Abstract
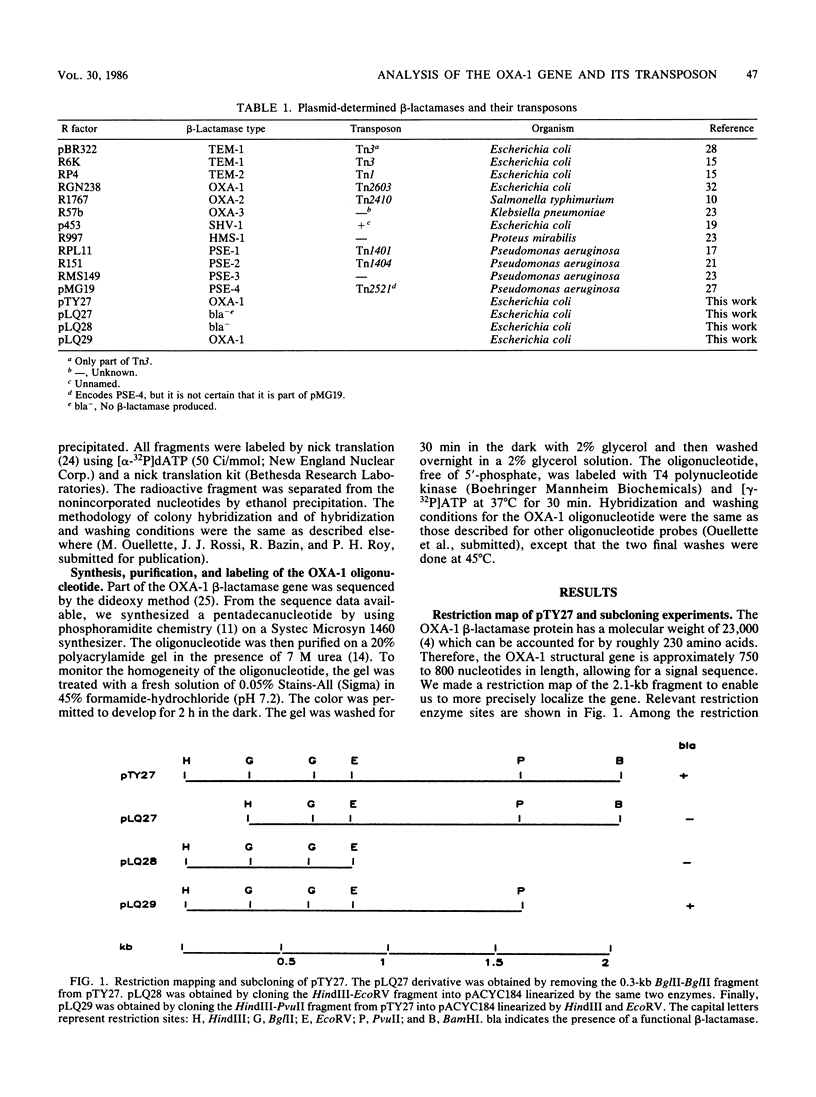
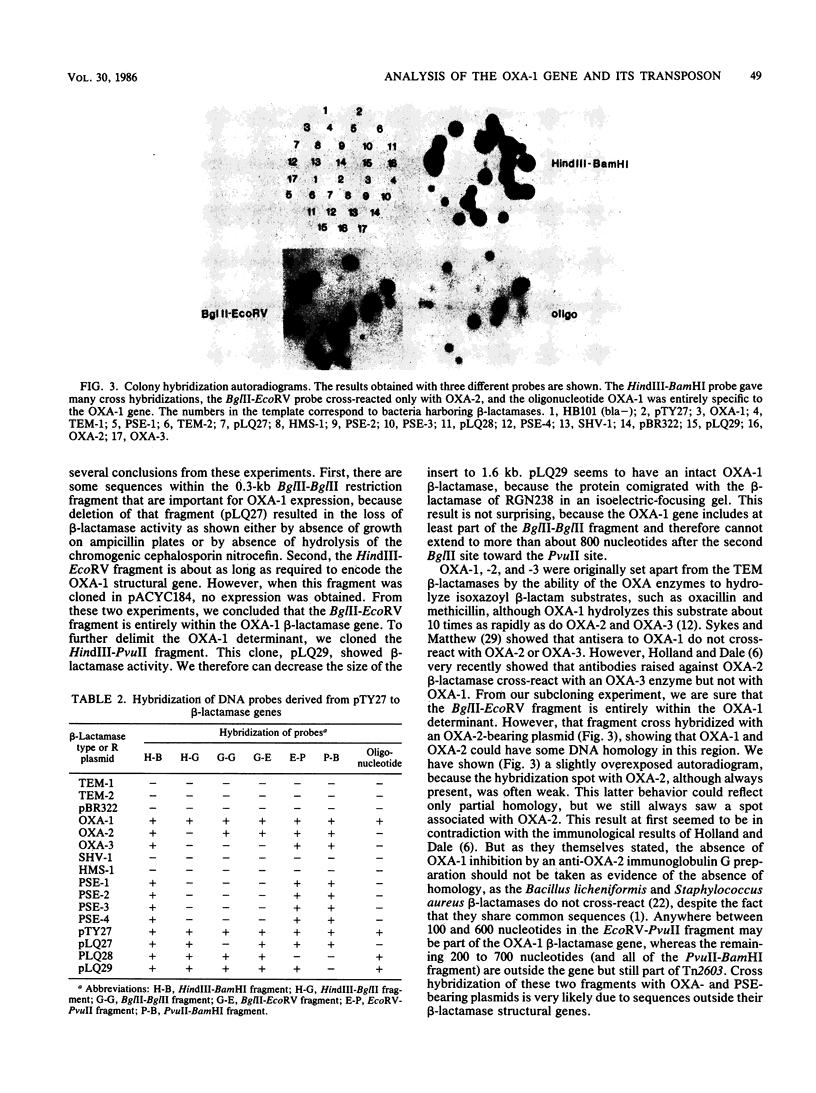
From recombinant clone pTY27, encoding an OXA-1 beta-lactamase gene, we performed subcloning experiments to more precisely delimit the gene. We describe the use as probes of six different restriction fragments known from subcloning experiments to be within the structural gene or part of the transposable element, Tn2603, flanking the OXA-1 determinant. We showed that the OXA-1 structural gene is slightly related to the OXA-2 determinant and also that sequences within Tn2603 are common to all the OXA- and PSE-producing strains tested. For epidemiological purposes, we began nucleotide sequencing of the OXA-1 determinant, and from preliminary sequence data we synthesized an oligonucleotide 15 bases in length, corresponding to a sequence within the OXA-1 gene. This oligonucleotide was found to be specific for the OXA-1 determinant, because it hybridized only to bacteria producing that beta-lactamase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ambler R. P. The structure of beta-lactamases. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):321–331. doi: 10.1098/rstb.1980.0049. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J. Plasmid-determined resistance to antimicrobial drugs and toxic metal ions in bacteria. Microbiol Rev. 1983 Sep;47(3):361–409. doi: 10.1128/mr.47.3.361-409.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges R. W., Datta N., Kontomichalou P., Smith J. T. Molecular specificities of R factor-determined beta-lactamases: correlation with plasmid compatibility. J Bacteriol. 1974 Jan;117(1):56–62. doi: 10.1128/jb.117.1.56-62.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland S., Dale J. W. Immunological comparison between OXA-2 beta-lactamase and those mediated by other R plasmids. Antimicrob Agents Chemother. 1985 Jun;27(6):989–991. doi: 10.1128/aac.27.6.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsu K., Inoue M., Mitsuhashi S. Transposition of the carbenicillin-hydrolyzing beta-lactamase gene. J Bacteriol. 1982 May;150(2):483–489. doi: 10.1128/jb.150.2.483-489.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- Kopecko D. J., Brevet J., Cohen S. N. Involvement of multiple translocating DNA segments and recombinational hotspots in the structural evolution of bacterial plasmids. J Mol Biol. 1976 Dec;108(2):333–360. doi: 10.1016/s0022-2836(76)80124-6. [DOI] [PubMed] [Google Scholar]
