Abstract
We demonstrate a highly sensitive method to characterize the structural composition and intracellular fate of polymeric DNA nanocomplexes, formed by condensing plasmid DNA with cationic polymers through electrostatic interactions. Rational design of more efficient polymeric gene carriers will be possible only with mechanistic insights of the rate-limiting steps in the nonviral gene transfer process. To characterize the composition and binding dynamics of nanocomplexes, plasmid and its polymer carrier within nanocomplexes were labeled with quantum dots (QDs) and fluorescent organic dyes, respectively, as a donor and acceptor pair for fluorescence resonance energy transfer (FRET). The high signal-to-noise ratio in QD-mediated FRET enabled precise detection of discrete changes in nanocomplex state at the single-particle level, against various intracellular microenvironments. The distribution and unpacking of individual nanocomplexes within cells could thus be unambiguously followed by fluorescence microscopy. QD-FRET is a highly sensitive and quantitative method to determine the composition and dynamic stability of nanocomplexes during intracellular transport, where barriers to gene delivery may be identified to facilitate gene carrier optimization.
Keywords: Non-viral gene delivery, Quantum-dots, FRET, Intracellular trafficking, Nanocomplexes
Introduction
As viral vectors encounter problems with toxicity and immunogenicity for gene therapy, nanoscale nonviral vectors offer an attractive alternative[1-4]. Cationic polymers which condense plasmid DNA (pDNA) through electrostatic interactions to form nanocomplexes have emerged as safer, though less efficient, options for gene transfer[5-7]. A critical barrier in nonviral gene delivery is the timely dissociation of the polymeric nanocomplexes within the cell to liberate the DNA for efficient gene transfer[8]. The stability of this electrostatic interaction between the polymer and pDNA must be optimized since either premature dissociation or overly stable binding would be detrimental to the overall transfection efficiency[7, 9, 10]. To rationally design an optimal polymeric gene carrier, there is a critical need to understand the intracellular trafficking and unpacking behavior of nanocomplexes[11-14].
Fluorescence-based methods have been widely used to study the physicochemical properties of nanocomplexes and intracellular trafficking mechanisms of nonviral gene delivery[15-17]. Fluorescently labeled DNA and its polymer or lipid carrier have often been used to determine the interactions between the DNA and its carrier[10, 15, 18-21]. Colocalization of the fluorescent markers may indicate that the plasmid and its carriers are associated, but the components must diffuse far enough away to be detected as distinctly separate. Therefore, such detection methods do not provide sufficient sensitivity to distinguish the interactions between plasmid and polymer and to detect the onset of dissociation.
FRET has been utilized to evaluate time-dependent intermolecular interactions at the nanometer scale. Condensation states of lipoplexes and polyplexes have been described with FRET, where DNA was doubly labeled[21, 22] or polymer and pDNA were separately labeled[23] with a pair of organic fluorophores. However, the former is dependent on stable coil-globule transition states of pDNA and both require additional ratiometric analysis. Moreover, organic fluorophores used in conventional FRET are often susceptible to photobleaching[24], hampering their use for time-lapse studies of intracellular trafficking.
QDs have emerged as efficient donors for FRET that overcome the complications in conventional FRET, such as spectral cross-talk and direct acceptor excitation[24], due to their unique photophysical properties such as broad absorption, narrow emission spectra, and high photostability[25-28]. QDs can be observed at the single particle level and tracked over an extended period of time in live cells with conventional methods such as confocal microscopy[29-31]. In this study, we demonstrate that QD-FRET, with the energy transfer pair of 605QD and Cy5[25], may be used as a sensitive and quantitative method to study the stability and composition of polymeric DNA nanocomplexes, as well as their intracellular fate for gene delivery.
Materials and Methods
QD-labeling of pDNA.
A 4.9 kb pDNA (pEGFP-C1, Clontech, Mountain View, CA) encoding for green fluorescent protein (GFP) driven by the cytomegalovirus promoter was labeled with streptavidin-functionalized 605QDs (Qdot 605, Quantum Dot Corp., Hayward, CA) via a biotin-streptavidin linkage. The DNA was biotinylated using a biotin peptide nucleic acid (pGeneGrip biotin, Genlantis, San Diego, CA) or PEO-psoralen-biotin (Pierce, Rockford, IL) and exposed to UV light (320-500 nm) for 30 min. Biotinylated DNA was purified from unreacted biotin cross-bridges by ethanol or isopropanol precipitation and centrifugation following standard protocols [32]. Biotinylated plasmid and streptavidin-functionalized 605QDs were kept at a ratio of at least 2:1 to ensure a complete conjugation of 605QDs to plasmid. The amount of QDs labeled per plasmid was estimated to be within the range of 0-3 by TEM image analysis as well as single molecule detection (SMD) by quantitative comparison of fluorescent burst frequencies between free QDs and QD-labeled pDNA.
