Abstract
ISWI proteins form the catalytic core of a subset of ATP-dependent chromatin remodelling activities in eukaryotes from yeast to man. Many of these complexes have been found to reposition nucleosomes, but with different directionalities. We find that the yeast Isw1a, Isw2 and Chd1 enzymes preferentially move nucleosomes towards more central locations on short DNA fragments whereas Isw1b does not. Importantly, the inherent positioning properties of the DNA play an important role in determining where nucleosomes are relocated to by all of these enzymes. However, a key difference is that the Isw1a, Isw2 and Chd1 enzymes are unable to move nucleosomes to positions closer than 15 bp from a DNA end whereas Isw1b can. We also find that there is a correlation between the inability of enzymes to move nucleosomes close to DNA ends and the preferential binding to nucleosomes bearing linker DNA. These observations suggest that the accessibility of linker DNA together with the positioning properties of the underlying DNA play important roles in determining the outcome of remodelling by these enzymes.
Nucleosomes are the fundamental subunits of eukaryotic chromatin. The assembly of DNA into chromatin fulfils important functions in both packaging DNA into nuclei and regulating access to genetic information. Crystallographic structures of nucleosomes provide a detailed picture of how DNA is bound to the surface of the histone octamer (1). However, in solution nucleosomes exhibit dynamic properties that include the ability to spontaneously relocate to different positions on DNA fragments (2–4). The positioning of nucleosomes has the potential to positively or negatively regulate access to DNA and consequently all genetic processes.
In addition to undergoing spontaneous thermal movements, nucleosomes can be repositioned by ATP-dependent chromatin remodelling enzymes. These enzymes consist of a catalytic subunit with a region of homology to the yeast Snf2 protein and a variable number of accessory subunits. Snf2 family proteins fall into distinct subfamilies. For example the ISWI subfamily is named after its founding member, the Drosophila ISWI protein (5). The ISWI protein was subsequently found to be a component of several distinct protein complexes that have the ability to alter chromatin structure in an ATP-dependent reaction (6). Related ISWI complexes have since been identified in a broad spectrum of eukaryotes from yeast to humans (7). These complexes have been found to function in a range of processes ranging from the regulation of transcription, and DNA replication to the maintenance of chromatin structure (7).
Characterisation of ISWI driven chromatin remodelling reactions has revealed that one outcome is the repositioning of nucleosomes along DNA (8–10). Although the redistribution of nucleosomes may represent the major means by which these complexes alter chromatin structure, this is not necessarily the case for other subfamilies of Snf2 proteins which have been shown to cause other transitions in chromatin structure (11–15). In addition, there are differences in the way that different ISWI containing complexes redistribute nucleosomes. For example, while the Drosophila ISWI containing complex NURF and its isolated catalytic subunit redistributes nucleosomes to positions closely related to those observed in thermal nucleosome redistribution reactions (4,16), the ACF, CHRAC and Isw2 complexes have been reported to preferentially move nucleosomes to positions closer to the centre of short DNA fragments (9,17,18).
Yeast Chd1p represents another, less well characterised subfamily of remodellers which shows genetic interactions with ISWI factors (19,20). This subfamily is also represented in multicellular eukaryotes and in Drosophila and yeast appears to be mainly monomeric (21,22).
The differences in the directionality with which nucleosomes are redistributed are likely to significantly influence the functions for which these complexes are used. In order to understand what these differences, we have systematically analysed the positions to which nucleosomes are redistributed in different contexts using yeast Isw1a, Isw1b, Isw2 and Chd1 remodelling enzymes. We find that the enzymes which relocate nucleosomes to more central locations preferentially engage with nucleosomes bearing linker DNA. This may explain why these enzymes are unable to move nucleosomes to positions close to DNA ends where linker DNA would be lost. In addition to this inability to move nucleosomes close to DNA ends we find that the inherent nucleosome positioning properties of the DNA play an important role in determining where nucleosomes are moved to.
EXPERIMENTAL PROCEDURES
Nucleosome binding
Binding of Isw1a and Isw1b was monitored in 10% glycerol, 50 mM Tris-Cl pH 8.0, 50 mM NaCl, 3 mM MgCl2 and 100 μg/ml BSA. For Chd1 reactions contained 2.5% Ficoll, 50 mM Tris-Cl pH 8.0, 50 mM NaCl and 3 mM MgCl2. All reactions contained 0.5 nM 5′32P labelled nucleosomes (concentrations were obtained by scintillation counting of nucleosomes and comparing the value to that of the parent DNA, whose concentration was obtained by absorbance at 260 nm). Reactions were set up on ice and electrophoresed through 0.2 ×TBE, 5% acrylamide gels for 4 hours at 150 V at 4 °C with running buffer recirculation.
Enzymes
Isw1a and Isw1b were purified from yeast strains (YTT1168 and YTT1167) in which the Ioc3p or Ioc2p factors respectively were expressed as fusions with the TAP double affinity tag (23). Chd1 and Isw2 were purified from Chd1-TAP or Isw2-TAP strains which were purchased from Euroscarf. Remodellers were purified by IgG and calmodulin affinity chromatography from 20 L yeast (24). Supplementary Figure 1 illustrates the purity of these complexes.
Nucleosomes
Histone octamers were assembled from individual Xenopus laevis histones expressed in bacteria (25). Octamers for high resolution repositioning assays contained a S47C mutation in H4 and C110A change in H3. These octamers were subsequently reacted with the thiol reactive EDTA derivative (EDTA-2-aminoethyl)2-pyridyl disulphide (26). Octamers were reconstituted into nucleosomes using PCR prepared DNA derived from the MMTV nucleosome A (4) or 601.3 (27) positioning sequences. Reconstitutions were performed at 1 μM concentration and pH 7.5 by stepwise dialysis from 2 M NaCl or KCl to 0.85 M, 0.65 M, 0.5 M and finally 0 M.
Mononucleosome repositioning assays
Nucleosome repositioning was carried out in 10 μl reactions containing 20 mM Tris-Cl pH 8, 50 mM KCl, 1 mM MgCl2, 1 mM MgATP, 1 pmol nucleosome (32P end labelled at a single 5′ DNA end) and various concentrations of enzyme as described in figure legends. After 20 minute incubation at 30 °C reactions were stopped by transfer to ice and addition of competitor DNA (0.1 μg/μl final) and additional salt (200 mM KCl final). One tenth of the reaction was electrophoresed through 0.2 ×TBE, 5% acrylamide gels for 3.5 hours at 300 V at 4 °C with running buffer recirculation; the remainder was subjected to site directed mapping to determine the exact nucleosome position (28). This involved the addition of 1 μl 40 μM ammonium ferrous sulphate and 5 μl each of 19.2 mM ascorbic acid and 0.2% hydrogen peroxide to each reaction followed by an 1 hour incubation on ice. DNA was extracted with phenol:chloroform:isoamyl alcohol (25:24:1), recovered by ethanol precipitation and resuspended in 5 μl formamide loading dye (80% formamide, 10 mM NaOH, 1 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue). Samples were run on 8% acrylamide sequencing gels containing 6 M urea, fixed with 10% methanol, 10% acetic acid, dried and exposed to image plates. Markers for sequencing gels were obtained by limited cleavage at G residues of the appropriate DNA template.
