Abstract
Human bystin is a cytoplasmic protein directly binding to trophinin, a cell adhesion molecule potentially involved in human embryo implantation. The present study shows that bystin is expressed in luminal and glandular epithelia in the mouse uterus at peri-implantation stages. In fertilized embryos, bystin was not seen until blastocyst stage. Bystin expression started during hatching and increased in expanded blastocyst. However, bystin disappeared from the blastocyst during implantation. After implantation bystin re-appeared in the epiblast. Targeted disruption of the mouse bystin gene, Bysl, resulted in embryonic lethality shortly after implantation, indicating that bystin is essential for survival of mouse embryos.
1. Introduction.
Bystin was originally identified in a human trophoblastic cell as a protein that forms a complex with trophinin and tastin [1]. Trophinin is an intrinsic membrane protein that mediates cell adhesion by homophilic trophinin-trophinin binding [1-6]. Tastin and bystin are cytoplasmic proteins required for trophinin to function as an efficient cell adhesion molecule. In humans, trophinin, tastin and bystin are expressed in the trophoblast and endometrial luminal epithelial cells at the utero-placental interphase or implantation site [4]. These proteins are found in the human placenta in early stages of pregnancy, but they disappear from the placenta later than 10 weeks pregnancy [4].
Genes encoding trophinin and tastin are only found in mammals [2, 7]. By contrast, the bystin gene, Bysl, is conserved across a wide range of eukaryotes including yeast, nematodes, insects, snakes, and mammals [1, 8-11]. The yeast Bysl orthologue, ENP1 or essential nuclear protein 1, is essential for vegetative growth of yeast [11]. Studies of Enp1 show that it plays a role in pre-ribosomal RNA splicing and ribosome biogenesis [12]. In Drosophila the bys gene, which is localized upstream of the ribosomal S6 kinase gene (hence termed “by S6”), is implicated in cell growth [9, 10]. In Drosophila embryos, bys mRNA is expressed in a tissue-specific manner during gastrulation [13]. In the larval wing disc, bys mRNA is expressed in the ventral and dorsal regions of the wing pouch, regions that give rise to epithelia and that adhere to one another after the wing disc everts. These studies suggest that bystin is generally involved in cell growth but also in cell adhesion, particularly in metazoans.
To determine the in vivo role of bystin in mouse embryo implantation, we analyzed the expression pattern of bystin protein in the mouse embryos and the uterus in the peri-implantation period. We also inactivated the Bysl gene in the mouse, and found that Bysl null embryos could implant in the uterus but failed to survive beyond embryonic day 6 (E6).
2. Materials and Methods.
2.1 Mouse bystin antibodies.
Rabbit antibodies were raised against a synthetic peptide, MEKLTEKQTEVETVC, in which cysteine was added for conjugation to keyhole limpet hemocyanin. Antibodies were purified by affinity chromatography on a protein A Sepharose column followed by a peptide-conjugated agarose column. Antibodies bound to the peptide column were eluted with 3M guanidine-HCl and dialyzed against cold PBS.
2.2 Transfection and immunocytochemistry.
A mouse bystin IMAGE cDNA clone containing a 1.7 kb (clone ID 4925307) open reading frame encoding full-length, 50 kDa bystin in pCMVSPORT6 was obtained from Invitrogen (Carlsbad, CA). COS cells grown on glass cover slips were transfected with the vector using LipofectAmine (Invitrogen). Transfected cells were fixed in 100% methanol and subjected to immunoperoxidase analysis using the affinity purified rabbit anti-bystin antibody described above. Peroxidase substrate AEC single solution was used for the color reaction (Zymed, South San Francisco, CA).
2.3 Immunohistochemistry of mouse tissues and embryos.
Frozen uteri were sectioned (12 mm) and mounted onto poly-L-lysine-coated glass slides and fixed with cold methanol for 15 minutes. Pre-implantation embryos were recovered from uteri, placed on the positively charged slide glass, and centrifuged for 1 minute in a cytocentrifuge. The embryo attached to the slide glass was fixed with cold methanol for 10 minutes. Uterine regions containing implanted embryos were identified by an enlarged uterine wall or decidual reaction, and frozen in O. C. T compound (Tissue Tek, Torrance, CA). Each section was placed on a positively charged glass slide, fixed in cold methanol, and stained with anti-bystin antibody described above. Stained tissue sections were lightly counter-stained with hematoxylin.
2.4 Western blot analysis.
Mouse uterine tissue was homogenized in PBS containing 1% NP-40 and a protease inhibitor mix (Roche Applied Science, Indianapolis, IN) and centrifuged. Supernatant proteins were subjected to Western blot analysis as described [14], except that the antibody was diluted with Can-Get-Signal solution (Toyobo, Osaka, Japan) to enhance antibody-antigen binding. Alpha-tubulin, an internal standard, was detected using a mouse monoclonal antibody (Sigma, St Louis, MO).