Cy5-labeling of chitosan.
The free amines on the chitosan polymer backbone (390 kDa, 83.5% deacetylated, Vanson, Redmond, WA) were labeled with N-hydroxy-succinimide (NHS)-functionalized Cy5 (Cy5-NHS, Amersham Biosciences, Piscataway, NJ). A solution of Cy5-NHS in dimethyl sulfoxide (DMSO) (1 mg/mL) was prepared and added gradually to chitosan (1 mg/mL) while stirring. The conjugation reaction mixture was maintained at pH 6.5 to keep chitosan soluble and stirred continuously for 12 h in the dark. To facilitate complete conjugation of Cy5 dye, the molar ratio of Cy5 to free amines of chitosan was kept at 1:20. Any remaining free dye was removed by dialysis against 1 L of 1% acetic acid in the dark to obtain only Cy5-chitosan. The level of Cy5 labeling of chitosan was 0.1-0.3% of primary amines and was determined by comparing fluorescence intensities of Cy5-chitosan to a standard curve from Cy5 dye.
QD-FRET DNA nanocomplexes formulation, characterization, and disruption.
Nanocomplexes were prepared from Cy5-chitosan and QD-DNA as previously reported[5]. Briefly, Cy5-chitosan (pH 5.5-5.7; 0.01-0.1% in 25 mM acetic acid solution) and QD-labeled plasmid (10 μg) added to 100 μl of 50 mM of sodium sulfate solution were both heated to 55°C separately. 100 μl of both solutions were quickly mixed under high vortexing for 30 s. Nanocomplexes were then used without further purification for all studies. Since the DNA concentration remains constant for all nanocomplex solutions, manipulating the chitosan concentration changes the N/P ratio — the theoretical ratio of protonated amines in the chitosan solution to the negative phosphates in the DNA solution. Size and zeta potential were measured with the Zetasizer 3000 (Malvern Instruments, Southborough, MA).
Disruption of nanocomplexes was accomplished by adding solutions containing 4 μg of heparin sodium salt (H9399, Sigma Aldrich, St. Louis, MO) and 0.05 U of chitosanase-RD (Bacillus sp. PI-7S, Seikagaku America, Ijamsville, MD) per μg of DNA encapsulated within nanocomplexes and incubating for at least 2 h at 37°C [5]. DNA retention was assessed by gel electrophoresis in 0.8% agarose in TAE with ethidium bromide. Gels were run at 100 V for 1 h and visualized under UV light (Versadoc 5000, Bio-Rad, Hercules, CA).
Single Molecule Detection.
A custom-made confocal spectrometer equipped with single molecule sensitivity was utilized for the study[25]. Samples within a focal volume were detected in a microcapillary (inner diameter = 100 μm) under a continuous flow of 1 μl/min driven by a syringe pump. A 488-nm argon laser served as the excitation source, focused through an oil immersion 100×/1.30 NA objective (Olympus America, Inc., Melville, NY). Photons emitted from 605QD and Cy5 were passed through a dichroic (505DCXR, Chroma Technology Corp. Rockingham, VT), followed by a 50 μm pinhole (Melles Griot Co, Irvine, CA) and a second dichroic (D625LP, Chroma Technology Corp, Brattleboro, VT). Signals from 605QD and Cy5 were filtered by band-pass filters (D605/20 and XF3034, respectively, Chroma Technology Corp, Brattleboro, VT) and collected by two independent avalanche photodiode detectors (Model SPCM-AQR-13, EG&G Canada, Vaudreuil, PQ, Canada) and a digital counter (National Instruments, Austin, TX). Fluorescent signals from both channels were integrated in 1-ms intervals. Labview (National Instruments, Austin, TX) was used to perform data analysis. The frequencies of fluorescent bursts were obtained by applying a threshold of 50 and 25 cps (counts per sec) for the 605QD and Cy5 channel, respectively. The average number of plasmids per nanocomplex was obtained by calculating: (Frequency of released pDNA)/(Frequency of nanocomplex), from separate preparations (n=5) with each providing three data points.
Fluorescent Microscopy.