RESULTS
Differences in the directionality of nucleosome redistribution by Isw1b, Isw1a, Isw2 and Chd1
In order to test the ability of the purified Isw1 and Chd1 enzymes to catalyze nucleosome sliding, recombinant Xenopus laevis histone octamers were assembled onto DNA fragments containing the MMTV nucleosome A positioning sequence flanked by different lengths of linker DNA. The DNA fragments used were designed such that the nucleosomes were initially flanked by 54 bp linkers on either side, off center (54 bp on one side and 18 bp on the other), or located at one end of a DNA fragment with 54 bp of linker DNA on one side and none on the other. The position of these nucleosomes was analyzed by native acrylamide gel electrophoresis following treatment with Isw1a, Isw1b or Chd1 and ATP. The Isw1a complex caused nucleosomes initially located at the end and off centre locations to run slower on the gel in an ATP dependent manner (Figure 1A lanes 1–8). However no obvious ATP dependent change was observed for the centrally located nucleosome (Figure 1A lanes 9–12). Conversely, treatment with the Isw1b complex and ATP caused the off-centre and centrally located nucleosomes to increase in mobility whereas no obvious change occurred with the end positioned nucleosome (Figure 1B). The activity of Chd1 was similar to Isw1a except less alteration to the 54A18 nucleosome was observed (Figure 1C).
Fig. 1.
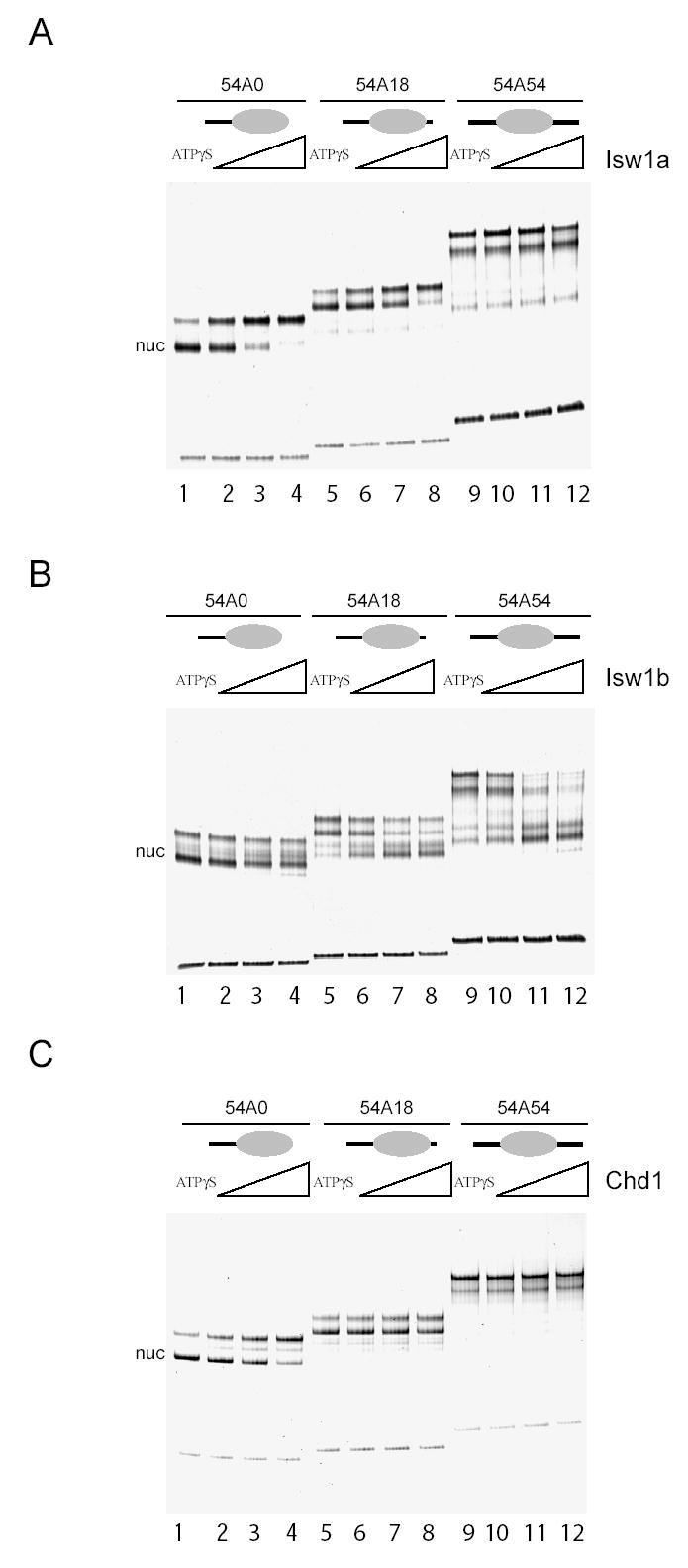
Native gel analysis of nucleosome repositioning catalysed by Isw1a, Isw1b and Chd1. Nucleosomes were assembled onto DNA fragments such that they were initially located at one end of a DNA fragment (54A0 nucleosomes), off centre (54A18 nucleosomes), or central (54A54 nucleosomes) prior to incubation with Isw1a (A), Isw1b (B) or Chd1 (C). Isw1a and Chd1 were more efficient at moving nucleosomes from terminal to more central positions whereas Isw1b moves nucleosomes with the opposite polarity. Approximately 1 pmol of nucleosomes were incubated with Isw1a and Isw1b at 40, 200 and 1000 fmoles; and 45, 90 and 180 fmoles Chd1 for 30 minutes at 30 °C. All reactions contained 1 mM ATP except those labelled ATPγS which contained 1 mM ATPγS and the maximum quantity of remodeller.
