Abstract
The structure of complex I from Yarrowia lipolytica was determined by three-dimensional electron microscopy. A random conical data set was collected from deep stain embedded particles. More than 14000 image pairs were analyzed. Through extensive classification combined with three-dimensional reconstruction, it was possible for the first time to show a much more detailed substructure of the complex. The peripheral arm is subdivided in at least six domains. The membrane arm shows two major protrusions on its matrix facing side and exhibits a channel like feature on the side facing the cytoplasm. Structures resembling a tether connecting the subunits near the catalytic center with the protrusions of the membrane arm provide a second connection between matrix and membrane domain.
Keywords: Complex I, NADH-dehydrogenase, mitochondria, Yarrowia lipolytica, 3D electron microscopy, image processing, structure
Introduction
Complex I is the first enzyme in the respiratory chain of mitochondria and of many bacteria. It is by far the largest and most complicated enzyme in the chain, yet the least understood (for review see e.g. special issues (BBA 1998) and(JBB 2001), (Hirst 2005), and (Brandt 2006)). Bacterial complex I consists of as few as 14 subunits with a total molecular mass of about 500 kDa while eukaryotic complex I exhibits a molecular mass of almost 1 MDa. In the yeast Yarrowia lipolytica, it consists of at least 38 different subunits (Abdrakhmanova et al. 2004). The subunits of complex I are encoded by both nuclear and mitochondrial DNA. All subunits of the so-called peripheral arm that protrudes into the mitochondrial matrix are encoded by nuclear DNA, whereas all seven mitochondrially coded subunits reside in the membrane arm. The approximate positions of two subunits, the 49 kDa subunit and the 30kDa subunit, have been determined by two-dimensional image analysis of electron micrographs of immuno-labeled complexes (Guénebaut et al. 1997; Zickermann et al. 2003). The locations of most other subunits are still highly speculative. Mitochondrial complex I contains 8 [FeS] clusters and one flavin mononucleotide (Ohnishi et al. 1998; Rasmussen et al. 2001). All these redox centers are localized in the peripheral arm of complex I where they form a ‘wire’ connecting the substrate binding sites (Hinchliffe and Sazanov 2005). The ubiquinone reducing catalytic core is located at the interface between the 49 kDa and the PSST subunit adjacent to iron-sulfur cluster N2 (Kashani-Poor et al. 2001). In the membrane arm, three of the membrane spanning subunits, ND4, ND5 and ND2 exhibit sequence homology with a class of Na+/H+ antiporters (Mathiesen and Hägerhäll 2002; Meloa et al. 2005). It has been speculated that these subunits are the actual proton pumping devices. Subunits ND4 and ND5 were shown to form the part of the membrane arm most distant from the peripheral arm (Sazanov and Walker 2000). This seems to suggest that the redox centers and the pumping modules are clearly separated from each other.
To date, the structural information about complex I is insufficient, and the mechanism of redox-linked proton pumping is still unknown. Several three-dimensional structures of holo-complex I from various species have been published previously (N. crassa: (Guénebaut et al. 1997) E. coli: (Guénebaut et al. 1998; Böttcher et al. 2002) bovine: (Grigorieff 1998) ). The first two structures were determined by the techniques of random conical tilting (Radermacher et al. 1986, 1987; Radermacher 1988). The reconstruction of bovine complex I was carried out by reference based alignment of untilted ice data relative to a low resolution negative stain reconstruction from a random conical data set. The last E. coli complex I reconstruction was determined using the technique of angular reconstitution (van Heel 1987). All published reconstructions except one (Böttcher et al. 2002) show the typical L-shape of the enzyme, where one arm of the L represents the matrix or peripheral arm and the other represents the membrane arm. Here we report the first reconstruction of complex I isolated from Yarrowia lipolytica, an aerobic yeast that has been modified such that efficient genetic manipulations of complex I are possible without compromising the viability of the yeast (Kerscher et al. 2001; Kerscher et al. 2002). The structure was determined using the random conical reconstruction technique. Complex I exhibits a large variation of apparent shapes when analyzed in the electron microscope and only tilting experiments are capable of differentiating between particles in different orientations and conformations. The technique is based on pairs of micrographs of each specimen area, one with the specimen tilted by a high angle and a second without tilt, so that each 0°-image has a tilted counterpart. By recording the tilt images first, the radiation damage to the data used for three-dimensional reconstruction is kept minimal. When the 0 °-degree images are classified, the tilt images corresponding to each class can be used to calculate the three-dimensional structure represented by the class. This volume data allows to separate variations caused by differences in orientation from variations in the three-dimensional structure.
A comparison of our results shows a variety of conformations; however, a reproducible architecture of complex I emerged at the same time. The peripheral arm of complex I consistently shows substructures and can be divided into distinct domains. The membrane arm now shows two major protrusions on the matrix side that appear connected to the redox active part of complex I. On the cytoplasmic side of the membrane arm, we found indications of a channel parallel to the membrane and a possible transmembrane channel close to the central protrusion. Conformational variability is observed in the domain positions on the peripheral arm and in the connections between catalytic subunits and membrane arm protrusions.
Materials and methods
Complex I purification
Complex I from Y. lipolytica was purified by his-tag affinity chromatography essentially as described in (Kashani-Poor et al. 2001). The final size exclusion chromatography was done in the presence of 0.025 % laurylmaltoside. Purity and subunit composition were checked by Tricine-SDS-Page (Schägger and von Jagow 1987) and dSDS-PAGE (Rais et al. 2004). Protein concentration was determined according to a modified Lowry protocol (Lowry et al. 1951).
Sample preparation for microscopy
The sample was diluted to 0.015 mg/ml in buffer containing 25 mM NaMOPS pH 7.2, 100 mM NaCl, 2 mM EDTA and 0.025 % laurylmaltoside, applied in 4μl droplets to carbon coated grids and deep stained with phosphotungstic acid at pH 7.2. 0.025 mg/ml Tobacco Mosaic Virus (TMV) was added to the preparation for magnification calibration. The deep staining was carried out according to Stoops et al. with modifications (Stoops et al. 1992; Radermacher et al. 2001; Ruiz and Radermacher 2006). The sample was blotted by touching the edge of the grid with #41 Whatman filter paper (Whatman Inc. Florham Park, NJ, USA) and then fast dried using a home built dry nitrogen blowing device.
Microscopy
Images were recorded in a Tecnai T12 electron microscope (FEI, Eindhoven, NL). The LaB6 filament of the microscope was operated in point mode to obtain optimum coherence and resolution (Ruiz et al. 2003). All the microscopy was done under low-dose conditions with a dose of less than 10el/A2 per image. Tilt pairs were collected at 0° and ~55° tilt, the tilt image being recorded first. The pairs were analyzed in an optical diffractometer and only those images showing no drift and resolutions better than 1 nm were used.