- Kratz J., Schmidt F., Wiedemann B. Characterization of Tn2411 and Tn2410, two transposons derived from R-plasmid R1767 and related to Tn2603 and Tn21. J Bacteriol. 1983 Sep;155(3):1333–1342. doi: 10.1128/jb.155.3.1333-1342.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew A., Harris A. M., Marshall M. J., Ross G. W. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J Gen Microbiol. 1975 May;88(1):169–178. doi: 10.1099/00221287-88-1-169. [DOI] [PubMed] [Google Scholar]
- Matthew M. Plasmid-mediated beta-lactamases of Gram-negative bacteria: properties and distribution. J Antimicrob Chemother. 1979 Jul;5(4):349–358. doi: 10.1093/jac/5.4.349. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros A. A. Beta-lactamases. Br Med Bull. 1984 Jan;40(1):18–27. doi: 10.1093/oxfordjournals.bmb.a071942. [DOI] [PubMed] [Google Scholar]
- Medeiros A. A., Cohenford M., Jacoby G. A. Five novel plasmid-determined beta-lactamases. Antimicrob Agents Chemother. 1985 May;27(5):715–719. doi: 10.1128/aac.27.5.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medeiros A. A., Hedges R. W., Jacoby G. A. Spread of a "Pseudomonas-specific" beta-lactamase to plasmids of enterobacteria. J Bacteriol. 1982 Feb;149(2):700–707. doi: 10.1128/jb.149.2.700-707.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa H., Mitsuhashi S. Tn2011, a new transposon encoding oxacillin-hydrolyzing beta-lactamase. Antimicrob Agents Chemother. 1983 Mar;23(3):407–412. doi: 10.1128/aac.23.3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent M. E., Hedges R. W. The nature of the genetic determinant for the SHV-1 beta-lactamase. Mol Gen Genet. 1979 Oct 1;175(3):239–243. doi: 10.1007/BF00397222. [DOI] [PubMed] [Google Scholar]
- O'Callaghan C. H., Morris A., Kirby S. M., Shingler A. H. Novel method for detection of beta-lactamases by using a chromogenic cephalosporin substrate. Antimicrob Agents Chemother. 1972 Apr;1(4):283–288. doi: 10.1128/aac.1.4.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POLLOCK M. R. STIMULATING AND INHIBITING ANTIBODIES FOR BACTERIAL PENICILLINASE. Immunology. 1964 Nov;7:707–723. [PMC free article] [PubMed] [Google Scholar]