It is known that when a histone octamer is positioned close to the end of a DNA fragment the mobility through a gel is greater than when the octamer is close to the centre of the same piece of DNA (3). Therefore these observations suggest that Isw1a and Chd1 relocate nucleosomes closer to the centre of the DNA (“end to centre” type sliding) and that Isw1b causes nucleosomes to move to locations that are near to, or at, the ends of the DNA (“centre to end” type sliding). To study this in more detail, site directed nucleosome mapping was used to determine the positions nucleosomes were relocated to at high resolution. Briefly, this involves the tethering of cysteaminyl EDTA to a recombinant histone octamer via a thiol group introduced at H4 cysteine 47 (29). The chelation of Fe2+ ions by this reagent provides a means of generating localized hydroxyl radicals that are capable of cleaving DNA but only within a range of less than 1.5 nm. The site at which the reagent is attached on histone H4 comes closest to DNA 2 bp from either side of the nucleosome dyad, meaning that the sites of cleavage can be used to determine nucleosome positions at base pair resolution.
In Figure 2A the positions to which Isw1a and Isw1b reposition nucleosomes on an DNA fragment designed to position nucleosomes initially at an off centre location with 54 bp of DNA on one side and 18 bp on the other. The untreated nucleosomes display a characteristic pattern of strong and weak DNA cleavage separated by 7 bp, indicating that nucleosomes assembled onto this DNA fragment were initially positioned predominantly at +70 relative to the MMTV transcription start site (Figure 2A lanes 1, 6, 10). Following incubation with Isw1b, the cleavage at the +70 location is reduced and new cleavage sites indicating predominant new locations at +22 and +27 are detected. These position nucleosomes close to the end of the fragment, consistent with the increased mobility following electrophoresis (Figure 1B). Nucleosomes are also redistributed to these positions following thermal equilibration (Figure 2A lanes 10 and 11). However, the positions following redistribution by Isw1b exhibit a subtle bias for the locations closest to the end of the fragment. The movement of nucleosomes to positions closely related to those used during thermal redistribution reactions is consistent with previous studies of NURF (16) and recombinant Drosophila ISWI protein (4). In contrast, Isw1a relocates this nucleosome to a series of more centrally located positions at +39, +42, +47 and +58 (Figure 2A lanes 5–8). This cluster of locations is too close together to be resolved by native gel electrophoresis (Figure 1A). Isw1a moves nucleosomes to a similar distribution of locations when the nucleosome is initially located at the end of the DNA fragment (Figure 2B lanes 1–3). The enzymes Chd1 and Isw2 also move nucleosomes to the same distribution of locations (Figure 2B lanes 4–9).
Fig. 2.
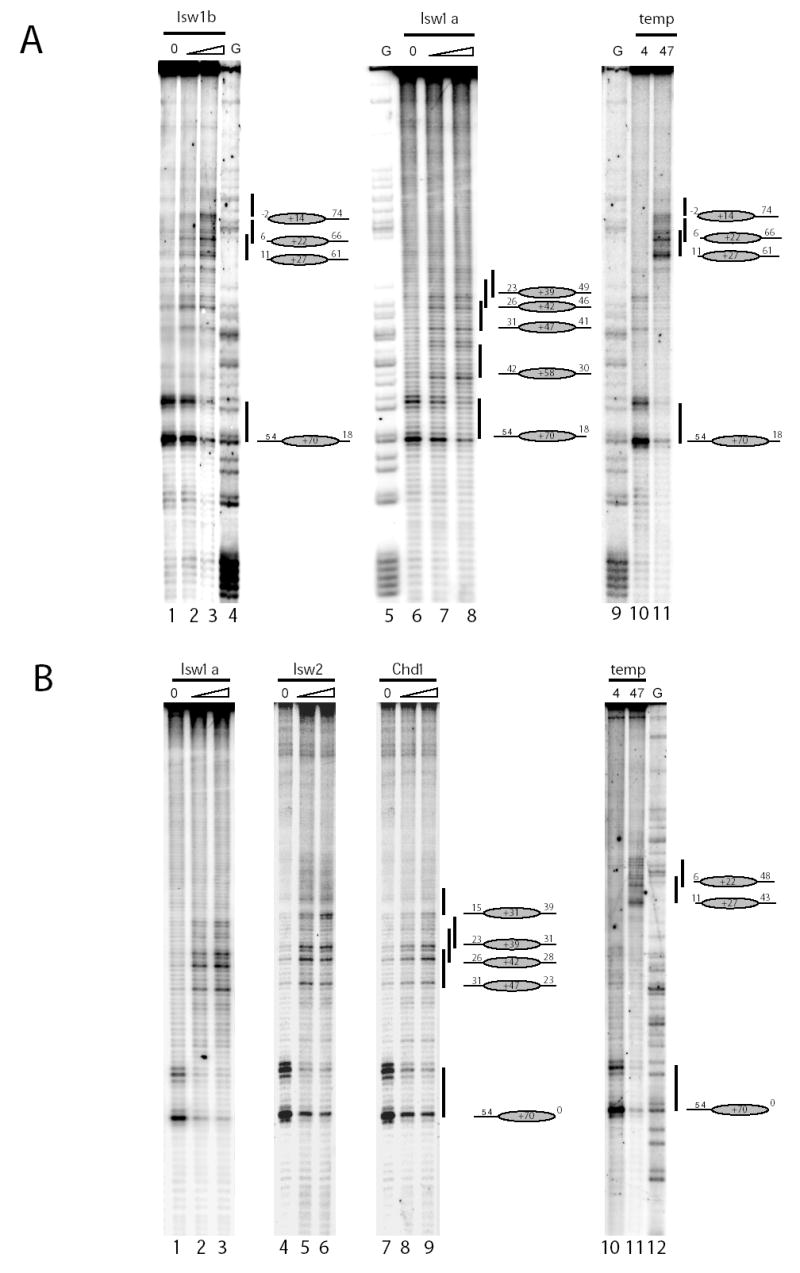
High resolution mapping of nucleosome repositioning by Isw1a, Isw1b, Chd1 and temperature. In order to precisely determine the positions to which nucleosomes were relocated following assembly on the off centre (54A18; fig 2A) or end positioned (54A0; fig 2B) fragments, repositioning assays were carried out using histone octamers derivatised with nucleosome mapping reagent (see methods). Nucleosomes were initially assembled predominantly at the +70 location (70bp upstream of the MMTV transcription start site). Following incubation of the 54A18 fragment with Isw1b (37.5 fmols Lane 2; 75 fmols Lane 3) or at high temperature (47 °C, 1 hour), nucleosomes were redistributed predominantly to the +22 and +27 locations. In contrast following incubations with Isw1a (160 fmols Lane 7; 320 fmols lane 8) nucleosomes were moved to the +39, +42, +47 and +52 positions. The 54A0 nucleosome was relocated by Isw1a (160 fmols Lane 2; 320 fmols Lane 3), Isw2 (1.6 fmols Lane 5; 3.2 fmols Lane 6) and Chd1 (3.6 fmols Lane 8; 7.2 fmols Lane 9) to positions at +39, +42 and +47. In addition Isw2 caused an increase in nucleosomes located at the +39 location (Lane 6). In contrast incubation of this nucleosome at 47 °C for 1 hour resulted in its relocation to the +22 and +27 sites. G tracks of the same DNA molecules were used to confirm the start positions of the nucleosomes and are shown in the lane marked G.