Image processing
SPIDER (version 5.0 with modifications) (Frank et al. 1981; Frank et al. 1996) and XMIPP (Marabini et al. 1996) were used for analyzing the data. The processing was done in two stages. First, 29 tilt pairs were evaluated yielding approximately 3600 pairs of particle images. Subsequently 81additional tilt pairs were processed resulting in a total of 14961 image pairs. The first set of micrographs was digitized on a Nikon Super Coolscan scanner (Nikon, Tokyo, JP) with a pixel size corresponding to 1.2Å on the specimen scale. After conversion to SPIDER format, the images were binned by a factor of 3 to a final pixel size of 3.6 Å. The magnification of the images was calibrated using the 23Å layer line of TMV. During scanning, a part of the blank negative area of each micrograph was included to serve as a reference value for transparency 1 in order to calculate correct optical densities. The particle coordinates were picked from tilt pairs using WEB (Frank et al. 1996). Images were then windowed from the micrographs and contrast normalized using the average density in a ring surrounding the particle. The contrast transfer function was determined according to (Radermacher et al. 2001), resulting in fitted values for the amplitude contrast, defocus and astigmatism for both the zero degree images and tilt images. The corrections were applied to the windowed single particle images by flipping the phases using a smooth function. All alignments were carried out using Radon transform alignment techniques, which allow for simultaneous translational and rotational alignment and do not depend on a definition of the rotation center (Radermacher 1994, 1997). The images first underwent reference free alignment (Marco et al. 1996). The resulting overall average was centered by translational alignment to a strongly low-pass filtered disk and used as a reference for aligning all the images. The aligned images underwent correspondence analysis (van Heel and Frank 1981; Frank and van Heel 1982; Bretaudiere and Frank 1986) followed by Diday’s methods of classification with moving centers (Diday 1971) and finally Hierarchical ascendant classification (HAC) (command CL CLA in SPIDER). The alignment proceeded with repeated rounds of multireference alignments, correspondence analysis, and classification. The number of references in each step varied between 9 and 12 depending on the clustering tree produced by CL CLA. Ten final classes were obtained from the first data set and preliminary reconstructions were calculated.
The second data set of 81 tilt pairs was scanned on an Intergraph SCAI scanner (Z/I Imaging Corporation, Huntsville, AL ) at 7μm pixel size, and binned by a factor of 3, resulting in a calibrated pixel size of 4.02 Å on the specimen scale. To keep consistency with the previous data sets, the images scanned on the SCAI scanner were interpolated up to 3.6Å pixel size after CTF correction. The combined image set was processed de-novo. A reference was created by reference free alignment (as described above). This reference was centered relative to a low-pass filtered image of a disk, then rotationally averaged, masked, low- and high-pass filtered and used as a centration reference for the complete data set. The ten class averages from the first data set of 3500 were centered with the same centration reference and used as references for multireference alignment of the full data set. This was followed by correspondence analysis and classification with Diday’s method of moving centers followed by HAC, resulting in 13 class averages. Seven classes, showing one major orientation relative to the carbon support and comprising 10885 images were processed further. These images were analyzed separately by multiple rounds of correspondence analysis followed by classification, as before. Eight classes were formed and three-dimensional reconstructions were calculated from the data set. Reconstructions from classes that contained more than 1000 particles were refined using Radon transform based alignment algorithms. (Radermacher 1994, 1997) until the resolution did not improve anymore. During all refinements, care was taken that the references used in each step of the refinement were sufficiently low-pass filtered to avoid creating an artificial high resolution structure. In addition, a reconstruction from all 10885 particles was calculated and refined before additional classification.
Further multivariate statistical analysis was carried out by applying a mask that focused on the matrix side of the membrane arm of the particle (delineated as a circle in Fig. 4). Several classifications and additional multireference alignments were carried out. As a final partitioning of the data set, 35 classes obtained by Diday’s methods of moving centers – without any additional hierarchical ascendant classification – were created. Reconstructions for all of the 35 classes were calculated, refined as above and compared.
Figure 4.

The eight classes after separate multireference alignment, correspondence analysis and classification. Class memberships: (1) 507, (2) 1921, (3) 1960, (4) 112, (5) 1674, (6) 2538, (7) 172, (8) 2002. Scale bar 10 nm. The circle in the left most image indicates the mask used for the classification into 35 classes shown in figure 8.
Because of the large number of classifications in the following we will number the reconstructions with M_n where n is the class number and M the number of classes, for example 8_1 will be class 1 from the partitioning into 8 classes.
The resolution of all reconstructions was determined by Fourier shell correlation using a cutoff criterion of 0.3, which in this study was conservative relative to the five times noise-correlation cutoff. The volumes were inspected slice by slice in WEB, and surface representations were generated using Chimera (Pettersen et al. 2004). If not stated differently, the threshold for surface representation was chosen such that any noise outside the structure just disappeared. This closely corresponds to the steepest density gradient at the particle surface
Results
We have analyzed the feasibility of imaging complex I using several negative stained preparations, as is common practice in our laboratory. Sometimes the heavy metals in the staining solution react either with the buffer or the sample rendering the electron microscopy samples useless for structural determination. We have tried five different staining solutions to visualize complex I: 1% uranyl acetate (UA), 2% phosphotunstic acid, pH 7 with NaOH (PTA), 2% ammonium molybdate pH 7 with NH4 (AM), 2% methyl amine tungstate (MAT) and Nanovan (NAN), both from Nanoprobes (data not shown). The effects of stain precipitation and sample preservation were judged visually from the homogeneity of the particles and the smoothness of the background. We obtained the best results with PTA and AM; however, PTA provided slightly better contrast and was used for the final analysis with a large data set. We also carried out three-dimensional reconstructions in ammonium molybdate preparations, using a smaller data set (data not shown). The results obtained were very similar to the results obtained with PTA.
One of the tilt pairs used to generate the random conical data set is shown in figure 1. Images of the untilted specimen showed a majority of L-shaped particles with substantial variability (figure 1 bottom). Some of the variations that appear as a change in angle of the two arms reflect orientational variations rather than structural differences. Some particles visible in the shape of a straight line in the 0°–image showed an L-shape in the tilted image.
Figure 1.