- Philippon A. M., Paul G. C., Jacoby G. A. Properties of PSE-2 beta-lactamase and genetic basis for its production in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1983 Sep;24(3):362–369. doi: 10.1128/aac.24.3.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond M. H., Bennett P. M., Choi C. L., Brown N., Brunton J., Grinsted J., Wallace L. The genetic basis of the spread of beta-lactamase synthesis among plasmid-carrying bacteria. Philos Trans R Soc Lond B Biol Sci. 1980 May 16;289(1036):349–359. doi: 10.1098/rstb.1980.0052. [DOI] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders J. R. Genetics and evolution of antibiotic resistance. Br Med Bull. 1984 Jan;40(1):54–60. doi: 10.1093/oxfordjournals.bmb.a071948. [DOI] [PubMed] [Google Scholar]
- Sinclair M. I., Holloway B. W. A chromosomally located transposon in Pseudomonas aeruginosa. J Bacteriol. 1982 Aug;151(2):569–579. doi: 10.1128/jb.151.2.569-579.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Nucleotide sequence of the ampicillin resistance gene of Escherichia coli plasmid pBR322. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3737–3741. doi: 10.1073/pnas.75.8.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Matsushita K., Yamamoto T. Genesis of a complex transposon encoding the OXA-1 (type II) beta-lactamase gene. Antimicrob Agents Chemother. 1985 Aug;28(2):227–234. doi: 10.1128/aac.28.2.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Yamamoto T., Sawai T. Evolution of complex resistance transposons from an ancestral mercury transposon. J Bacteriol. 1983 Mar;153(3):1432–1438. doi: 10.1128/jb.153.3.1432-1438.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Baba R., Yamagishi S. Physical and functional mapping of Tn2603, a transposon encoding ampicillin, streptomycin, sulfonamide, and mercury resistance. Mol Gen Genet. 1981;181(4):464–469. doi: 10.1007/BF00428737. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tanaka M., Nohara C., Fukunaga Y., Yamagishi S. Transposition of the oxacillin-hydrolyzing penicillinase gene. J Bacteriol. 1981 Feb;145(2):808–813. doi: 10.1128/jb.145.2.808-813.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Watanabe M., Matsumoto K., Sawai T. Tn2610, a transposon involved in the spread of the carbenicillin-hydrolyzing beta-lactamase gene. Mol Gen Genet. 1983;189(2):282–288. doi: 10.1007/BF00337818. [DOI] [PubMed] [Google Scholar]