The finding that the Isw1 protein when associated with different accessory subunits in the Isw1a and Isw1b complexes moves nucleosomes with different directionalities illustrates that it is not the catalytic subunit alone that is responsible for this. While the locations to which nucleosomes are moved by Isw1b are clearly related to the most favourable locations available on these fragments, the mechanism for selecting the sites used by Isw1a, Chd1 and Isw2 is less clear. Nethertheless, the observation that these enzymes use the same subset of locations suggests that sequence or structural properties of the DNA fragment may contribute to the selection of sites to which nucleosomes are repositioned.
Isw1a moves nucleosomes to a subset of locations no closer than 15 bp from a DNA end
In order to investigate what underlies the selection of these more central positions, nucleosome repositioning by Isw1a was investigated on a series of nucleosomes with successively shorter DNA extensions. Nucleosomes were redistributed predominantly to the +39, +42 and +47 locations on the template with a 54 bp extension (Figure 2B lanes 1–3). As the length of the DNA extension is reduced to 44 and 38 bp, there is a progressive decrease in the proportion of nucleosomes accumulating at the +39 and +42 locations while nucleosomes are still relocated to the +47 position efficiently (Figure 3 lanes 4–9). It is also notable that the proportion of nucleosomes accumulating at the +58 location increases as the length of the linker DNA is reduced. For the fragments with 34 and 31 bp extension nucleosomes that have relocated 12 bp to the +58 location are the major new species observed following remodelling. It is possible that this 12 bp movement represents a minimal distance that the mechanical action of the Isw1a ATPase motor can move a nucleosome. However, it is also possible that this is the first location that is sufficiently stable to be detected.
Fig. 3.
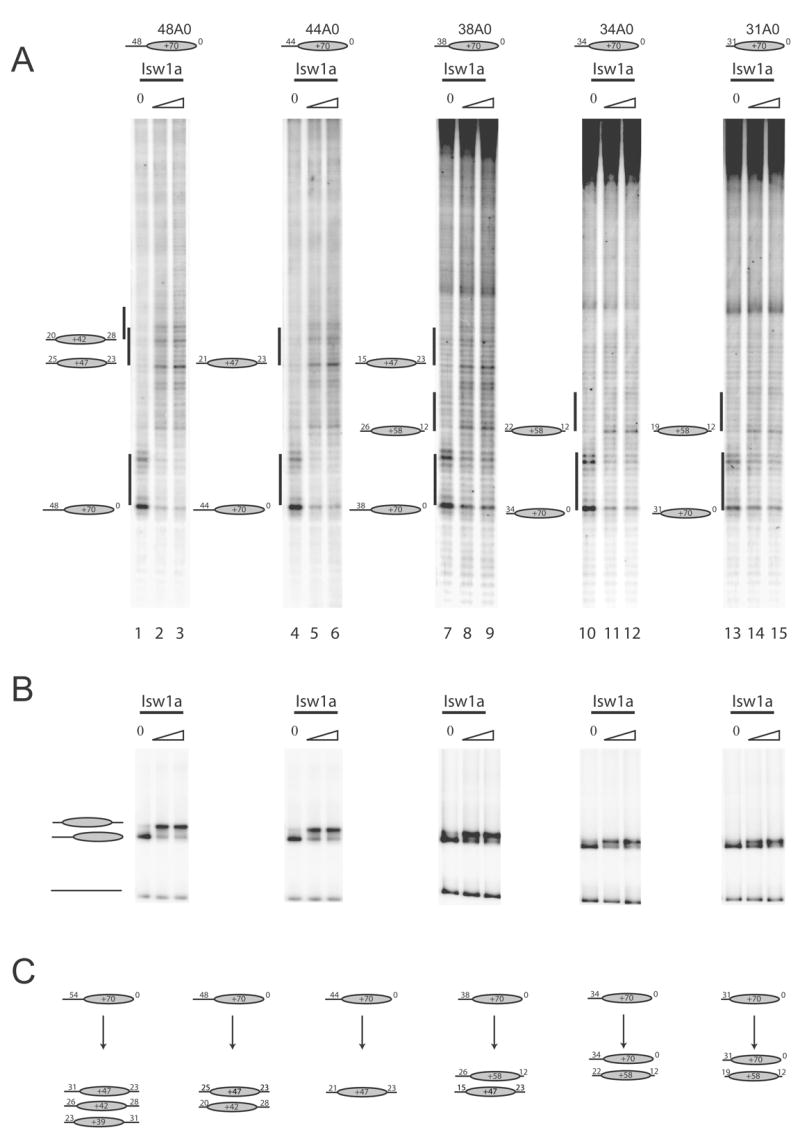
Isw1a moves nucleosomes to a subset of locations no closer than 15 bp from a DNA end. In order to systematically investigate the positions to which nucleosomes are redistributed by Isw1a, remodelling reactions were performed on a series of fragments with successively shorter DNA extensions. The amounts of Isw1a used were 36 fmoles (lanes 14, 17, 20, 21); 107 fmoles (lanes 11, 15, 18, 21, 25); 320 fmoles Isw1a (lane 12). 10% of each remodelling reaction was loaded on a 5% native gel (B). The positions to which nucleosomes were redistributed are illustrated schematically in (C).
Overall, it was found that reducing the length of the DNA extension caused nucleosomes to be moved to locations progressively closer to the starting position. It is also notable that the efficiency of redistribution decreases as the length of linker DNA is reduced, with a greater proportion of nucleosomes remaining at their original location on the shorter fragments.
The nearest to a DNA end that we have observed Isw1a relocate nucleosomes is 15 bp, (Figure 3 lanes 7–9). However, the range over which DNA ends influence positioning varies depending on the context. For example, the +47 location is disfavoured despite being 31bp from a DNA end in the presence of a 34 or 54bp downstream linker (Supplementary figure 2 Lanes 6–13).