2.5 Construction of targeting vector.
A genomic clone of mouse Bysl was isolated from a mouse genomic P1 plasmid library (Genome Systems, St. Louis, MO) by polymerase chain reaction (PCR) amplification using the forward primer 5′-TGGAGCCTGGTTCAAAGGTAGA-3′ and the reverse primer 5′-ATGCAGTGGCTTTTAGACCGG-3′. The plasmid DNA was digested with Hind III. A 9.3 kb fragment, which hybridized with a mouse bystin cDNA probe was subcloned at the Hind III site of pBluescript II KS+ for targeting vector construction. For positive selection a 2.9-kb EcoRV-XhoI fragment including Bysl exons 3-6 was replaced with a PGK-neo cassette in which the neomycin resistance gene is driven by the phosphoglycerate kinase promoter. The HSV-TK cassette including the Herpes simplex thymidine kinase gene was ligated at the 3′ end of the targeting vector for negative selection. Mouse embryonic stem (ES) D3 cells derived from 129/SvJ mice [15] were transfected with the linearized vector (20 mg) by electroporation at 400 V, 250 mF, in a GenePulser (BioRad, Hercules, CA). Transfected cells were selected in medium containing 150 mg/ml geneticin (GibcoBRL, Gaithersburg, MD) and 2mM ganciclovir as described [14]. Screening of 296 ES clones by Southern blot identified 11 homologous recombinants. Two clones showing a modal chromosome number of 40 were used to inject blastocysts, which were transferred to foster mothers to generate chimeric mice. Chimeras were mated with 129/SvJ mice and also with C57BL6 mice to obtain lines carrying a mutated Bysl allele. Mice were housed in the animal care facility at the Burnham Institute according to NIH and institutional guidelines concerning care and use of laboratory animals.
2.6 Southern blot of mouse genomic DNA.
Genomic DNA prepared from ES clones or mouse tail biopsies was digested with EcoRI and separated by electrophoresis on 0.7% agarose gels. DNAs were transferred to a NYTRAN PLUS membrane (Schleicher & Schuell, Keene, NH) and fixed under ultraviolet light (1,200 mjoules). A DNA fragment encoding the 5’ region outside of the targeting vector was labeled with [α-32P] dCTP and used as a probe. Filters were prehybridized at 35°C for 5 hours in 6 x SSPE containing 50% formamide, 5x Denhardt′s solution, 0.1% SDS, and 0.2 mg/ml denatured salmon sperm DNA, and then hybridized in the same solution containing the [32P]-labeled probe at 35°C for 20 hours. Filters were washed three times at 35°C for 10 minutes in 6X SSPE containing 0.1% SDS and subjected to autoradiography.
2.7. Genotyping of mice and mouse embryos.
Genotyping was carried out by PCR using genomic DNA isolated from tail biopsies as described [14] or from preimplantation embryo as described below. Primers for the Neo gene were: forward 5’-TAAGCTATCAGAGATGAAGT-3’ and reverse 5’-GCTAATACCAGCACCATCAC-3’. Amplifications were carried out in a Thermal Cycler (Perkin Elmer, model 2400) by denaturation at 94°C for 10 min, followed by 35 cycles for tail or 55 cycles for embryo of denaturation at 94°C for 30 sec, annealing at 58°C for 60 sec, extension at 72°C for 45 sec, and further extension at 72°C for 10 min.
Analysis of the Bysl gene in pre-implantation embryos was carried out by nested PCR directed at the sequence within exon 5. Primers for the first PCR were: forward, 5’-CTTCCCCGAGTGCGAGATGACATT-3’ and reverse, 5’-TCCATTCGCACCCTCACGACCA-3’. Amplifications were carried out by denaturation at 94°C for 3 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 sec, extension at 72°C for 30 sec, and further extension at 72°C for 7 min. Nested PCR was carried out using the following primers: forward 5’-AAGGTCATCCCTGCACTGTC-3’ and reverse 5’-CAGCAATGTCATCTCGCACT-3’. Amplification was carried out in the same conditions as the first PCR reaction, except that annealing was at 58°C. Reaction products were separated on agarose gels and stained with ethidium bromide. DNA bands were visualized under UV light and photographed.
Genomic DNA from pre-implantation stage embryos was prepared as follows. Timed pregnancies were determined defining E0.5 as the morning of plug. Embryos were flushed from the pregnant female uterus at E2.5, E3.5, and E4.5. Each embryo was placed in a tube containing 20 ml PBS, mixed with 20 ml DexPat (Pan-Vera, Madison, WI), boiled for 10 min, and centrifuged to remove DexPat. Each supernatant was mixed with 4 ml 3M Na-acetate and 100 ml ethanol and left at -20 °C for 20 hours to precipitate genomic DNA.
Genotypings of E5.5, E6.5, and E7.5 embryos were carried out using genomic DNA isolated from frozen sections of implanted embryos. The uterus showing positivity for the decidual reaction was frozen and sections were cut. Sections were placed on a slide, briefly stained with hematoxylin, and embryonic cells were collected using 21 gauge needles. Genomic DNA was prepared using DexPat as described above.
