutathione is a reductant important for detoxification, maintaining a reducing environment, and reducing protein disulfide bonds (35).
Comparison of Genes Regulated with Age and in Response to Oxidative Stress.
To evaluate the relationship between aging and the oxidative stress response, we compared age-dependent changes in transcript levels to those caused by paraquat-induced oxidative stress. We defined paraquat-regulated ESTs as those displaying a greater than 1.8-fold change in at least three of the six 12-, 25-, and 34-h paraquat arrays. This criterion was passed by the 246 ESTs that represented 236 genes. When combined with 132 age-regulated ESTs, there were 329 ESTs whose transcript levels were significantly altered with age, oxidative stress, or both. The expression patterns of 295 of these 329 ESTs were analyzed and categorized into 20 clusters (Fig. 4; ref. 20). The other 34 were excluded from the analysis, because they were missing 6 or more of the 21 data points.
Genes That Are Primarily Regulated with Age.
Clusters 1, 2, 3, 4, 11, and 12 consist of genes primarily regulated with age. These include a homolog of selenocysteine methyltransferase in cluster 1, FBA in cluster 2, and ATP synthase β-subunit in cluster 3 (Fig. 4). In addition, the homolog of aconitase, not included in the 20 clusters, is down-regulated primarily with increasing age. Expression patterns of these genes suggest that free radicals have little effect on regulating the transcript levels of these metabolic genes, which may not play a significant role in free-radical generation.
Genes That Are Primarily Regulated in Response to Oxidative Stress.
Clusters 8, 9, 14, and 16 to 20 include genes primarily regulated by paraquat treatment. Genes in clusters 8 and 9 show a decrease in transcript levels in response to paraquat treatment. A group of genes involved in fatty acid metabolism are in these clusters, including fatty acid desaturase, fatty acid CoA ligase, and acetyl-CoA carboxylase. Because fatty acid metabolism is a major source of free radicals in the cell, reduction of transcript levels of these genes may reflect a defense mechanism in response to oxidative stress by reduction of free-radical generation. None of the age-regulated genes seem to be directly involved in fatty acid metabolism. Thus, it seems that this defense mechanism is not used by aging flies to confront the accumulated reactive oxygen species.
The ESTs in clusters 16 to 20 show an increase in transcript levels with paraquat treatment. A number of genes in these clusters encode proteins functioning in antioxidant defense, such as glutathione S-transferases and a homolog of triglyceride lipase. Some of them seem to be regulated coordinately. For example, a homolog of glutathione S-transferase and a homolog of hydrooxyacylglutathione hydrolase, potentially involved in detoxification, were found side by side in cluster 16, indicating a high similarity in the transcript patterns of these two genes. This similarity may be caused by a coordinated regulation of these two genes under oxidative stress, because the hydrooxyacylglutathione hydrolase uses glutathione as a cofactor to detoxify 2-oxoaldhydes in the cell (36).
Genes That Were Regulated with Both Age and Oxidative Stress.
There are 47 ESTs, representing 42 genes, that are coregulated with age and oxidative stress, including clusters 5, 6, 7, and 15 (Figs. 2 and 4). The total number of coregulated genes accounted for 33% (42:127) of all of the age-regulated genes and 18% (42:236) of all of the paraquat-regulated genes, suggesting that free radicals play a significant role in regulating transcription in aging. Most of the coregulated ESTs encode proteins with unknown functions or with functions whose roles in aging and/or oxidative stress responses are not clear. Some of the coregulated ESTs, however, fell into four functional classes. The ESTs in clusters 5, 6, and 7 displayed a decrease in transcript levels, both with increasing age and in response to oxidative stress. Three functional groups of genes are prominent in these three clusters. One functional group includes two genes encoding serine proteases in cluster 6 and one gene encoding a homolog of carboxypeptidase A in cluster 7, suggesting those free radicals that are accumulated during aging reduce protein turnover (37). The second functional group includes three genes, encoding an Acp and a homolog of MS2 in cluster 7 and an Acp in cluster 5, which are involved potentially in reproductive capacity. Additional proteases, such as metalloendopeptidase and serine protease, and additional reproduction-related genes, such as another homolog of MS2 and ornithine carboxylase (the later is involved in spermidine synthesis) in cluster 8, were down-regulated in response to oxidative stress but not aging. It thus seems that reproduction and protein degradation are important sources for free-radical production. A defense mechanism used in aging and oxidative stress response seems to reduce transcript levels of proteases and genes involved in reproduction. The third functional group included the ubiquinone binding protein in cluster 7, which encoded enzymes involved in energy production in mitochondria. Given that most of the free radicals in the cell are generated in the mitochondria (9), one way to reduce the production of free radicals might be the down-regulation of mitochondrial enzymes whose activities generate free radicals. Cluster 15 consists of genes up-regulated with both age and paraquat treatment, including genes that are involved in detoxification and homologs of arsenite-translocating ATPase and glutathione S-transferase D1. It thus seems that part of the aging process involves the up-regulation of genes encoding chaperones or detoxification agents in response to oxidative stress and the down-regulation of genes responsible for both energy production and free-radical generation.