Epifluorescent images were captured with a fluorescence microscope (BX-51, Olympus America, Inc., Melville, NY) equipped with a 100-W mercury arc lamp and an intensified Retiga CCD (Qimaging, Burnaby, BC, Canada). Excitation light was filtered (475AF40, Omega Optical Inc, Brattleboro, VT) and transmitted through an oil immersion 100×/NA 1.30 objective. Emission from 605QD and Cy5 were collected and filtered through appropriate filters (595AF60 and 670DF40, respectively; Omega Optical Inc, Brattleboro, VT) and dichroics (500 DRLP and 595DRLP, respectively; Omega Optical Inc, Brattleboro, VT). Image analysis and colocalization was performed with IPLab (Scanalytics, Rockville, MD).
Transmission Electron Microscopy.
Nickel grids (300 mesh) were cleaned with ethanol and then coated with Formvar prior to usage. A droplet of an aqueous suspension of nanocomplexes was placed on top of the grid and left to stand for 15 min while being capped to prevent evaporation. After deposition, excess nanocomplexes were blotted off with a filter paper, and the grid was allowed to dry for another 15 min. Nanocomplexes were then imaged with a Philips EM 420 TEM (Philips Electron Optics, Eindhoven, The Netherlands).
Spectrofluorometry.
Fluorescence emission spectra (550-700 nm) of QD-FRET nanocomplexes upon excitation at 488 nm were scanned by a spectrofluorometer (Spex FluoroLog-3, Horiba Jobin Yvon, Edison, NJ).
Cellular Transfection and Labeling for Confocal Microscopy.
HEK293 cells (ATCC, Manassas, VA) were seeded and grown on Labtech II chambered coverglass (4 × 104 cells/well; 4-well, Nalge Nunc International, Rochester, NY) or on coverslips in 6-well plates (15 × 104 cells/well) in complete medium (MEM supplemented with 10% FBS, 2 mM L-glutamine, 50 U/ml penicillin, and 50 U/ml streptomycin). Duplicate wells of 60-70% confluent cells were transfected with QD-FRET nanocomplexes containing 1-4 μg pEGFP-C1 plasmid in 0.5-1.0 ml reduced-serum Opti-MEM (Gibco Invitrogen, Carlsbad, CA) for 4 h at 37°C. At 24 and 48 h post-transfection, cells were fixed with 4% paraformaldehyde and stained with DAPI (Molecular Probes, Eugene, OR) prior to laser confocal microscopy (LSM 510 Meta, Carl Zeiss, Thornwood, NY). For live cell imaging at 72 h post-transfection, Hoechst nuclear stain was incubated with the cells during the final 15 min immediately prior to observation by laser confocal microscopy.
To quantify the bioactivity of plasmid after QD-labeling, HEK293 cells were transfected with unlabeled or QD-labeled plasmid complexed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA), following the manufacturer’s protocol. After 24 h, cells were identified for GFP expression and counted on a flow cytometer (FACSCalibur, Becton Dickinson), and the results were analyzed with Cell Quest software.
Results and Discussion
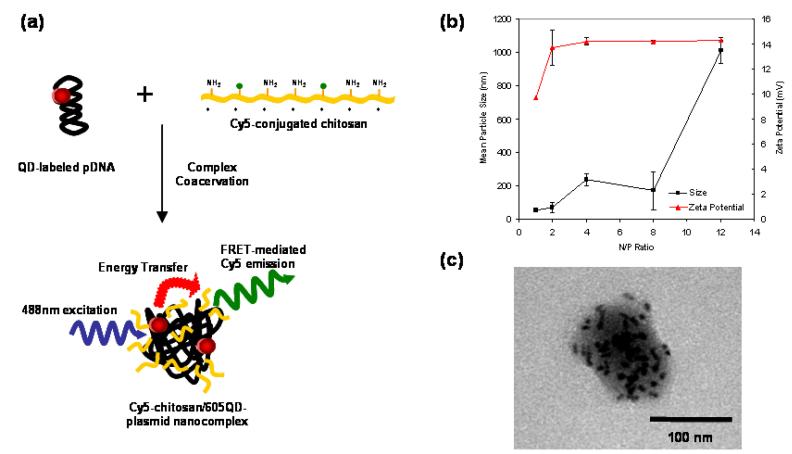
Serving as a model cationic polymer in this study, chitosan, a biodegradable natural polysaccharide consisting of repeated glucosamine units, has been shown to be an effective polycationic gene carrier by condensing pDNA through electrostatic interactions[5]. Cy5-chitosan/605QD-pDNA nanocomplexes, henceforth named QD-FRET nanocomplexes, were formulated from Cy5-chitosan and 605QD-labeled pDNA using a complex coacervation method (Figure 1a)[5]. The size and zeta potential of QD-FRET nanocomplexes were measured by photon correlation spectroscopy and laser Doppler anemometry, respectively, over a range of N/P ratios (Figure 1b).. The N/P ratio represents the theoretical molar ratio of protonated amines in the chitosan solution to the negative phosphates in the DNA solution. At high N/P ratios, aggregation of nanocomplexes is the likely cause of the larger particle size and distribution. At low N/P ratios, the particle size (∼60 nm) may reflect small clusters of QD-pDNA loosely associated with chitosan when compared to the size of QD-pDNA alone (∼24 nm), which is consistent with that of functionalized QDs. The presence of QD bound to pDNA may weaken the condensation by chitosan at low N/P ratios compared to unlabeled pDNA, though such effects are not fully understood. More importantly, the size and zeta potential for nanocomplexes at a N/P ratio of 4 were reproducible and comparable to reported values [5], indicating that the labeling of plasmid and polymer did not significantly affect the physical properties of these nanocomplexes.