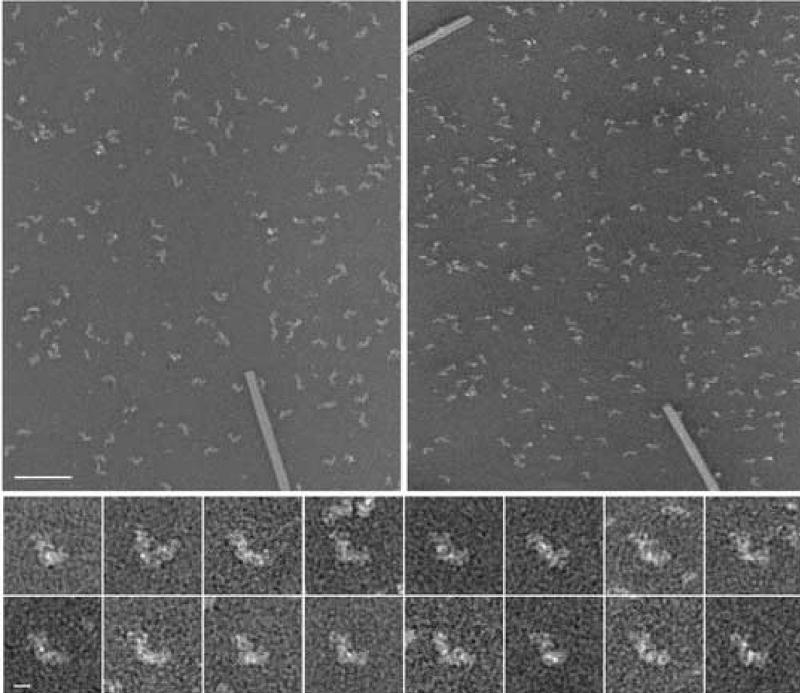
Tilt pair of complex I from Y. lipolytica, prepared in deep stain (PTA). For a reconstruction from a random conical data set, pairs of micrographs of each specimen area are recorded, the first with a high tilt angle and the second without tilt. The particles from 0° micrographs are used for alignments and classification and three-dimensional reconstructions are calculated from the corresponding particles extracted from the tilt micrographs. The right image was recorded at a tilt angle of 55°, the left image at 0°tilt. TMV was used for the calibration of the magnification. Tilt axis horizontal, Scale bar 100 nm. Bottom: Series of aligned 0° images, Scale bar 10 nm.
The particles were subjected to multiple rounds of alignment and classification (see methods). The variations found among the particles are illustrated in the factor maps of the final correspondence analysis of the complete data set (figure 2). The classification following this correspondence analysis yielded 13 classes (figure 3). The particle occurs in two major views which we term “flip-view” (peripheral arm up, classes 13_1, 13_8 to 13_13) and “flop-view” (peripheral arm horizontal, class 13_4 ). Class averages 13_2, 13_3, and 13_5 to 13_7 look blurred, likely caused by mixtures of views and damaged particles in these classes. Surprisingly, the flip-view was much more dominant than the flop-view. This prevalence could be caused by a difference in affinity of the two sides of the molecule to the carbon surface.
Figure 2.

Correspondence analysis was carried out with a total of 12 factors. Shown are visual representation of factor maps 1 versus 2 (left) and 3 versus 4 (right) of the complete data set. The two maps present an overview over the major variations found in the data set.
Figure 3.
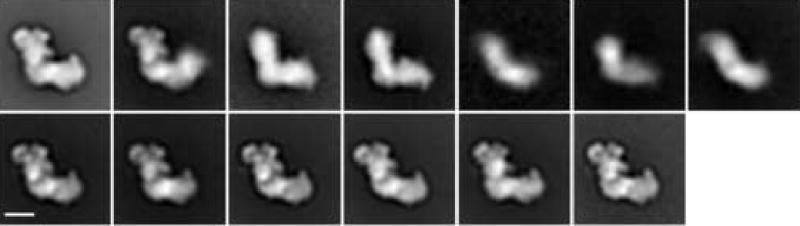
Class-averages of the complete data set calculated from the run of correspondence analysis shown in fig. 2. Classes are shown in the order of their class number. Class memberships: (class number followed by number of particles): (1) 1552, (2) 686, (3) 580, (4) 920, (5) 131.00, (6) 431, (7) 1092, (8) 1352, (9) 1580, (10) 1672, (11) 1975, (12) 2328, (13) 427.
Classes 13_2 to 13_7 were further analyzed and processed separately. The data set underwent additional rounds of multireference alignment and classification and was again divided into 12 classes. Visual inspection showed that merging into any fewer classes was not justified. The largest class corresponded to the flop view similar to 13_4 and contained 780 particles. Three-dimensional reconstructions were calculated for all classes, but all classes, except the largest, showed obviously distorted or incomplete particles(data not shown). Thus, we concentrated the major effort of the analysis on the “flip-view” data set.
The “flip view” data set, containing a total of 10885 particles ( Classes 13_1, and 13_8 to 13_13 in figure 3), underwent additional rounds of correspondence analysis, classification and multireference alignment. The last classification, with a combination of Diday’s method of moving centers and hierarchical ascendant classification, resulted in 8 classes (figure 4). Initial three-dimensional reconstructions were calculated for each class and the projection angles and positions of the five largest classes were refined. For comparison, a reconstruction from the full “flip-view” set of 10885 particles was calculated and refined. We will refer to this data set as the full set reconstruction. The resolutions of all reconstruction were determined after refinement using the 0.3 Fourier shell correlation criterion and are shown in table 1.
Table 1.
Number of particles and resolutions for the combined 8-class data set and the five larges classes in this data set.
| No. of particles | Resolution | |
|---|---|---|
| Combined set | 10885 | 24 Å |
| Class 8_2 | 1921 | 16.5 Å |
| Class 8_3 | 1960 | 22 Å |
| Class 8_5 | 1674 | 20.5 Å |
| Class 8_6 | 2538 | 18.5 Å |
| Class 8_8 | 2002 | 18.5 Å |
The reconstructions were calculated from a random conical data set and subsequently refined by reference based angular refinement using the techniques described in (Radermacher 1994, 1997). Before refinement, each volume had a missing cone with a half angle of approximately 35°. With the equations for three-dimensional image points in a conical reconstruction (Radermacher 1980; Radermacher and Hoppe 1980) this leads to a reduction in resolution in z-direction by a factor of 1.317. In connection with an analysis of data used in the study of the three-dimensional structure of the ryanodine receptor (Wagenknecht and Radermacher 1995), it has been observed that even if a particle has a predominantly preferred orientation, the average range of wobbling of a particle around this position was 15° and could not be distinguished in the classification of the 0° projections. A similar variation in position can be found in the current data set. As a result, during the angular refinement, the missing cone was filled completely in the full set reconstruction and partially in the reconstructions of the five largest classes. Thus, the effect of a missing cone is absent in the full set reconstruction and substantially smaller than 1.3 in the reconstructions from the five largest classes.
Surface representations of the five largest classes, 8_2, 8_3, 8_5, 8_6 and 8_8 are shown in figure 5. Variations can be most clearly seen in the peripheral arm (depicted vertically) and in the central protuberance on the matrix side of the membrane arm. The reconstruction of the combined set shows a shape that, as expected, represents an intermediate image between the reconstructions of the five classes (fig.6). The peripheral arm measured from the top of the particle to the bottom of the membrane arm has a length of ~220 Å. The length of the matrix arm is approximately the same. If one assumes an overall thickness of 40 Å for the inner mitochondrial membrane, the peripheral arm extends out of the membrane by about 145 Å.