2.8 Adhesion of mouse blastocysts to HB-EGF-expressing cells.
Binding of mouse blastocysts to 32D cells, which express membrane bound form HB-EGF, was tested as described [16]. Briefly, Bysl heterozygotes were mated and blastocysts were flushed from the uteri of pregnant females on E4.5. Hatched and well-expanded blastocysts were placed in a Petri dish containing HT medium, and 32D cells (1×105 cells/ml) were added to the blastocysts. Each blastocyst was inspected for the binding of 32D cells under phase contrast microscope and scored.
3. Results.
3.1 Expression of bystin in mouse uterus and pre-implantation mouse embryos.
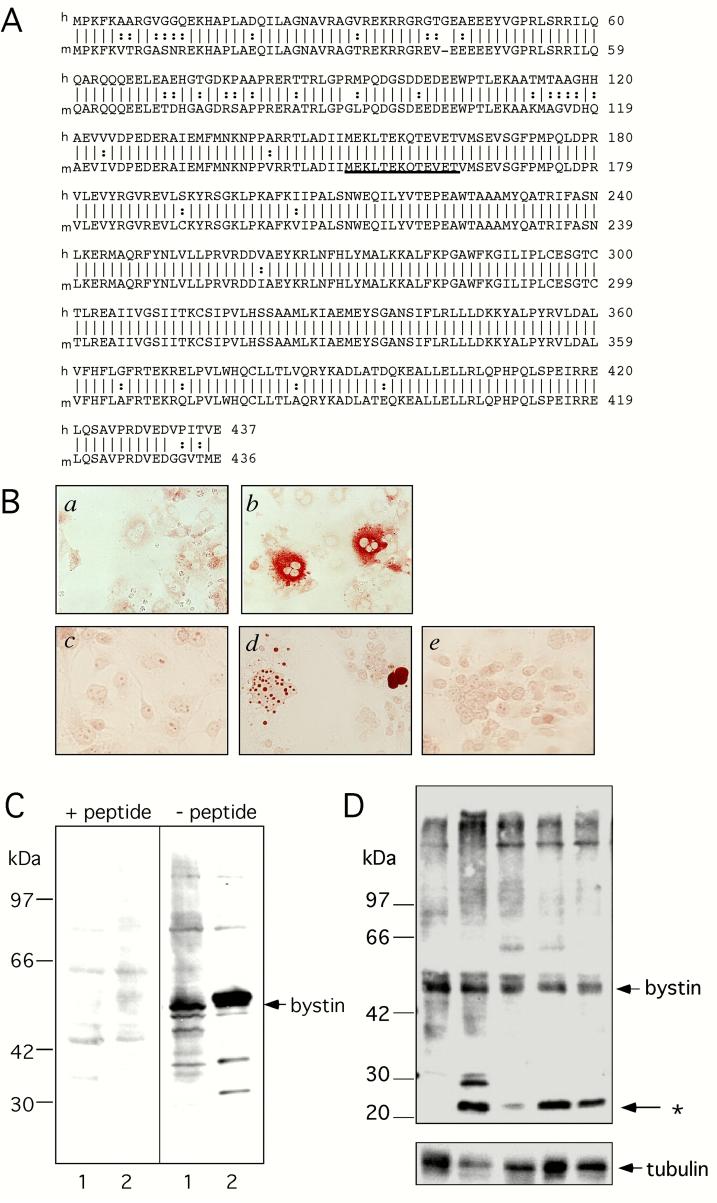
Fig. 1A shows the peptide sequences of full-length human and mouse bystins. A polyclonal antibody was raised against a synthetic peptide of underlined sequence. COS cells were transfected with a mammalian expression vector encoding the full-length mouse bystin cDNA and reacted with affinity purified anti-bystin antibody. The antibody did not react or very weakly reacted with mock-transfected COS cells (Fig. 1B-a). Under normal culture conditions, presumably when most COS cells are at G1 phase, bystin was detected in the cytoplasm (Fig. 1B-b). When transfected COS cells were arrested at G1 phase by serum starvation following stimulation with serum to induce mitosis, immunostaining was seen in nuclei and nucleoli (Fig. 1B-d). This observation suggests that bystin enters the nucleus before mitosis and moves to nucleoli. In the presence of the peptide used for immunogen, immunocytochemistry of transfected COS cells with the same antibody showed no staining (Fig. 1B-e). These results verified the specificity of anti-bystin antibody in immunocytochemistry used in this study.
Fig. 1.
Characterization of anti-bystin antibody by immunocytochemistry and Western blot. A, Peptide sequences of full-length human (upper) and mouse (lower) bystin proteins. The underlined sequence was used for antibody production. B. Immunohistochemistry of COS cells transfected with vector alone (a, c) or a vector encoding mouse bystin (b, d, e). Cells were reacted with an anti-bystin antibody with (e) or without (a-d) the peptide used as immunogen. C, Western blot analysis of FLAG-tagged human bystin. Each lane represents an untransfected (lane 1) and a transfected (lane 2) 293T cell lysate. The filter was reacted with an anti-bystin antibody with (left) or without (right) the epitope peptide. D. Western blot analysis of mouse uterus tissues. Lanes 1 and 2: non-pregnant. Lanes 3-5, pregnant day 3.5 (lane 3), day 4.5 (lane 4) and day 5.5 (lane 6).