Global View of Regulation at Transcript Levels in Aging.
We found that the aging process in D. melanogaster is accompanied by reduction of transcript levels for genes involved in reproduction, metabolism, and protein turnover. Some genes that encode detoxification agents and chaperones are up-regulated, whereas others down-regulate with aging. One-third of the age-regulated genes show significant changes in response to oxidative stress. These changes suggest a response to free radicals that accumulated with increasing age accounts for a significant part of age-regulated changes in transcript levels. The response to oxidative stressors that accumulated during aging includes up-regulating genes to counteract oxidative damage and down-regulating genes whose activities increase endogenous oxidant levels. However, more than 60% of the age-regulated genes show no or little response to oxidative stress; therefore, free radicals are not the only causal factor in aging. Additionally, more than 80% of paraquat-regulated genes show no or small change in transcript levels in aging, suggesting free radicals that accumulated during aging do not elicit a full response to oxidative stress.
Lee et al. (12, 13) have used a similar genome-wide approach to examine changes associated with aging in the mouse. They examined expression patterns of 6,347 genes, representing 5–10% of the mouse genome, in the aging muscle and brain. Without ready access to the sequences of all of the ESTs surveyed in the mouse studies and in our study, a direct comparison among all ESTs is difficult. We compared, however, age-regulated genes identified in mouse in the previous studies (about 110 genes in each of the mouse studies) and in fly in this study (127 genes). We did not find any genes that are regulated in the same direction in aging of these two species. One gene, FBA, was found up-regulated in aging mouse neocortex but down-regulated in aging flies, suggesting differences in aging-related regulation at the level of individual genes. Common themes of aging-related regulation were observed, however, at the level of cellular functions. Down-regulation of genes in energy metabolism and protein turnover was observed both in fly and mouse, although different sets of genes were involved. Of genes encoding proteins as detoxification agents and chaperones, some displayed an increase and some a decrease in transcript levels with aging in fly and mouse. Taken altogether, these results suggest that these two species alter their cellular processes in a similar way during aging but do so through the regulation of distinct genes.
Genes that have significant changes in their RNA levels with increasing age can serve as molecular markers for aging. Some of these genes may encode molecules that are causal factors in aging. Alternatively, regulation of certain genes may reflect changes as a consequence of aging. The characterization of changes in gene expression per se does not allow one to distinguish between these two types of genes. However, this study singles out candidates for further analysis. Characterizations of these genes and the biological pathways involved could facilitate our understanding of aging and longevity at the genetic level and help us gain insight into the mechanisms of aging.
Acknowledgments
We thank C. Kenyon, J. DeRisi, Y.-M. Chan, S. Abdelilah, and S. Greenwood for comments on the manuscript and members of the Jan lab for discussions. We are grateful to Y. Hong and L. Ackerman for help on computer work, T. Kornberg for use of the arrayer, H. Bennett and P. Cifuentes for help with microarray experiments, and H. R. Qian for help on statistical analysis. S.Z. thanks H. Y. Chang for support and encouragement. S.Z. is supported by the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation Fellowship Grant DRG-1453. L.Y.J. and Y.N.J. are investigators at Howard Hughes Medical Institute.