Fig. 1.
QD-FRET nanocomplex formulation and physical characteristics. (a) pDNA and chitosan were labeled with 605QD and Cy5, respectively. Condensation of DNA and chitosan by complex coacervation formed QD-FRET nanocomplexes. Upon excitation at 488 nm, QD-FRET-mediated Cy5 emission (pseudo-colored green) indicates a compact and intact nanocomplex. (b) Size (circle) and zeta potential (triangle) of nanocomplexes were not altered by QD or Cy5 labeling. (c) TEM image of a typical nanocomplex with electron-dense QDs conjugated to multiple encapsulated plasmids.
In this study, we focused on an N/P ratio of 4 because these nanocomplexes are relatively unimodal in size, exhibit a high colloidal stability[5], and can induce transgene expression through oral administration in vivo[1]. TEM (Figure 1c) confirmed that the size and morphology of the QD-FRET nanocomplexes were consistent with unlabeled nanocomplexes as well. In Figure 1c, multiple QD-labeled plasmids were encapsulated within a single nanocomplex and appeared as dark elliptically-shaped objects due to the high electron density of QDs.
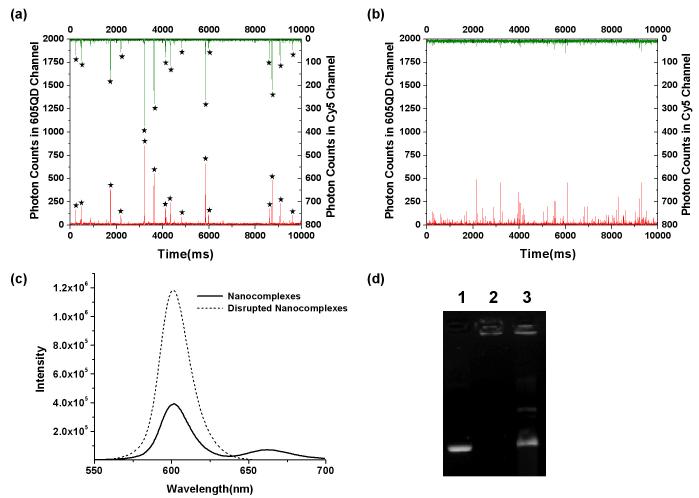
Investigation of the structural integrity of the nanocomplex was conducted with a custom-made single-molecule detection (SMD) confocal spectroscopy system[25, 33]. Figure 2a and 2b demonstrate a typical 10-sec fluorescence signal from QD-FRET nanocomplexes before and after treatment with heparin, a competing polyanion for DNA[34], and chitosanase, a degradative enzyme[5]. When nanocomplexes formed, bursts of fluorescence from both 605QD (Figure 2a, red/bottom) and Cy5 (Figure 2a, green/top) were simultaneously obtained. The presence of the FRET-mediated Cy5 signals indicated the tight association between chitosan and pDNA. In contrast, FRET was no longer observed in Figure 2b, when the nanocomplexes were disrupted after treatment with heparin and chitosanase. As a result, the FRET-mediated Cy5 signal may be considered as a digital “on” and “off”, representing intact and dissociated nanocomplexes, respectively, and obviating the need for further colocalization or FRET efficiency analysis. In parallel, an ensemble fluorescence measurement using spectrofluorometry also showed the absence of Cy5 emission and the recovery of 605QD signal intensity after disruption of nanocomplexes (Figure 2c). Gel electrophoresis confirmed that the nanocomplexes were disrupted physically by heparin and chitosanase (Figure 2d). The slight retardation of the band in lane 3 was likely due to biotinylation and degraded fragments of chitosan still associated with the plasmid.