Western blot analysis of 293T cells detected a 50 kDa band, which became undetectable after pre-incubation of the antibody with the peptide used for immunogen (Fig. 1C, lanes 1). Immunoblots from lysates of 293T cells transfected with cDNA encoding FLAG-tagged human bystin showed a strong band of slightly lower mobility than endogenous bystin, which became undetectable after pre-incubation with the peptide (Fig. 1C, lanes 2). These results indicate that the antibody detects both human and mouse bystin proteins. Western blot analysis of mouse uterus with the same antibody showed a 50kDa band and additional bands (Fig. 1D). When the antibody was pre-incubated with the peptide used for immunogen, the 50kDa and 25 kDa bands disappeared (data not shown), suggesting that they represent bystin. The presence of 50 kDa bystin indicates that human bystin that we identified previously was truncated and lacked part of the 5’ region, as commented on by Chen et al., [13]. It is presently unknown whether 25kDa band shown by an asterisk in Fig. 1D represents a bystin variant or a fragment produced by proteolysis. Nonetheless, the result suggests that bystin is expressed in both non-pregnant and pregnant mouse uteri.
Immunohistochemistry of non-pregnant mouse uteri showed signals in the luminal epithelium and glandular epithelium (Fig. 2A-a). When the same antibody was incubated with the peptide used for the immunogen, signals disappeared (data not shown), strongly suggesting that staining represents bystin. Bystin was seen in vacuole-like vesicles at the luminal and abluminal sides of the epithelia. By contrast, immunostaining of pregnant uteri showed uniform cytoplasmic staining with greater signals at the apical side of the luminal epithelia (Fig. 2A-b). Glandular epithelia were uniformly stained with the antibody. These observations suggest that the presence of embryos affects bystin localization in these epithelial cells.
Fig. 2.
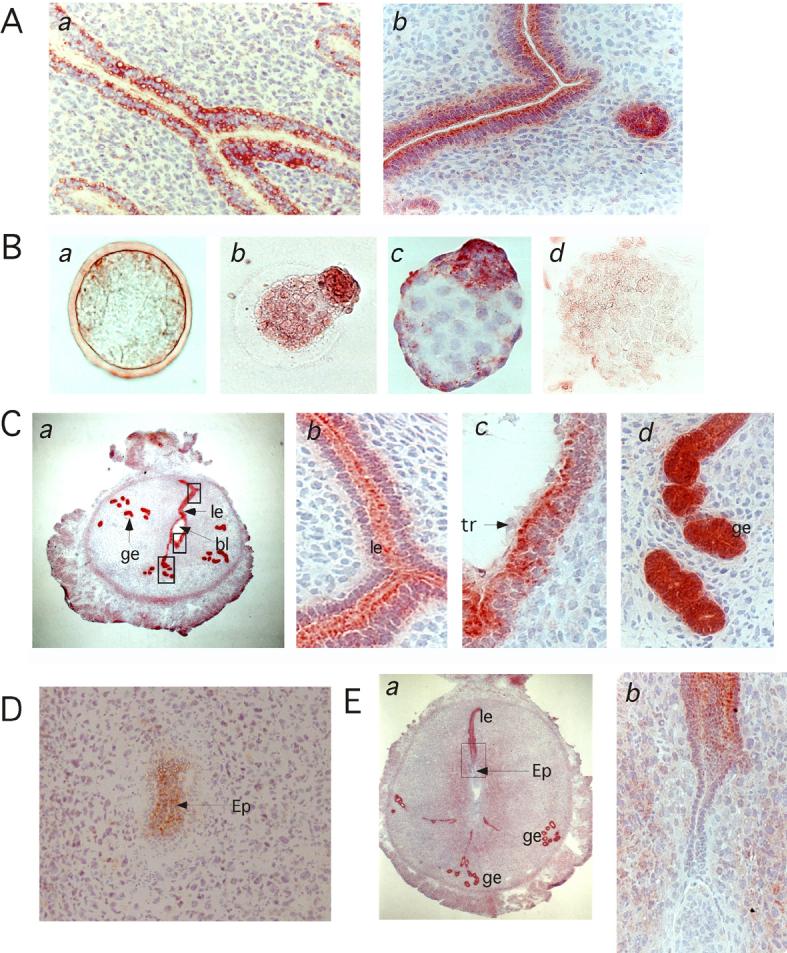
Immunohistochemistry of mouse uteri and embryos before, during and after implantation. A, mouse uterus from non-pregnant (a) and 4.5 day pregnant (b) mice. B, Pre-implantation stage mouse embryos. Morulae (a), blastocysts during hatching (b), a hatched and expanded blastocyst (c), and a hatched and one day cultured blastocyst (d). C, Day 4.5 pregnant mouse uterus with implanting blastocyst. Bystin was seen in the uterine luminal epithelial cells (le) and glandular epithelial cells (ge), at low (a) and high (b-d) magnification. Note that in luminal epithelial cells, bystin localizes to the apical side (b). In luminal epithelial cells adjacent to the blastocyst, bystin localizes to the apical side, whereas trophectoderm cells do not show bystin signals (c). Glandular epithelia cells strongly express bystin, whereas no polarization of bystin staining is seen in these cells (d). D, Implanted embryo at the epiblast stage or at day 5.5. E. Implanted embryo and uterus at day 6.5. ge, glandular epithelia; le, luminal epithelia; bl, blastocyst; tr, trophectoderm; ep, epiblast.