Isw1a repositions nucleosomes to thermodynamically favourable locations
In addition to the exclusion of nucleosomes from regions close to the DNA ends, the data in figures 2 and 3 show that Isw1a, Isw2 and Chd1 move nucleosomes to discrete locations that are not located at the geometric centre of the DNA fragment. We next sought to investigate what underlies the selection of these more central locations. We first characterised the positions to which nucleosomes relocate during thermal incubation in more detail. Nucleosomes were assembled onto series of DNA fragments designed to form nucleosomes with progressively shorter DNA extensions. On nucleosomes with 48 bp and 44 bp extensions we observed nucleosomes being redistributed to the positions at +22 +27 as reported previously (4)(Figure 4A and B lanes 2–6). In order to determine the location of less favourable positions, thermal redistribution was carried out on shorter fragments where these most favourable locations were no longer present. Reduction of the DNA extensions to 38 and 36 bp revealed new locations at +31, +39, +42 and +47 (Figure 4A and B lanes 8–13). It is notable that of these less strongly preferred positions, the +39, +42 and +47 locations were also observed during Isw1a driven redistribution. This supports the hypothesis that the positioning properties of the DNA contribute to the sites selected by Isw1a. In fact, the positions observed following redistribution by Isw1a appear to result from rearrangement between favourable locations with the exception that locations too close to a DNA ends are excluded.
Fig. 4.
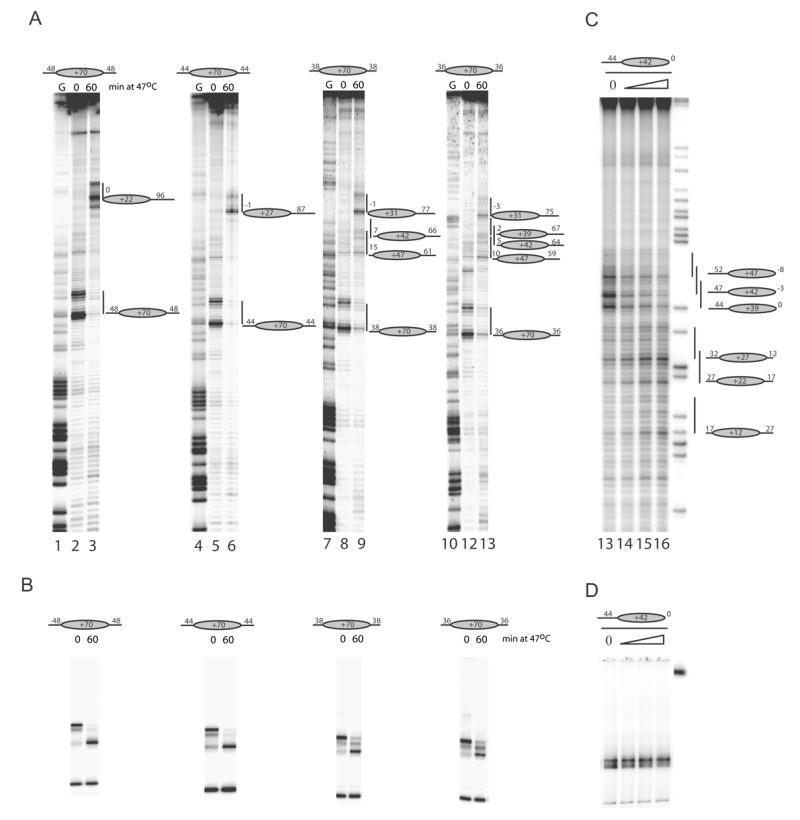
Isw1a relocates nucleosomes to thermodynamically favourable locations. Thermal redistribution of histone octamers with symmetrical linkers of 36, 38, 44 or 48 bp was analysed by site directed mapping (A) and native gels (B). Nucleosomes with shorter linker DNAs were observed to move to the +39, +42 and +47 locations that are also used by Chd1, Isw2 and Isw1a (See Figure 2). Nucleosomes deposited predominantly at the +42 location are found to be repositioned to the thermally favoured locations at +27 and +22 by Isw1a using site directed mapping (C) and native gel electrophoresis (D). Amounts of Isw1a used were 0, 36, 107 or 320 fmols (lanes 13–16). Note that DNA is labelled at the upstream end in (C), but on the downstream side in (A).
A prediction of this hypothesis would be that if DNA fragments were designed on which the most highly preferred +22 and +27 locations were sufficiently far from DNA ends, then they would be occupied following redistribution by Isw1a. Figure 4C shows that this is indeed true. Nucleosomes initially located predominantly at the +39 position with a 44 bp DNA extension are redistributed to the highly favourable positions at +27 and +22 (Figure 4C lanes 13–16). Similarly it would be anticipated that the bias against an unfavoured location would be reduced by increasing the linker DNA that flanks it. This is illustrated for the +70 location. Although nucleosomes are deposited during assembly at this location, it is not favoured following redistribution on a DNA fragment with an asymmetric extension. However, extension of linker DNA on the downstream side of this location such that it is placed more centrally progressively increase occupancy at this site (Supplementary Figure 2).
Chromatin remodelling enzymes display a preference for interacting with linker DNA that correlates with the orientation in which they reposition nucleosomes
It has previously been proposed that a factor contributing to the preference of the Isw2 remodelling enzyme for the repositioning of nucleosomes to central locations could be the preference of this enzyme for interaction with nucleosomes containing additional linker DNA (30). If this model is generally applicable to other remodelling enzymes that move nucleosomes to more central locations, then it would be anticipated that any enzyme displaying a preference for the relocation of nucleosomes to more central locations would also display this property. In order to extend the repertoire of remodelling enzymes for which this type of investigation has been performed, we investigated the binding of Isw1a, Isw1b and Chd1 to nucleosomes with minimal and extended linker DNA. Isw1a and Chd1, which both preferentially relocate nucleosomes to more central locations, were both found to display a preference for interacting with nucleosomes containing linker DNA. In contrast Isw1b, which can move nucleosomes to DNA ends, was able to interact with nucleosomes in a way that was not influenced by the presence of linker DNA (Figure 5). These observations provide additional support for the model proposed by Bartholomew and co-workers in which the preference of the remodelling enzymes for nucleosomes bearing linker DNA facilitates the removal of nucleosomes from DNA ends (18).
Fig. 5.