Figure 5.
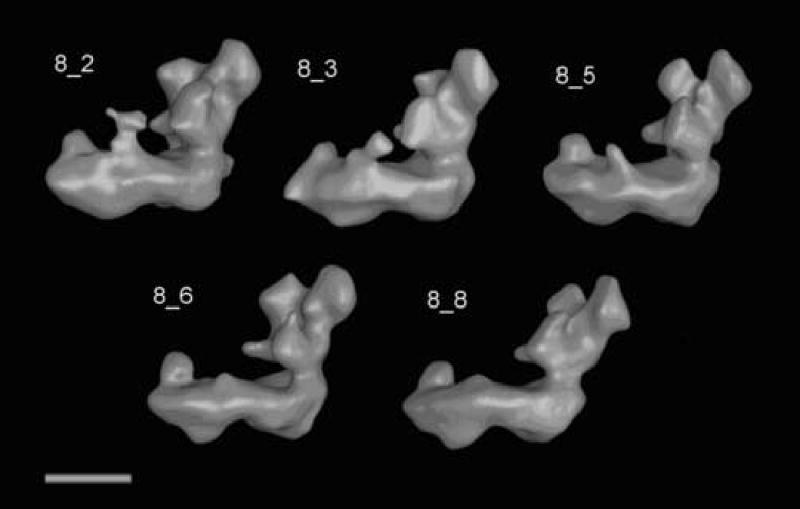
Surface representation of the reconstruction from the five largest classes of the “flip”-view analysis. The classes shown are top left to bottom right: 8_2, 8_3, 8_5, 8_6, 8_8.
Although a number of interesting details that can be observed in a finer classification are lost in the full set reconstruction, the structure shows features common to all reconstructions. The peripheral arm can be divided into at least six distinct domains (numbered 1–6). It should be noted that domain 4 is not part of domain 5, although both are connected. The peripheral arm has two strong connections to the matrix side of the membrane arm (C1 and C2). C1 is the stronger connection, as can be seen when the surface threshold is increased (C2 disappears first). Connections C1 and C2 are also visible in the volumes shown in Fig. 5. Only in reconstruction 8_6 is C2 quite weak, while both are very clearly visible in the other four reconstructions. The membrane arm in this reconstruction shows a central protuberance (labeled CMP for central membrane arm protuberance) and a distal protuberance (labeled DMP for distal membrane arm protuberance) with a height of approximately 40 Å. At the back of the membrane arm, an indentation can be observed, labeled I. When viewed from the top, the membrane arm appears curved. Increasing the threshold level shows that domain 5 in the peripheral arm is connected to domains 3 and 4 by two connections labeled C3 and C4. C4 is the stronger of the two connections. When the same analysis is applied to the volume 8_2, which has the highest resolution of all the reconstructions (16.5 Å), the same connections are visible (figure 7). In addition, there is a connection from domain 5 to domain 1 (C5). This connection varies in strength among all the reconstructions. At the higher threshold used in figure 7, the indentation I indicated in figure 6 forms a groove (G) along the side of the membrane arm that faces the cytoplasm.
Figure 7.
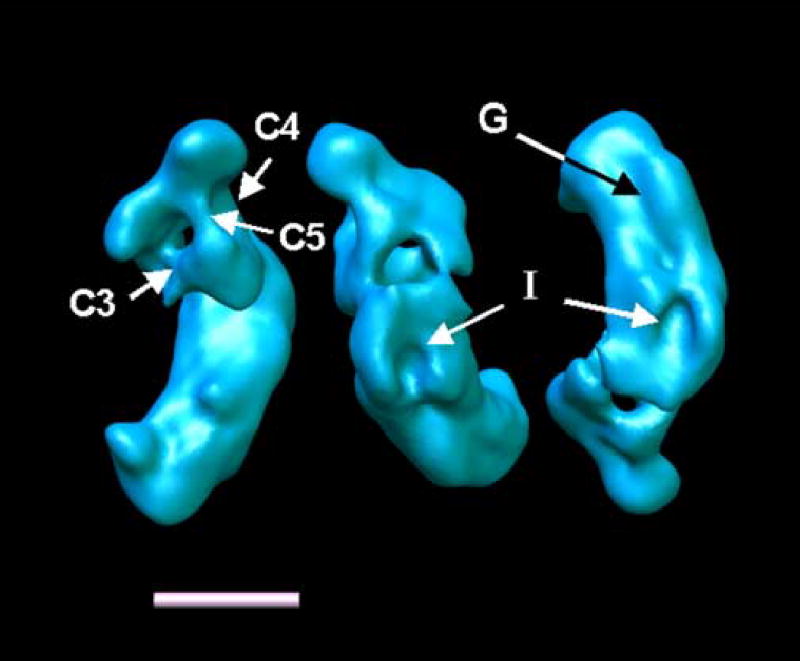
Reconstruction 8_2 at a contour level that reveals the connections between the domains in the peripheral arm. C5 is a connection that can be observed in many reconstructions with varying density. I Indentation (same as in Fig. 8). G groove along the cytoplasmic side of the membrane arm. Shown are: left: a top view of complex I, center: a view of the bottom of the back of the matrix arm, right: a view of the cytoplasmic side of the membrane arm.
Figure 6.