Immunocytochemistry of fertilized mouse eggs from the single to the 16-cell stage showed no antibody signals (data not shown). Weak signals were detected in morula stages (Fig. 2B-a). Bystin immunostaining increased in the blastocyst during hatching (Fig. 2B-b). Hatched and expanded blastocysts were strongly positive for bystin (Fig. 2B-c). When a hatched blastocyst was cultured in vitro for one day, bystin staining was reduced (Fig. 2B-d). In extended cultures on fibronectin-coated plate, bystin was not found in blastocyst-derived cells (data not shown). These results show that bystin is expressed in pre-implantation stage mouse embryos, but that bystin disappears from the blastocyst after culturing in vitro.
During blastocyst implantation, uterine epithelial cells surrounding the blastocyst showed strong bystin signals (Fig. 2C-abc). Glandular epithelial cells also express bystin, whereas bystin was uniformly localized in the cytoplasm (Fig. 2C-d). In implanting embryos, bystin was not detected in trophectoderm cells. Trophoblastic giant cells and ectoplacental cone were also negative for bystin (Fig. 2C-c). In luminal epithelial cells adjacent to the blastocyst, bystin was localized to the apical side of the epithelium (Fig. 2C-c).
Shortly after implantation in E5.5 embryos, bystin appeared in the epiblast (Fig. 2D), in contrast to the loss of bystin seen in extended blastocyst culture in vitro. However, bystin was not seen in E6.5 embryos (Fig. 2E-ab). Also on E6.5, bystin disappeared from the luminal epithelia surrounding the embryo, whereas luminal epithelia or glandular epithelia distant from the implantation site continued to express bystin. Immunohistochemistries of mouse embryos older than E15 and adult mice showed negative or weak signals in a variety of organs (data not shown), suggesting that bystin is expressed widely and weakly in a variety of cell types in the mouse embryos and adult mice.
3.2 Targeted disruption of Bysl gene in the mouse.
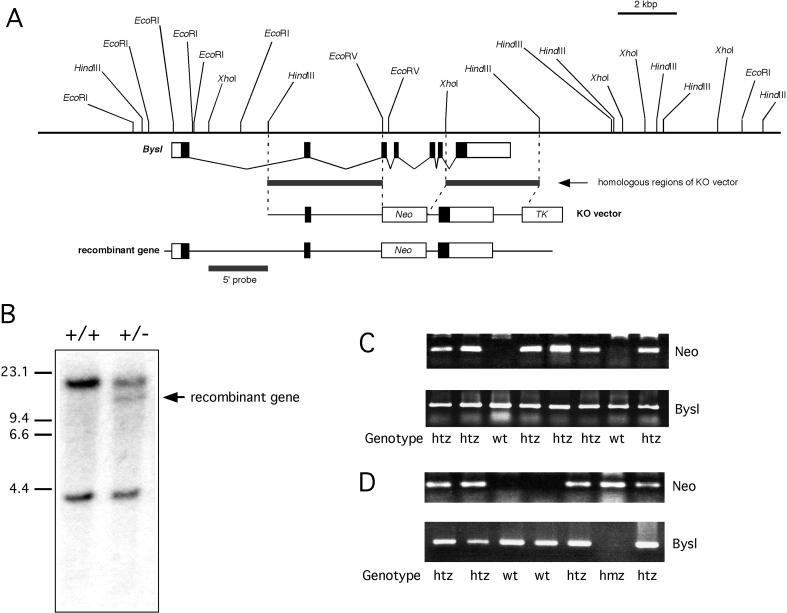
To determine the in vivo role of bystin in the mouse, Bysl was disrupted by homologous recombination (Fig. 3A). Since bystin contains many potential phosphorylation sites for protein kinases, we replaced these potentially modified regions, which were encoded primarily by exons 3-6, with the Neo gene. Following transfection of ES cells derived from the 129/SvJ strain [16] with the target vector, no morphological or proliferative abnormalities were observed in homologously recombined Bysl (+/-) ES clones (Fig. 3B) under normal culture conditions.
Fig. 3.