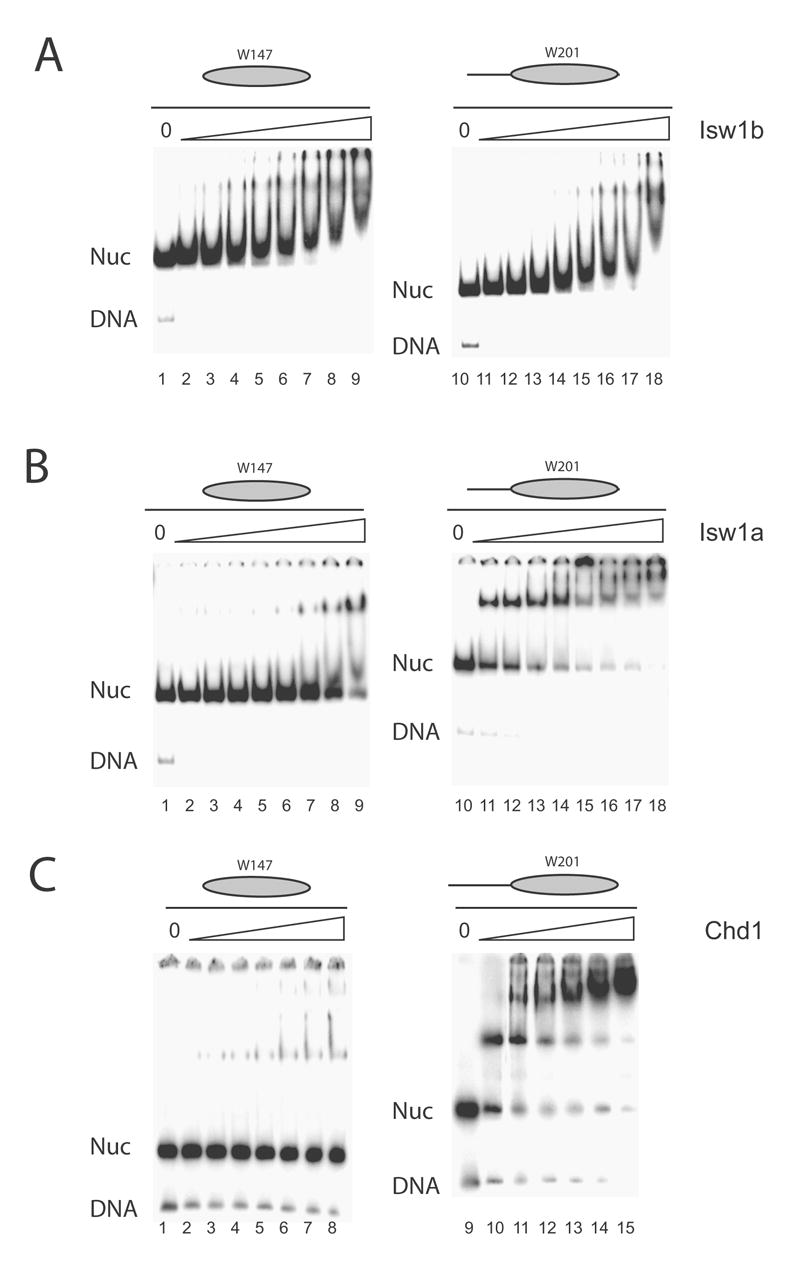
Remodelling enzymes that relocate nucleosomes to more central locations preferentially bind nucleosomes bearing linker DNA. Nucleosomes were assembled onto 147 bp DNA (W147) or with 54 bp linker DNA on one side (W201). The binding of Isw1b (A), Isw1a (B) and Chd1 (C) to these templates was investigated by native gel electrophoresis. While Isw1b bound to both templates similarly, Isw1a and Chd1 preferentially engage with nucleosome bearing linker DNA. Remodeller concentrations were varied between 0.74 nM and 12.8 nM for Isw1a; between 2.6 nM and 45 nM for Isw1b; and between 1.4 nM and 24 nM for Chd1.
DISCUSSION
We have characterised nucleosome redistribution by the Isw1a, Isw1b, Isw2 and Chd1 remodelling complexes. Although they share the same Isw1p catalytic subunit, the Isw1a and Isw1b complexes redistribute nucleosomes with different apparent directionalities: the Isw1a complex removes nucleosomes from locations within about 15 bp from DNA ends whereas Isw1b does not. We also find that the Chd1 protein is able to move nucleosomes and does so with the same directionality as Isw1a and Isw2. Importantly, we find that the inherent nucleosome positioning properties of the underlying DNA play an important role in determining the destinations to which nucleosomes are moved by all of these enzymes. This is consistent with previous studies suggesting that enzymes which do not remove nucleosomes from positions close to DNA ends, move nucleosomes to thermodynamically favourable locations (4,16). However, for enzymes that move nucleosomes with the opposite directionality this is not the case. Instead, it has been reported that the most favourable locations are not selected (18,30). We believe that the reason for confusion on this issue is that the outcome of remodelling reactions performed using enzymes such as Isw2 involves a compromise between the selection of thermodynamically favoured locations and the inability to move nucleosomes to positions close to DNA ends. This means that while the positions selected are favourable in comparison to the surrounding locations, they need not necessarily represent the most stable nucleosome positioning sequence on a given DNA fragment.
In order to illustrate how enzymes such as Isw2 select more central locations, we generated a plot illustrating the location and relative preference for the different locations along MMTV DNA that we have detected (Figure 6A). Remodelling enzymes might cause redistribution between these sites according to a number of different schemes. For example, an enzyme able to rapidly associate and dissociate from nucleosomes along an idealised DNA with no preferential nucleosome positioning properties would redistribute nucleosomes along the DNA fragment uniformly as shown in Figure 6B trace 1. However, certain DNA motor proteins have been observed to undergo a prolonged dissociation and reassociation reaction upon reaching a DNA end (31). Such behaviour would result in an idealised distribution similar to that shown in Figure 6B trace 2. Superposing this with the nucleosome positional preference shown in Figure 6A results in a distribution of nucleosomes with a bias towards DNA ends (Figure 6C). This is very similar to the pattern of redistribution observed by Isw1b (Figure 2A) and previously observed for NURF and ISWI (4,16). Note that while the enzyme itself exhibits no preference for either end of the DNA, the structural properties of the DNA fragment result in the preferential accumulation of nucleosomes at the end with more favourable nucleosome positioning sequences.
Fig. 6.