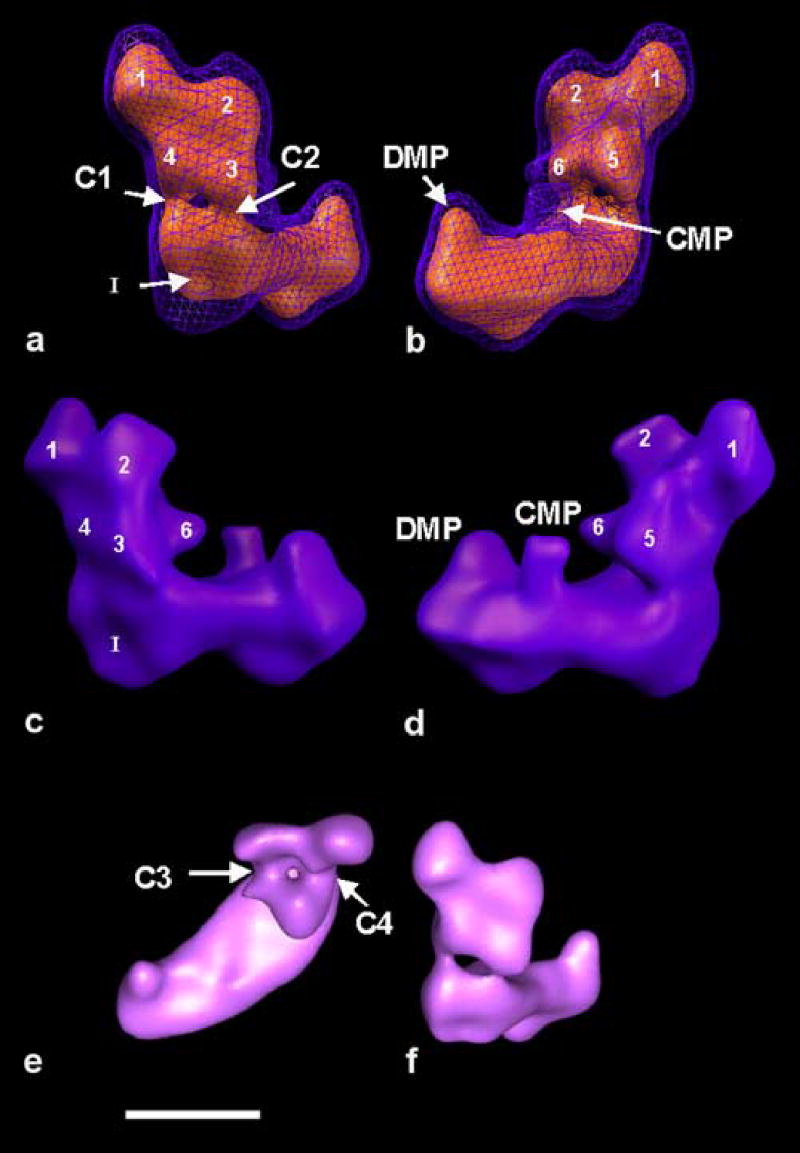
The reconstruction from the full “flip-view” set. Indicated are the common features found in the majority of reconstructions. Distinguishable domains are numbered. DMP distal membrane arm protuberance, CMP central Membrane arm protuberance. C1-C4 connections. a-d) show the particle with the membrane arm approximately parallel to the horizontal. a) shows the particle rotated with the membrane arm pointing into the image, so that the back-side of the matrix arm is best visible. b) is rotated around the z-axis by approximately 180° relative to a). c) and d) are side views of the particle, related by ~180° and approximately perpendicular to their preferred orientation on the specimen support. e) shows a top view of the particle, as seen from the side of the mitochondrial matrix. f) for comparison the same surface level as seen from the back of the matrix arm. a) and b) show a combination of two threshold levels, the blue mesh includes a calculated volume of ~1.3 MDa and the red surface ~670 kDa. At limited resolution, these measurements are usually up to 30% larger than the actual enclosed protein volume. In b) and c) the same surface threshold was used as for the blue mesh in a) and b). Scale bar 10 nm.
A comparison of the five reconstructions (figure 5) shows as the most obvious difference a variation in the CMP. In the reconstruction shown in figure 5a, this protuberance shows an extra mass similar to the representation in figure 6b. When the surface threshold is lowered to a level that also shows the surrounding noise, CMP first connects to domain 5. Under similar conditions, CMP connects to DMP in the volume shown in figure 5c. The last two volumes (figure 5 d and e) show only a weak indication of CMP.
In earlier two-dimensional studies, a thin connection between the major protuberance of the peripheral arm and the protuberances of the membrane arm had been observed occasionally (Zickermann et al 2003). A close inspection of the two-dimensional averages published in connection with the early three-dimensional reconstruction of complex I from N. crassa (Guénebaut et al. 1997) shows hints of this connection. In one of our preliminary reconstructions of complex I (unpublished) a faint, very low density was seen that may have been this connection. In the reconstructions from our five classes this density exhibits a high variability. A mass connecting at the same time DMP, CMP and domain 5 becomes visible only when the surface threshold is lowered to a level where spurious noise features outside the molecule become visible. Therefore, we reanalyzed the data set using correspondence analysis, with a mask that focused on the area between the peripheral and the membrane arm (s. figure 4) and repeated the classification. This time only Diday’s moving centers classification was used, without subsequent hierarchical ascendant classification (HAC). Using HAC, classes were merged in a way that seemed not justified when visually inspected. Of the 71 classes created by Diday’s method of moving centers, we analyzed the first 35 classes containing a minimum of 62 particles each. Classes with fewer particles were discarded because they would have had insufficient data for a three-dimensional analysis. The total number of particles represented in this data set was 10167. We calculated all 35 structures, and refined the projection orientations by 10 iterations of the reference based projection refinement. The number of particles per class and final resolution, determined by Fourier shell correlation are shown in table 2.
Table 2.
Number of particles and resolutions (Fourier shell correlation with cutoff of 0.3) for the reconstructions of the 35 classes.
| Particles per class | Resolution | Particles per class | Resolution | ||
|---|---|---|---|---|---|
| Class 35 _ 1 | 1139 | 33.9 Å | Class 35 _ 19 | 182 | 41.1 Å |
| Class 35 _ 2 | 876 | 33.8 Å | Class 35 _ 20 | 162 | 41.1 Å |
| Class 35 _ 3 | 767 | 36.0 Å | Class 35 _ 21 | 161 | 41.1 Å |
| Class 35 _ 4 | 724 | 33.9 Å | Class 35 _ 22 | 151 | 38.4 Å |
| Class 35 _ 5 | 643 | 38.4 Å | Class 35 _ 23 | 144 | 41.1 Å |
| Class 35 _ 6 | 524 | 38.4 Å | Class 35 _ 24 | 109 | 38.4 Å |
| Class 35 _ 7 | 507 | 32.0 Å | Class 35 _ 25 | 99 | 41.1 Å |
| Class 35 _ 8 | 507 | 38.4 Å | Class 35 _ 26 | 92 | 41.1 Å |
| Class 35 _ 9 | 368 | 36.0 Å | Class 35 _ 27 | 84 | 41.1 Å |
| Class 35 _ 10 | 358 | 33.9 Å | Class 35 _ 28 | 75 | 52.4 Å |
| Class 35 _ 11 | 341 | 36.0 Å | Class 35 _ 29 | 74 | 48.0 Å |
| Class 35 _ 12 | 323 | 38.4 Å | Class 35 _ 30 | 73 | 48.0 Å |
| Class 35 _ 13 | 278 | 36.0 Å | Class 35 _ 31 | 72 | 48.0 Å |
| Class 35 _ 14 | 257 | 36.0 Å | Class 35 _ 32 | 72 | 41.1 Å |
| Class 35 _ 15 | 219 | 41.1 Å | Class 35 _ 33 | 69 | 52.4 Å |
| Class 35 _ 16 | 206 | 38.4 Å | Class 35 _ 34 | 67 | 44.3 Å |
| Class 35 _ 17 | 196 | 48.0 Å | Class 35 _ 35 | 62 | 57.6 Å |
| Class 35 _ 18 | 186 | 41.1 Å |
We compared the results in three-dimensions (figure 8). The L-shape is in common to all reconstructions. The peripheral arm in all structures can be divided into the same distinct domains as shown above. The most interesting variations are in the shape and connections of the membrane arm protrusions. The reconstructions shown in figure 9 start with 35_1 in the upper left corner and continue line by line until reconstruction 35_35. In panel 1 we see a rather massive central membrane protuberance, connected to the main protuberance on the peripheral arm. Similar connections can be seen in reconstructions 35_16, 35_18 and maybe 35_24. A connection between the central and distal membrane arm protuberances can be observed in panel 35_14. Most central protuberances visible in these surface representations have a component that leans either more to the distal protuberance or to the peripheral arm. In many cases, when the threshold defining the surface is lowered, this protuberance grows and makes connections to the peripheral arm comparable to the one seen in some of the panels shown here.