Production of Bysl gene knockout mouse. A, Genomic DNA map of the Bysl gene, and the Bysl targeting vector. B, Southern blot of homologous recombinant ES clones. Genomic DNA digested by EcoRI was probed by the 5’ DNA probe shown in A. C, Genotyping by PCR for Neo gene and Bysl gene of genomic tail DNA from pups produced from the cross between Bysl (+/-) parents. D, Genotyping by PCR for Neo gene and Bysl gene of mouse embryos produced from Bysl(+/-) parents.
Two Bysl (+/-) ES clones were used for the blastocyst injection. Chimeric males were back-crossed with SvJ129 female mice and with C57BL/6 mice. Genotyping of more than 2,000 3 week-old pups (Fig. 3C) from two Bysl gene knockout lines showed no Bysl null offspring under either genetic background.
Genotyping of pre-implantation stage embryos (Fig. 3D) recovered from uteri revealed the presence of Bysl(-/-) embryos (Table 1). In these embryos, no abnormal morphologies were seen. Furthermore all blastocysts showed positive binding by 32D cells expressing a membrane bound form of HB-EGF [16], indicating that deletion of a large part of the Bysl gene did not affect adhesion of a blastocyst through the EGF receptor, suggesting that loss of bystin does not affect binding between the EGF receptor and HB-EGF. Genotyping of post implantation E8.5 embryos revealed a reduction in the number of Bysl(-/-) embryos, suggesting that lethality occurred after E3.5 and before E8.5 (Table 1).
Table I.
Genotyping of offspring resulted from the cross between Bysl+/- females and Bysl+/- males.
| Developmental stages. | Genotypes | |||
|---|---|---|---|---|
| +/+ | +/- | -/- | ||
| E2.5 | 8 | 15 | 5 | |
| E3.5 | 11 | 23 | 13 | |
| E4.5 morulae | 13 | 20 | 9 | |
| E4.5 blastocyst | 8 | 16 | 9 | |
| E5.5 | 6 | 11 | 3 | 4 e. d.1 |
| E6.5 | 2 | 8 | 2 | 4 e. d. |
| E7.5 | 7 | 15 | 1 | 5 e. d. |
| E8.5 | 1 | 4 | 32 | 1 e. d. |
| E10.5 | 10 | 25 | 0 | |
| Post natal 3 weeks | 48 | 76 | 0 | |
Empty deciduas. Genotype of embryos not available.
Embryos were degenerated
Bysl(+/-) females mated with Bysl(+/-) males showed no macroscopic signs of dead embryos in the uterus, suggesting embryonic lethality occurred before embryonic growth taking place or shortly after implantation. Morphological examination of frozen sections of mouse uterus showed degenerated embryos at E6.5, and their genotypes were identified as Bysl(-/-) (Fig. 4). These observations strongly suggest that Bysl(-/-) embryos die after implantation, at around E5.5-6.5.
Fig. 4.
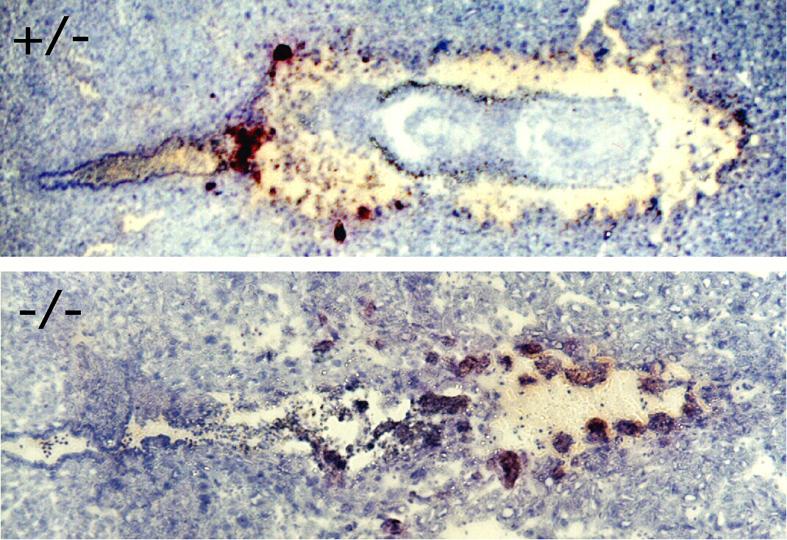
Histology of implanted E6.5 mouse embryos resulted from Bysl(+/-) parents. A pair of litter mates with each genotype Bysl(+/-) (upper) and Bysl(-/-) (lower) embryos is shown. Note that Bysl (-/-) embryo was degenerated. Tissue sections were stained with hematoxylin.