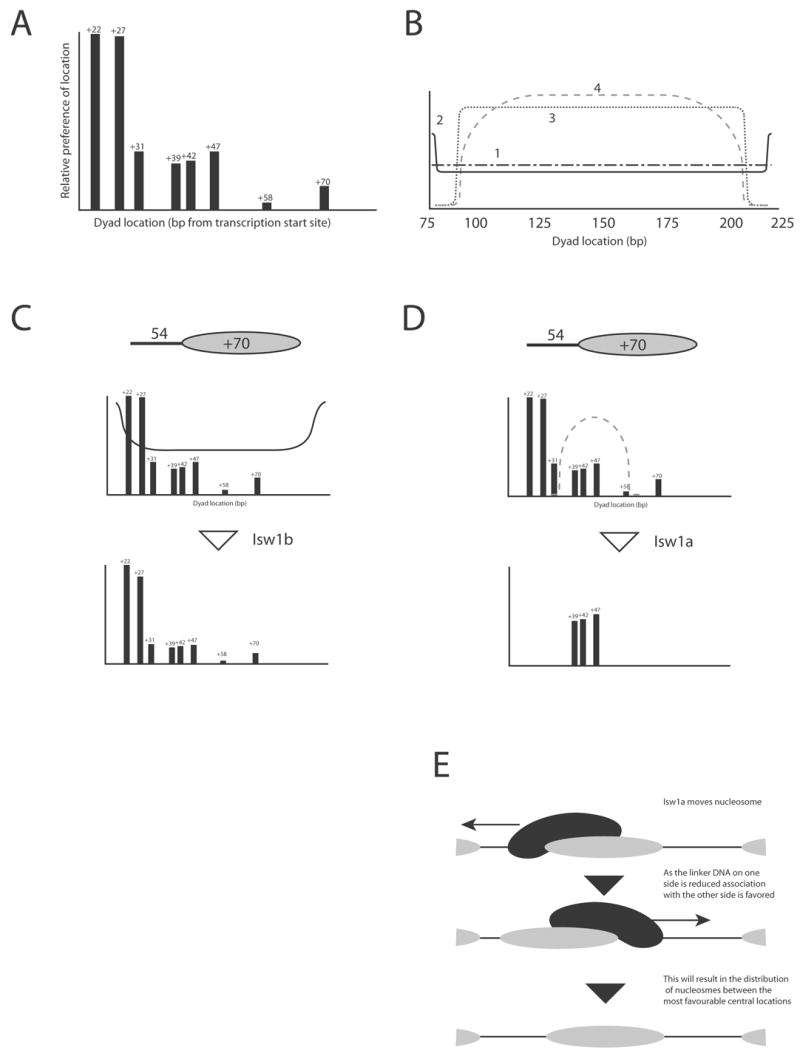
Superpostion of nucleosome positioning preferences with enzyme specific distribution patterns models the outcome of remodelling reactions on many DNA fragments. (A) The hierarchy of favourable nucleosome locations over the MMTV NucA region was estimated from analysis such as that shown in Figure 4. (B) Idealised nucleosome distribution patterns for a 200 bp DNA fragment with no nucleosome positioning properties. Trace 1 remodeler distributes nucleosomes at uniform velocity; Trace 2 remodeler undergoes a delay at DNA ends; Trace 3 remodeler is excluded from 15 bp close to DNA ends; Trace 4 enzyme exhibits progressive bias a against positions from 45 to 15 bp from DNA end. Supersition of Trace 2 with the positioning preferences for a DNA fragment with upstream extensions of 54 bp results with a close match to the experimental observed repositioning with Isw1b (C). Supersition of Trace 4 with the positioning preferences of the same DNA fragment with results with a close match to the experimental observed repositioning with Isw1a (D). Scaling of trace B to fit other DNA fragments used shows that this fit holds true for all of the DNA fragments studied (Supplementary Figure 3). Note that while the distribution patterns of the enzymes are asymmetrical, the positioning properties of the DNA can result in asymmetrical nucleosome distributions. (E) Illustrates how a requirement for engagement with linker DNA could result in the redistribution between favourable locations that are central with respect to barriers such as the ends of a short DNA fragment, or adjacent nucleosomes.
In contrast, the inability to move nucleosomes closer than about 15 bp from DNA ends gives rise to an idealised distribution illustrated in Figure 6B trace 3. However, the 15 bp exclusion limit we found for Isw1a is a lower limit, and DNA ends were observed to reduce occupancy at otherwise favourable locations over distances of at least 34 bp (data not shown). Such behaviour is represented by the idealised distribution in Figure 6B trace 4. If this trace is scaled to fit a DNA fragment with a 54 bp extension on one side, it can be superposed with the nucleosome positional preferences shown in Figure 6A to provide a means of modelling the outcome of Isw1a redistribution on this fragment (Figure 6D). The modelled outcome fits well with that obtained experimentally (Figure 2B lanes 1–3). If a similar process is performed on a DNA fragment with a shorter extension, then the idealised trace has to be adjusted to meet the new dimensions of the DNA fragment (Supplementary Figure 3C), but again, the outcome of superposition is in good agreement with the experimental data. In fact this holds true for all fragments we have studied (Supplementary Figure 3). This correlation strengthens support for the hypothesis that redistribution between thermally favourable locations that do not encroach upon DNA ends underlies the repositioning of nucleosomes by Isw1a. However, it is important to point out that this model remains qualitative because we have not quantitatively measured either the free energy of nucleosome positioning (32) or the energetic penalty associated with Isw1a moving nucleosomes close to DNA ends. Nonetheless, it provides a basis for understanding the outcome of remodelling carried out by enzymes such as Isw1a.
While most of the data we have presented has been obtained using Isw1a, we believe that the principles involved are likely to be applicable to other enzymes that act with this apparent directionality. For example, the Isw2 remodelling complex has previously been found to redistribute nucleosomes to positions no closer than 13 bp from DNA ends (30). This is very similar to the limit of 15 bp we have observed for Isw1a (Figure 3). In the case of Isw2 it has been proposed that preferential binding to nucleosomes containing linker DNA may underlie the exclusion of nucleosomes from locations close to DNA ends (18). The rationale for this is that if a remodelling enzyme requires contact with linker DNA on one side of a nucleosome in order to move it in that direction, the DNA available will be reduced as the nucleosome approaches the end of the fragment making it difficult to move any further (30) (Figure 6E). Our observation that the Isw1a and Chd1 activities preferentially bind to nucleosomes bearing linker DNA, but the Isw1b complex does not (Figure 5) provide further support for this model and suggest it may be generally applicable. In fact published observations suggest that it applies for ACF, and Mi-2 (16,33). Our observation that positioning sequences also contribute to the process explains why nucleosomes are not always moved to the geometric centre of relatively short mononucleosomal DNA fragments.
Interestingly, Bartholomew and co-workers previously observed that while extending linker DNA to 20 bp most significantly improves binding of Isw2 to nucleosomes, additional linker DNA extending to over 60 bp has more subtle effects (30). The strong requirement for a short length of linker DNA together with an extended region over which more subtle effects are observed is consistent with the broad range over which we observe that DNA ends can influence nucleosome positioning by Isw1a.
We also report here the yeast Chd1 protein can slide nucleosomes and does so with a directionality to move nucleosomes away from DNA ends in a manner that is very similar to the Isw2 and Isw1a complexes. This similarity in action may be related to the fact that these complexes perform partially redundant functions in vivo (19).