Figure 8.

The 35 reconstructions. Clearly visible is the variable density connected to the central membrane arm protuberance. Scale bar 10 nm
Figure 9.
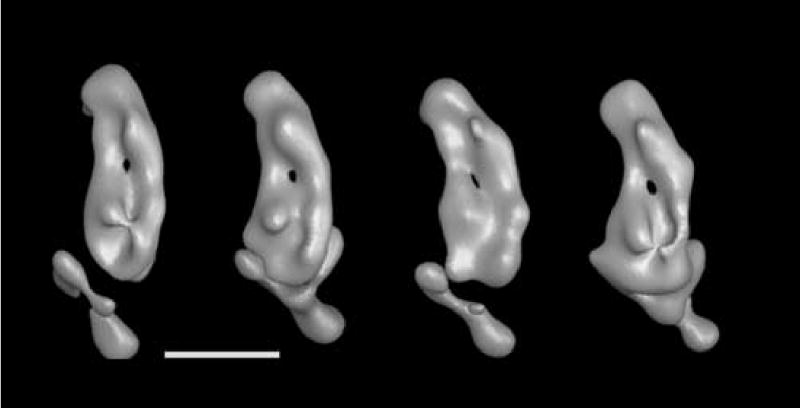
From left to right: Volumes 35_2, 35_13, 35_15 and 35_20 from figure 8 shown here at a higher threshold value that reveals the groove in the cytoplasmic side of the membrane arm and the possible channel through the membrane arm. All views are from the cytoplasmic side of the membrane arm.
When the reconstructions are closely inspected and the surface threshold is varied, one can observe dense features on the cytoplasmic side of the membrane arm in volumes 35_13, 35_15 and 35_20 that form a groove parallel to the membrane as seen in reconstruction 8_2 (figure 7, right panel), and exhibit an access portal that closely corresponds to the indentation indicated in figure 6. As the threshold is increased further, a channel opens through the membrane arm in the vicinity of the central membrane arm protuberance (Fig. 9). This may be an indication for the location of subunits ND4/5 or ND2 all of which may form the proton pumping unit.
Discussion
We have calculated three-dimensional reconstructions of complex I from Yarrowia lipolytica with a highest resolution of 16.5 Å for one of the particle classes (class 8_2 in table 1) and between 18.5 to 22 Å for the other four major classes. It is interesting to note that the highest resolution was not obtained for the class with the largest number of images, and may reflect instead a higher homogeneity of class 8_2.
Complex I is a highly variable structure which may explain why the detailed substructure has not been seen previously when either the data set was smaller, or a lack of tilt images precluded an extensive structural classification. The matrix arm in the reconstruction of bovine complex I (Grigorieff 1998), the only attempt at a reconstruction of complex I in a frozen hydrated preparation, showed almost no subdomains. While as a first reference a random conical reconstruction of a stain embedded sample was used in this study, the lack of available techniques at the time precluded a classification of the randomly oriented ice data into classes that would have separated possible conformational differences from orientational differences. The unusual U-shaped structure of the E. coli complex I (Böttcher et al. 2002), reconstructed without any tilting from a heterogeneous specimen, is inconsistent with all previous reconstructions of complex I from N. crassa (Guénebaut et al. 1997), E. coli (Guénebaut et al. 1998), bovine (Grigorieff 1998) and the structure presented here. Recently, it has been suggested that the U-shaped structure may be an artifact originating from the purification conditions (Sazanov et al. 2003). In addition, valid classification techniques to separate any conformational variations from orientation variation when no tilts are available are in their infancy at best and the lack of better techniques may have led to an artifact. The reconstructions by Guénebaut et al. agree well with our results, although they showed substantially less detail mainly because the data set was smaller. The current detailed structure was possible to obtain only because we carried out extensive classifications of a large data set and reconstructed volumes for each class using the tilted images of the random conical data.
The variability in the complex I structure that renders structure determination a difficult task may be related to the function of this large membrane protein and fits well with redox-induced conformational changes that were detected by cross-linking studies (Belogrudov and Hatefi 1994) (Mamedova et al. 2004). This study showed that NAD(P)H prevented the cross-linking with a zero length cross-linker between the hydrophilic subunits, while cross-linking was observed in the presence of NAD+. It was suggested that these conformational changes may extend to subunits in the membrane arm and activate a proton pumping mechanism.
For the first time, we see a structure of the peripheral arm of eukaryotic complex I divided into distinct domains. Recently an envelope structure of the peripheral arm of bacterial complex I from Thermus thermophilus was determined by X-ray crystallography (Hinchliffe and Sazanov 2005). It is not possible to compare this structure directly with the one presented here because the peripheral arm of the eukaryotic enzyme comprises a number of additional accessory subunits that add up to a mass of at least 100 kDa. It seems likely that the three domains seen in the Y-shaped bacterial structure correspond to the three major domains 1, 2 and 5 of the Y. lipolytica structure (Fig. 6), however, domains 1, 2 and 5 cannot be assigned with certainty to the core subunits present in the complexes from both sources. The 2D averages of Y. lipolytica complex I decorated with monoclonal antibodies against the 49 kDa (Zickermann et al. 2003) give some indications that the subunit is in the upper two thirds of the peripheral arm. When compared with the structure presented here, for example, one of the epitopes of the antibody against the 49 kDa subunit could be located on either domain 5, 6 or 3 in figure 6. A three-dimensional reconstruction of the complex I/antibody complex will be required for unambiguous localization of the epitope.
The connection between the peripheral arm and the protrusions on the membrane arm is easier to visualize by volume rendering techniques than by surface representation. When viewed with 3D stereo volume rendering methods, this connection is easier to see, and traces of it can be made out in many of the reconstructions. The variability that we see in this connection is partially a problem of specimen purification and preservation but may also be related to the functional principle of complex I. It seems clear, that the protrusions seen on the matrix side of the membrane arm are connected to redox active part in the peripheral arm by this tether. Suboptimal purification or preservation techniques could only have damaged this connection, but they could not have created it. It is interesting to note that this connection, together with a potential channel close to the central protrusion in the membrane arm could provide a pathway, whereby conformational changes induced by the redox chemistry occurring in the peripheral arm could be transferred to subunits in the membrane arm. The putative transporter subunits ND4 and ND5 have previously been assigned to the center and peripheral end of the membrane arm (Sazanov and Walker 2000). It would not be surprising, if these subunits were located near the two membrane arm protrusions that we observe.