4. Discussion.
The present study demonstrates that Bysl is essential for mouse embryo survival. Bysl null embryos survive and develop during pre-implantation stages (Table 1). Global gene expression analysis of delayed implantation model identified genes differentially expressed between dormant and activated blastocysts, and Bysl was one of the genes activated by estrogen [17]. However, successful implantation of bystin null embryos and absence of bystin protein in the trophectoderm of an implanting blastocyst (Fig. 2C-c) suggest that bystin does not play an essential role in the initial attachment of a blastocyst to the uterine epithelia in the mouse. Given that trophinin null mouse embryos also implant [14], the present study indicates that neither trophinin nor bystin play a significant role in this process in the mouse. The present study, however, clearly shows that bystin is essential for mouse embryo survival after implantation. Similar embryonic lethality but with incomplete penetrance was detected in trophinin null embryos [14], suggesting that trophinin and bystin may function together after implantation.
Bystin’s role in mammalian cell growth was previously suggested by a study of brain injury in rat [18], which showed that bystin is overexpressed in reactive astrocytes after injury. The embryonic lethality of Bysl null embryos and bystin expression in the epiblast shown in this study (Fig. 2D) strengthen the hypothesis that bystin is essential for cell growth in mammals, as epiblast or embryonic stem cells should proliferate rapidly after implantation.
Bystin expression in the epiblast (Fig. 2D) is consistent with a report showing that Bysl is one of 216 genes commonly expressed in embryonic, neural and hematopoietic stem cells. Thus Bysl is a specific stem cell marker. Bysl is also included in a gene cluster of stem cell markers on mouse chromosome 16 [19]. The present study therefore supports the idea that Bysl may play an important role in embryonic stem cells.
Studies of ribosomal biogenesis in yeast have shown that the bystin orthologue Enp1 is required for synthesis of 40S ribosomal subunits by functioning in their nuclear export [12, 20, 21]. Formation of eukaryotic ribosomes occurs predominantly in the nucleoli, but late maturation steps take place in both the nucleoplasm and cytoplasm [22]. The presence of bystin in the cytoplasm during G1 phase (Fig. 1B-b) and in the nucleus in G2 phase (Fig. 1B-d) suggests a role of bystin in cellular processes following mitosis, such as ribosomal biogenesis in nucleoli, as has been reported for ENP1, the bystin yeast orthologue [11, 12, 19, 20]. The observation of bystin in the nucleoli (Fig. 1B-d) supports this hypothesis.
Although information relevant to the function of human BYSL is limited except for its potential involvement in human embryo implantation, the publicly available database indicates that its expression is elevated in many human cancers. In particular, bystin is overexpressed in prostate cancer cells at perineural invasion [23]. Microarray analysis also identified tastin, another trophinin-associated protein, as a prominently overexpressed gene in prostate cancer [24]. Perineural invasion likely provides a microenvironment conferring a survival advantage for prostate cancer cells. Furthermore, network reverse engineering, which enables indentification of co-regulated genes from genome-wide expression profiles identified BYSL as the most co-regulated gene to the Myc proto-oncogene in humans [25]. Since overexpression of c-Myc in the mouse prostate caused prostate cancer [26], bystin’s function in cell proliferation may be closely connected with c-Myc.
In higher organisms, many cellular functions are controlled by cell-cell interactions. It is possible that bystin participates in cytoplasmic signaling in a manner sensitive to cell adhesion. It is tempting to speculate that some human cancers may be promoted by a mechanism similar to trophoblast invasion and proliferation in embryo implantation. Further studies on bystin as well as trophinin and tastin will provide a better understanding of these processes and may suggest therapeutic strategies against human cancers.
Acknowlegement.
This study was supported by NIH grant HD34108 and DoD grant W81XWH-04-1-0917. The authors thank Dr. Elise Lamar for editing the manuscript, and Shuk-Man Wong for technical assistance.
References.
- [1].Suzuki N, Zara J, Sato T, Ong E, Bakhiet N, Oshima RG, Watson KL, Fukuda MN. A novel cytoplasmic protein, bystin, interacts with trophinin, tastin and cytokeratin, and may be involved in trophinin mediated cell adhesion between trophoblast and endometrial epithelial cells. Proc. Natl. Acad. Sci. USA. 1998;95:5027–5032. doi: 10.1073/pnas.95.9.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Fukuda MN, Sato T, Nakayama J, Klier G, Mikami M, Aoki D, Nozawa S. Trophinin and tastin, a novel cell adhesion molecule complex with potential involvement in embryo implantation. Genes Dev. 1995;9:1199–1210. doi: 10.1101/gad.9.10.1199. [DOI] [PubMed] [Google Scholar]
- [3].Fukuda MN, Nozawa S. Trophinin, tastin, and bystin: a complex mediating unique attachment between trophoblastic and endometrial epithelial cells at their respective apical cell membranes. Sem. Reprod. Endocrinol. 1999;17:229–34. doi: 10.1055/s-2007-1016230. [DOI] [PubMed] [Google Scholar]