Although our observations are made using short DNA fragments on which DNA ends are encountered at a far higher frequency than would be expected in a physiological setting, we believe that they reflect important mechanistic differences in the way these complexes function. The behaviour of remodelling enzymes as they encounter a DNA end may be relevant to nucleosome remodelling in proximity to other barriers which in vivo are most likely to be adjacent nucleosomes or bound transcription factors (Figure 6E). Supporting this, many of the enzymes that move nucleosomes to more central locations have the ability to space arrays of nucleosomes (19,21,23). Although some enzymes that have the ability to move nucleosomes to positions adjacent to DNA ends have also been reported to be able to space nucleosomes, they appear to be less efficient in this assay (23). If the variable range over which we observe DNA ends influence nucleosome positioning also applies in the context of a nucleosome spacing reaction, we anticipate that these enzymes would be capable of establishing nucleosome spacings over the range 15 to at least 34 bp.
In biological contexts, movement of nucleosomes away from barriers may act to organise chromatin, while enzymes that move nucleosomes close to barriers may be more disruptive. An example where this may hold true is provided by the S. cerevisiae MET16 promoter (34). Here, Isw1a is involved in establishing nucleosome positioning that is refractory to transcription initiation. When expression is induced, the repositioning of a nucleosome by Isw1b appears to play an important role in regulating the amount of RNA Pol II that is able to enter productive elongation. Further studies will be required to establish whether remodelling enzymes that remove nucleosomes from barriers generally act to organise chromatin.
Our observations suggest that the inherent nucleosome positioning properties of DNA fragments play a role in determining the outcome of remodelling reactions. This means that the underlying structural properties of DNA are likely to establish a context that plays an important role in determining the outcome of remodelling reactions carried out by ISWI and Chd1 complexes. The observation that nucleosomes are shuttled between relatively favourable locations also indicates that, like many classical enzymes, these remodellers act to accelerate the otherwise slow redistribution of their substrate between energetically favoured states.
Supplementary Material
Footnotes
We thank Toshi Tsukyama for strains YTT1167 and YTT1168 and members of the TOH lab for useful discussion and comments on the manuscript. This work was supported by the Wellcome Trust.
References
- 1.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. J Mol Biol. 2002;319(5):1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 2.Beard P. Cell. 1978;15:955–967. doi: 10.1016/0092-8674(78)90279-9. [DOI] [PubMed] [Google Scholar]
- 3.Meersseman G, Pennings S, Bradbury EM. Embo J. 1992;11(8):2951–2959. doi: 10.1002/j.1460-2075.1992.tb05365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flaus A, Owen-Hughes T. Mol Cell Biol. 2003;23(21):7767–7779. doi: 10.1128/MCB.23.21.7767-7779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elfring LK, Deuring R, McCallum CM, Peterson CL, Tamkun JW. Molecular and Celluar Biology. 1994;14(4):2225–2234. doi: 10.1128/mcb.14.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Becker PB, Horz W. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 7.Tsukiyama T. Nat Rev Mol Cell Biol. 2002;3(6):422–429. doi: 10.1038/nrm828. [DOI] [PubMed] [Google Scholar]
- 8.Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. Cell. 1999;97(7):833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 9.Langst G, Bonte EJ, Corona DF, Becker PB. Cell. 1999;97(7):843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 10.Fazzio TG, Tsukiyama T. Mol Cell. 2003;12(5):1333–1340. doi: 10.1016/s1097-2765(03)00436-2. [DOI] [PubMed] [Google Scholar]
- 11.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. Science. 2004;303(5656):343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 12.Kassabov SR, Zhang B, Persinger J, Bartholomew B. Molecular Cell. 2003;11(2):391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 13.Lorch Y, Cairns BR, Zhang M, Kornberg RD. Cell. 1998;94(1):29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 14.Schnitzler G, Sif S, Kingston RE. Cell. 1998;94(1):17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 15.Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T. Mol Cell. 2003;12(6):1599–1606. doi: 10.1016/s1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang JG, Hamiche A, Wu C. Embo J. 2002;21(6):1406–1413. doi: 10.1093/emboj/21.6.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eberharter A, Ferrari S, Langst G, Straub T, Imhof A, Varga-Weisz P, Wilm M, Becker PB. Embo J. 2001;20(14):3781–3788. doi: 10.1093/emboj/20.14.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zofall M, Persinger J, Bartholomew B. Mol Cell Biol. 2004;24(22):10047–10057. doi: 10.1128/MCB.24.22.10047-10057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Genes Dev. 1999;13(6):686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alen C, Kent NA, Jones HS, O’Sullivan J, Aranda A, Proudfoot NJ. Mol Cell. 2002;10(6):1441–1452. doi: 10.1016/s1097-2765(02)00778-5. [DOI] [PubMed] [Google Scholar]
- 21.Lusser A, Urwin DL, Kadonaga JT. Nat Struct Mol Biol. 2005;12(2):160–166. doi: 10.1038/nsmb884. [DOI] [PubMed] [Google Scholar]
- 22.Tran HG, Steger DJ, Iyer VR, Johnson AD. Embo J. 2000;19(10):2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vary JC, Jr, Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T. Mol Cell Biol. 2003;23(1):80–91. doi: 10.1128/MCB.23.1.80-91.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. Nat Biotechnol. 1999;17(10):1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 25.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. J Mol Biol. 1997;272(3):301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 26.Bruno M, Flaus A, Owen-Hughes T. Methods Enzymol. 2004;375:211–228. doi: 10.1016/s0076-6879(03)75014-9. [DOI] [PubMed] [Google Scholar]
- 27.Lowary PT, Widom J. J Mol Biol. 1998;276(1):19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 28.Flaus A, Richmond TJ. Methods Enzymol. 1999;304:251–263. doi: 10.1016/s0076-6879(99)04015-x. [DOI] [PubMed] [Google Scholar]
- 29.Flaus A, Luger K, Tan S, Richmond TJ. Proc Natl Acad Sci U S A. 1996;93(4):1370–1375. doi: 10.1073/pnas.93.4.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Embo J. 2004;23(10):2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dillingham MS, Wigley DB, Webb MR. Biochemistry. 2000;39(1):205–212. doi: 10.1021/bi992105o. [DOI] [PubMed] [Google Scholar]
- 32.Drew HR. J Mol Biol. 1991;219(3):391–392. doi: 10.1016/0022-2836(91)90179-a. [DOI] [PubMed] [Google Scholar]
- 33.Brehm A, Langst G, Kehle J, Clapier CR, Imhof A, Eberharter A, Muller J, Becker PB. Embo J. 2000;19(16):4332–4341. doi: 10.1093/emboj/19.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morillon A, Karabetsou N, O’Sullivan J, Kent N, Proudfoot N, Mellor J. Cell. 2003;115(4):425–435. doi: 10.1016/s0092-8674(03)00880-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