Many questions remain unanswered at this time and require further investigation. Among them are the locations of the subunits in the domains seen in the peripheral arm, and if the connection we observe connects alternatively the distal and central membrane arm protuberance or the catalytic subunits and the central membrane arm protrusion, or if this connection can be continuous.
The structure of complex I presented here shows for the first time distinct substructures that provide a solid framework for future assignments of subunits to specific domains. The variations that we observe are consistent with a functional model of complex I that links redox chemistry to changes in conformation. The observed connections between the peripheral arm and the membrane arm could provide a path for linking the electron transfer reaction from NADH to ubiquinone to an active transport mechanism within the membrane arm.
Acknowledgments
This work was supported by grants NIH RO1 GM068650 and Deutsche Forschungsgemeinschaft, SFB 472 (to U.B and V.Z.). The work has benefited from grant NIH RO1 GM069551 (to T.R.)
References
- Abdrakhmanova A, Zickermann V, Bostina M, Radermacher M, Schagger H, Kerscher S, Brandt U. Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica. Biochim Biophys Acta. 2004;1658:148–156. doi: 10.1016/j.bbabio.2004.04.019. [DOI] [PubMed] [Google Scholar]
- BBA . Special Issue. Biochimica et Biophysica Acta. 1998;1364:89–296. [Google Scholar]
- Belogrudov G, Hatefi Y. Catalytic sector of complex I (NADH:ubiquinone oxidoreductase): subunit stoichiometry and substrate-induced conformation changes. Biochemistry. 1994;33:4571–4576. doi: 10.1021/bi00181a018. [DOI] [PubMed] [Google Scholar]
- Böttcher B, Scheide D, Hesterberg M, Nagel-Steger L, Friedrich T. A novel, enzymatically active conformation of the Escherichia coli ADH:ubiquinone oxidoreductase (complex I) Journal of Biological Chemistry. 2002;277:17970–17977. doi: 10.1074/jbc.M112357200. [DOI] [PubMed] [Google Scholar]
- Brandt U. Energy converting NADH:quinone oxidoreductase (Complex I) Ann Rev Biochem. 2006;75 doi: 10.1146/annurev.biochem.75.103004.142539. in press. [DOI] [PubMed] [Google Scholar]
- Bretaudiere JP, Frank J. Reconstitution of molecule images analysed by correspondence analysis: a tool for structural interpretation [published erratum appears in J Microsc 1987 May;146(Pt 2):222] Journal of Microscopy. 1986;144:1–14. doi: 10.1111/j.1365-2818.1986.tb04669.x. [DOI] [PubMed] [Google Scholar]
- Diday E. La methode de nuees dynamiques. Rev Stat Appl. 1971;19:19–34. [Google Scholar]
- Frank J, van Heel M. Correspondence analysis of aligned images of biological particles. Journal of Molecular Biology. 1982;161:134–137. doi: 10.1016/0022-2836(82)90282-0. [DOI] [PubMed] [Google Scholar]
- Frank J, Shimkin B, Dowse H. SPIDER - A Modular Software System for Electron Image Processing. Ultramicroscopy. 1981;6:343–358. [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. Journal of Structural Biology. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- Grigorieff N. Three-dimensional structure of bovine NADH:ubiquinone oxidoreductase (complex I) at 22 A in ice. Journal of Molecular Biology. 1998;277:1033–1046. doi: 10.1006/jmbi.1998.1668. [DOI] [PubMed] [Google Scholar]
- Guénebaut V, Vincentelli R, Mills D, Weiss H, Leonard KR. Three-dimensional Structure of NADH-dehydrogenase from Neurospora crassa by Electron Microscopy and Conical Tilt Reconstruction, in. Journal of Molecular Biology. 1997:409–418. doi: 10.1006/jmbi.1996.0753. [DOI] [PubMed] [Google Scholar]
- Guénebaut V, Schlitt A, Weiss H, Leonard K, Friedrich T. Consistent structure between bacterial and mitochondrial NADH:ubiquinone oxidoreductase (complex I) Journal of Molecular Biology. 1998;276:105–112. doi: 10.1006/jmbi.1997.1518. [DOI] [PubMed] [Google Scholar]
- Hinchliffe P, Sazanov LA. Organization of iron-sulfur clusters in respiratory complex I. Science. 2005;309:771–774. doi: 10.1126/science.1113988. [DOI] [PubMed] [Google Scholar]
- Hirst J. Energy transduction by respiratory complex I--an evaluation of current knowledge. Biochem Soc Trans. 2005;33:525–529. doi: 10.1042/BST0330525. [DOI] [PubMed] [Google Scholar]
- JBB . Special Issue. Journal of Bioenergetics and Biomembranes. 2001;53:155–266. [Google Scholar]
- Kashani-Poor N, Kerscher S, Zickermann V, Brandt U. Efficient Large Scale Purification of his-tagged Proton Translocating NADH:Ubiquinone oxireductase (complex I) from the Strictly Aerobic Yeast Yarrowia lipolytica. Biochimica et Biophysica Acta. 2001;1504:363–370. doi: 10.1016/s0005-2728(00)00266-8. [DOI] [PubMed] [Google Scholar]
- Kerscher S, Dröse S, Zwicker K, Zickermann V, Brandt U. Yarrowia lipolytica, a yeast genetic system to study mitochondrial complex I. Biochimica et Biophysica Acta. 2002;1555:83–91. doi: 10.1016/s0005-2728(02)00259-1. [DOI] [PubMed] [Google Scholar]
- Kerscher SJ, Eschemann A, Okun PM, Brandt U. External alternative NADH:ubiquinone oxidoreductase redirected to the internal face of the mitochondrial inner membrane rescues complex I deficiency in Yarrowia lipolytica. Journal of Cell Science. 2001;114:3915–3921. doi: 10.1242/jcs.114.21.3915. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Mamedova AA, Holt PJ, Carroll J, Sazanov LA. Substrate-induced conformational change in bacterial complex I. J Biol Chem. 2004;279:23830–23836. doi: 10.1074/jbc.M401539200. [DOI] [PubMed] [Google Scholar]
- Marabini R, Masegosa IM, San Martin MC, Marco S, Fernandez JJ, de la Fraga LG, Vaquerizo C, Carazo JM. Xmipp: An Image Processing Package for Electron Microscopy. Journal of Structural Biology. 1996;116:237–240. doi: 10.1006/jsbi.1996.0036. [DOI] [PubMed] [Google Scholar]
- Marco S, Chagoyen M, de la Fraga LG, Carazo JM, Carrascosa JL. A Variant to the Random Approximation of the Reference-free Alignment Algorithm. Ultramicroscopy. 1996;66:5–10. [Google Scholar]
- Mathiesen C, Hägerhäll C. Transmembrane topology of the NuoL, M and N subunits of NADH:quinone oxidoreductase and their homologues among membrane-bound hydrogenases and bona fide antiporters. Biochimica et Biophysica Acta. 2002;1556:121–132. doi: 10.1016/s0005-2728(02)00343-2. [DOI] [PubMed] [Google Scholar]