- [4].Suzuki N, Nakayama J, Shih IM, Aoki D, Nozawa S, Fukuda MN. Expression of trophinin, tastin, and bystin by trophoblast and endometrial cells in human placenta. Biol. Reprod. 1999;60:621–7. doi: 10.1095/biolreprod60.3.621. [DOI] [PubMed] [Google Scholar]
- [5].Nakayama J, et al. Implantation-Dependent Expression of Trophinin by Maternal Fallopian Tube Epithelia during Tubal Pregnancies: Possible Role of Human Chorionic Gonadotrophin on Ectopic Pregnancy. Am. J. Pathol. 2003;163:2211–9. doi: 10.1016/S0002-9440(10)63579-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Aoki R, Fukuda MN. Recent Molecular approaches to elucidate the mechanism of embryo implantation: trophinin, bystin and tastin as molecules invoved in the initial attachment of blastocyst to the uterus in humans. In: Giudice LC, Dey SK, editors. Sem. Reprod. Med., “Implantation”. Thieme Medical Publishers; New York: 2000. p. 265271. [DOI] [PubMed] [Google Scholar]
- [7].Pack S, Tanigami A, Ledbetter DH, Sato T, Fukuda MN. Assignment of trophoblast/endometrial epithelium cell adhesion molecule trophinin gene TRO to human chromosome bands Xp11.22→p11.21 by in situ hybridization. Cytogenet. Cell Genet. 1997;79:123–124. doi: 10.1159/000134698. [DOI] [PubMed] [Google Scholar]
- [8].Trachtulec Z, Forejt J. Synteny of orthologous genes conserved in mammals, snake, fly, nematode, and fission yeast. Mamm. Genome. 2001;12:227–31. doi: 10.1007/s003350010259. [DOI] [PubMed] [Google Scholar]
- [9].Stewart MJ, Denell R. Mutations in the Drosophila gene encoding ribosomal protein S6 cause tissue overgrowth. Mol. Cell. Biol. 1993;13:2524–2535. doi: 10.1128/mcb.13.4.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Watson KL, Konrad KD, Woods DF, Bryant PJ. Drosophila homolog of the human S6 ribosomal protein is required for tumor suppression in the hematopoietic system. Proc. Natl. Acad. Sci. USA. 1992;89:11302–11306. doi: 10.1073/pnas.89.23.11302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Roos J, Luz JM, Centoducati S, Sternglanz R, Lennarz WJ. ENP1, an essential gene encoding a nuclear protein that is highly conserved from yeast to humans. Gene. 1997;185:137–146. doi: 10.1016/s0378-1119(96)00661-0. [DOI] [PubMed] [Google Scholar]
- [12].Chen W, Bucaria J, Band DA, Sutton A, Sternglanz R. Enp1, a yeast protein associated with U3 and U14 snoRNAs, is required for pre-rRNA processing and 40S subunit synthesis. Nucleic Acids Res. 2003;31:690–9. doi: 10.1093/nar/gkg145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Stewart MJ, Nordquist EK. Drosophila Bys is nuclear and shows dynamic tissue-specific expression during development. Dev. Genes Evol. 2005;215:97–102. doi: 10.1007/s00427-004-0447-8. [DOI] [PubMed] [Google Scholar]
- [14].Nadano D, et al. Significant differences between mouse and human trophinins are revealed by their expression patterns and targeted disruption of mouse trophinin gene. Biol. Reprod. 2002;66:313–21. doi: 10.1095/biolreprod66.2.313. [DOI] [PubMed] [Google Scholar]
- [15].Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J. Embryol. Exp. Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- [16].Raab G, Kover K, Paria BC, Dey SK, Ezzell RM, Klagsbrun M. Mouse preimplantation blastocysts adhere to cells expressing the transmembrane form of heparin-binding EGF-like growth factor. Development. 1996;122:637–45. doi: 10.1242/dev.122.2.637. [DOI] [PubMed] [Google Scholar]
- [17].Hamatani T, Daikoku T, Wang H, Matsumoto H, Carter MG, Ko MS, Dey SK. Global gene expression analysis identifies molecular pathways distinguishing blastocyst dormancy and activation. Proc. Natl. Acad. Sci. U S A. 2004;101:10326–31. doi: 10.1073/pnas.0402597101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sheng J, et al. Bystin as a novel marker for reactive astrocytes in the adult rat brain following injury. Eur. J. Neurosci. 2004;20:873–84. doi: 10.1111/j.1460-9568.2004.03567.x. [DOI] [PubMed] [Google Scholar]
- [19].Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 2002;298:597–600. doi: 10.1126/science.1072530. [DOI] [PubMed] [Google Scholar]
- [20].Grandi P, et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–15. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- [21].Schafer T, Strauss D, Petfalski E, Tollervey D, Hurt E. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. Embo J. 2003;22:1370–80. doi: 10.1093/emboj/cdg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kressler D, Linder P, de La Cruz J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol. Cell Biol. 1999;19:7897–912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ayala GE, Dai H, Li R, Ittmann M, Thompson TC, Rowley D, Wheeler TM. Bystin in perineural invasion of prostate cancer. Prostate. 2006;66:266–72. doi: 10.1002/pros.20323. [DOI] [PubMed] [Google Scholar]
- [24].Dhanasekaran SM, et al. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- [25].Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–90. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- [26].Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors Cancer. Cell. 2003;4:223–38. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]