Abbreviations
- EST
expressed sequence tag
- Cy
fluorescent dye
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260496697.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260496697
References
- 1.Johnson F B, Sinclair D A, Guarente L. Cell. 1999;96:291–302. doi: 10.1016/s0092-8674(00)80567-x. [DOI] [PubMed] [Google Scholar]
- 2.Orr W C, Sohal R S. Science. 1994;263:1128–1130. doi: 10.1126/science.8108730. [DOI] [PubMed] [Google Scholar]
- 3.Lin Y J, Seroude L, Benzer S. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- 4.Parkes T L, Elia A J, Dickinson D, Hilliker A J, Phillips J P, Boulianne G L. Nat Genet. 1998;19:171–174. doi: 10.1038/534. [DOI] [PubMed] [Google Scholar]
- 5.Migliaccio E, Giorgio M, Mele S, Pelicci G, Reboldi P, Pandolfi P P, Lanfrancone L, Pelicci P G. Nature (London) 1999;402:309–313. doi: 10.1038/46311. [DOI] [PubMed] [Google Scholar]
- 6.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. Nature (London) 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 7.Morris J Z, Tissenbaum H A, Ruvkun G. Nature (London) 1996;382:536–539. doi: 10.1038/382536a0. [DOI] [PubMed] [Google Scholar]
- 8.Harman D. J Gerontol. 1956;2:298–300. doi: 10.1093/geronj/11.3.298. [DOI] [PubMed] [Google Scholar]
- 9.Beckman K B, Ames B N. Physiol Rev. 1998;78:547–581. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 10.Storz G, Tartaglia L A, Farr S B, Ames B N. Trends Genet. 1990;6:363–368. doi: 10.1016/0168-9525(90)90278-e. [DOI] [PubMed] [Google Scholar]
- 11.Um H D, Orenstein J M, Wahl S M. J Immunol. 1996;156:3469–3477. [PubMed] [Google Scholar]
- 12.Lee C K, Klopp R G, Weindruch R, Prolla T A. Science. 1999;285:1390–1393. doi: 10.1126/science.285.5432.1390. [DOI] [PubMed] [Google Scholar]
- 13.Lee C K, Weindruch R, Prolla T A. Nat Genet. 2000;25:294–297. doi: 10.1038/77046. [DOI] [PubMed] [Google Scholar]
- 14.Ly D H, Lockhart D J, Lerner R A, Schultz P G. Science. 2000;287:2486–2492. doi: 10.1126/science.287.5462.2486. [DOI] [PubMed] [Google Scholar]
- 15.White K P, Rifkin S A, Hurban P, Hogness D S. Science. 1999;286:2179–2184. doi: 10.1126/science.286.5447.2179. [DOI] [PubMed] [Google Scholar]
- 16.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 17.Ashburner M. Drosophila: A Laboratory Handbook. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Iribarren A M, Sproat B S, Neuner P, Sulston I, Ryder U, Lamond A I. Proc Natl Acad Sci USA. 1990;87:7747–7751. doi: 10.1073/pnas.87.19.7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arking R, Buck S, Berrios A, Dwyer S, Baker G T I. Dev Genet (Amsterdam) 1991;12:362–370. doi: 10.1002/dvg.1020120505. [DOI] [PubMed] [Google Scholar]
- 20.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harnett D L. Statistical Methods. Reading, MA: Addison–Wesley; 1982. [Google Scholar]
- 22.Finch C E. Longevity, Senescence, and the Genome. Chicago: Univ. of Chicago Press; 1990. [Google Scholar]
- 23.Hsin H, Kenyon C. Nature (London) 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- 24.Wolfner M F. Insect Biochem Mol Biol. 1997;27:179–192. doi: 10.1016/s0965-1748(96)00084-7. [DOI] [PubMed] [Google Scholar]
- 25.Aarts M G, Hodge R, Kalantidis K, Florack D, Wilson Z A, Mulligan B J, Stiekema W J, Scott R, Pereira A. Plant J. 1997;12:615–623. doi: 10.1046/j.1365-313x.1997.00615.x. [DOI] [PubMed] [Google Scholar]
- 26.Neufeld T P, Rubin G M. Cell. 1994;77:371–379. doi: 10.1016/0092-8674(94)90152-x. [DOI] [PubMed] [Google Scholar]
- 27.Garrett R H, Grisham C M. Biochemistry. Orlando, FL: Saunders College Publishing; 1995. [Google Scholar]
- 28.Walford R L, Harris S B, Weindruch R. J Nutr. 1987;117:1650–1654. doi: 10.1093/jn/117.10.1650. [DOI] [PubMed] [Google Scholar]
- 29.Yan L J, Levine R L, Sohal R S. Proc Natl Acad Sci USA. 1997;94:11168–11172. doi: 10.1073/pnas.94.21.11168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feder M E, Hofmann G E. Annu Rev Physiol. 1999;61:243–282. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 31.Hassett C, Aicher L, Sidhu J S, Omiecinski C J. Hum Mol Genet. 1994;3:421–428. doi: 10.1093/hmg/3.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thatcher D R. Biochem J. 1980;187:875–883. doi: 10.1042/bj1870875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen B P. Trends Microbiol. 1999;7:207–212. doi: 10.1016/s0966-842x(99)01494-8. [DOI] [PubMed] [Google Scholar]
- 34.Won K A, Schumacher R J, Farr G W, Horwich A L, Reed S I. Mol Cell Biol. 1998;18:7584–7589. doi: 10.1128/mcb.18.12.7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pickett C B, Lu A Y. Annu Rev Biochem. 1989;58:743–764. doi: 10.1146/annurev.bi.58.070189.003523. [DOI] [PubMed] [Google Scholar]
- 36.Bito A, Haider M, Hadler I, Breitenbach M. J Biol Chem. 1997;272:21509–21519. doi: 10.1074/jbc.272.34.21509. [DOI] [PubMed] [Google Scholar]
- 37.Cole K R, Kumar S, Trong H L, Woodbury R G, Walsh K A, Neurath H. Biochemistry. 1991;30:648–655. doi: 10.1021/bi00217a009. [DOI] [PubMed] [Google Scholar]