- Meloa AMP, Loboa SAL, Sousaa FL, Fernandesa AS, Pereiraa MM, Hreggvidssond GO, Kristjanssond JK, Saraivaa LM, Teixeiraa M. A nhaD Na+/H + antiporter and a pcd homologues are among the Rhodothermus marinus complex I genes. Biochimica et Biophysica Acta. 2005:95–103. doi: 10.1016/j.bbabio.2005.06.003. [DOI] [PubMed] [Google Scholar]
- Ohnishi T, Sled VD, Yano T, Yagi T, Burbaev DS, Vinogradov AD. Structure-function studies of iron-sulfur clusters and semiquinones in the NADH-Q oxidoreductase segment of the respiratory chain. Biochim Biophys Acta. 1998;1365:301–308. doi: 10.1016/s0005-2728(98)00082-6. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A Visualization System for Exploratory Research and Analysis. Journal of Computational Chemistry. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Radermacher M. Max-Planck-Institut für Biochemie. Technische Universität München; Martinsried/FRG: 1980. Dreidimensional Rekonstruktion bei kegelförmiger Kippung im Elektronenmikroskop. [Google Scholar]
- Radermacher M. Three-dimensional reconstruction of single particles from random and nonrandom tilt series. Journal of Electron Microscopy Technique. 1988;9:359–394. doi: 10.1002/jemt.1060090405. [DOI] [PubMed] [Google Scholar]
- Radermacher M. Three-dimensional reconstruction from random projections: orientational alignment via Radon transforms. Ultramicroscopy. 1994;53:121–136. doi: 10.1016/0304-3991(94)90003-5. [DOI] [PubMed] [Google Scholar]
- Radermacher M. Radon transform techniques for alignment and 3D reconstruction from random projections. Scanning Microscopy. 1997;11:171–177. [Google Scholar]
- Radermacher M, Hoppe W. Properties of 3D Reconstruction from Projections by Conical Tilting Compared to Single Axis Tilting. In: Brederoo P, Boom G, editors. 7th European Congress on Electron Microscopy. Seventh European Congress on Electron Microscopy Foundation: The Hague, Netherlands; 1980. pp. 132–133. [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. A New 3-Dimensional Reconstruction Scheme Applied to the 50s Ribosomal Subunit of E.Coli. Journal of Microscopy. 1986;141:Rp1–Rp2. doi: 10.1111/j.1365-2818.1986.tb02693.x. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Wagenknecht T, Verschoor A, Frank J. Three-dimensional reconstruction from a single-exposure, random conical tilt series applied to the 50S ribosomal subunit of Escherichia coli. Journal of Microscopy. 1987;146:113–136. doi: 10.1111/j.1365-2818.1987.tb01333.x. [DOI] [PubMed] [Google Scholar]
- Radermacher M, Ruiz T, Wieczorek H, Grueber G. The structure of the V1-ATPase determined by three-dimensional electron microscopy of single particles. Journal of Structural Biology. 2001;135:26–37. doi: 10.1006/jsbi.2001.4395. [DOI] [PubMed] [Google Scholar]
- Rais I, Karas M, Schagger H. Two-dimensional electrophoresis for the isolation of integral membrane proteins and mass spectrometric identification. Proteomics. 2004;4:2567–2571. doi: 10.1002/pmic.200400829. [DOI] [PubMed] [Google Scholar]
- Rasmussen T, Scheide D, Brors B, Kintscher L, Weiss H, Friedrich T. Identification of two tetranuclear FeS clusters on the ferredoxin-type subunit of NADH:ubiquinone oxidoreductase (complex I) Biochemistry. 2001;40:6124–6131. doi: 10.1021/bi0026977. [DOI] [PubMed] [Google Scholar]
- Ruiz T, Radermacher M. Three-dimensional analysis of single particles by electron microscopy: sample preparation and data acquisition., in. In: Taatjes DJ BTM, editor. In Methods in Molecular Biology: Cell imaging techniques: methods and protocols. Humana Press Inc.; Totowa: 2006. [DOI] [PubMed] [Google Scholar]
- Ruiz T, Mechin I, Bar J, Rypniewski W, Kopperschlager G, Radermacher M. The 10.8-A structure of Saccharomyces cerevisiae phosphofructokinase determined by cryoelectron microscopy: localization of the putative fructose 6-phosphate binding sites. Journal of Structural Biology. 2003;143:124–134. doi: 10.1016/s1047-8477(03)00140-0. [DOI] [PubMed] [Google Scholar]
- Sazanov LA, Walker JE. Cryo-electron crystallography of two sub-complexes of bovine complex I reveals the relationship between the membrane and peripheral arms. J Mol Biol. 2000:455–464. doi: 10.1006/jmbi.2000.4079. [DOI] [PubMed] [Google Scholar]
- Sazanov LA, Carroll J, Holt P, Toime L, M FI. A Role for Native Lipids in the Stabilization and Two-dimensional Crystallization of the Escherichia coli NADH-Ubiquinone Oxidoreductase (Complex I) Journal of Biological Chemistry. 2003;278:19483–19491. doi: 10.1074/jbc.M208959200. [DOI] [PubMed] [Google Scholar]
- Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Analytical Biochemistry. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Stoops JK, Kolodziej SJ, Schroeter JP, Bretaudiere JP, Wakil SJ. Structure-function relationships of the yeast fatty acid synthase: negative-stain, cryo-electron microscopy, and image analysis studies of the end views of the structure. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6585–6589. doi: 10.1073/pnas.89.14.6585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heel M. Angular reconstitution: a posteriori assignment of projection directions for 3D reconstruction. Ultramicroscopy. 1987;21:111. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- van Heel M, Frank J. Use of multivariate statistics in analysing the images of biological macromolecules. Ultramicroscopy. 1981;6:187–194. doi: 10.1016/0304-3991(81)90059-0. [DOI] [PubMed] [Google Scholar]
- Wagenknecht T, Radermacher M. Three-dimensional architecture of the skeletal muscle ryanodine receptor. FEBS Letters. 1995;369:43–46. doi: 10.1016/0014-5793(95)00581-s. [DOI] [PubMed] [Google Scholar]
- Zickermann V, Bostina M, Hunte C, Ruiz T, Radermacher M, Brandt U. Functional Implications from an Unexpected Position of the 49-kDa Subunit of NADH:Ubiquinone Oxidoreductase. The Journal of Biological Chemistry. 2003;278:29072–29078. doi: 10.1074/jbc.M302713200. [DOI] [PubMed] [Google Scholar]