Fig. 2.
Detection of QD-FRET between 605QD-pDNA and Cy5-chitosan. By single molecule detection, representative fluorescent bursts from 605QD (red/bottom) and Cy5 (green/top) of (a) nanocomplexes and (b) disrupted nanocomplexes after treatment with heparin and chitosanase. Stars denote coincident 605QD and Cy5 detection, indicating FRET from individual intact nanocomplexes. Complexation or disruption of the nanocomplexes are indicated by the digital “on” and “off” of FRET-mediated Cy5 emission. (c) By spectrofluorometry, intact nanocomplexes exhibited FRET-mediated Cy5 emission centered at 670 nm, while disrupted nanocomplexes exhibited no Cy5 emission and recovery of 605QD signal intensity. (d) DNA retention by gel electrophoresis showing GFP plasmid (lane 1), nanocomplexes (lane 2), and nanocomplexes treated with heparin and chitosanase (lane 3). Nanocomplexes remained stable, but readily release plasmid after disruption with treatment.
The combination of chitosanase and heparin was necessary to completely digest the chitosan and disrupt nearly all the nanocomplexes in this study. Although heparin typically disrupts nanocomplexes when evaluated by gel retention, most of the nanocomplexes treated with heparin alone remained associated (>90%) when evaluated by SMD. This difference is likely due to the application of an electromotive force to separate the DNA from the polycation in gel electrophoresis, whereas fluorescence methods such as QD-FRET do not require external forces. Hence, the combination of QD-FRET and SMD may offer higher sensitivity to characterize nanocomplex stability.
Of note, comparing the relatively high signal intensity of 605QD fluorescence bursts, obtained from the nanocomplexes (Figure 2a, red), with that of single 605QDs (data not shown) or disrupted nanocomplexes indicated that each nanocomplex contained more than one 605QD or multiple plasmids. The difference in fluorescent burst frequency in the 605QD channel before and after disruption was used to estimate the average number of plasmids encapsulated per nanocomplex. Further analysis of the increased frequency of QD bursts upon disruption resulted in an estimated ∼30 plasmids per nanocomplex, which was consistent with that determined by TEM imaging. This QD-FRET approach thereby enables quantitative analysis of the composition of DNA nanocomplexes simply by fluorescence spectroscopy.
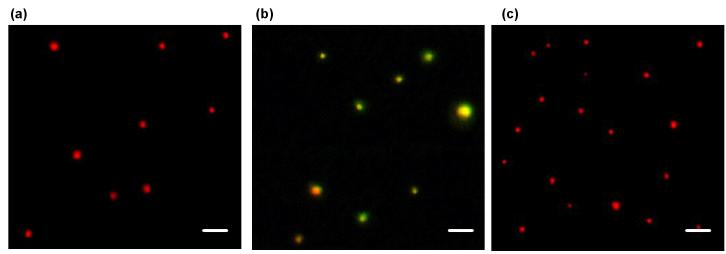
Analysis of single QD-FRET nanocomplexes was also performed with fluorescence microscopy following previously established procedures[35]. Figure 3b is a representative image of nanocomplexes combining signals from both 605QDs and Cy5, shown as red and green, respectively. Emission from Cy5 (green spots in Figure 3b) was readily detected and consistently colocalized (yellow/orange) with 605QD fluorescence, indicating efficient energy transfer from donor to acceptor due to their close association as nanocomplexes. Images of 605QD-pDNA (Figure 3a) were similar to that of disrupted QD-FRET nanocomplexes (Figure 3c) where both images do not exhibit any Cy5 signal, providing further evidence that energy transfer only happens with compact nanocomplexes.
Fig. 3.
Fluorescent image analysis of single QD-FRET nanocomplexes. Fluorescence images of (a) 605QD-labeled pDNA, (b) nanocomplexes, and (c) disrupted nanocomplexes. 605QD and FRET-mediated Cy5 signals are pseudo-colored in red and green, respectively. Colocalization is displayed in yellow/orange and indicates the tight association of plasmid and polymer within intact QD-FRET nanocomplexes. After treatment with heparin and chitosanase, nanocomplexes were disrupted and no longer exhibited FRET or colocalized signals. Scale bar: 1 μm.
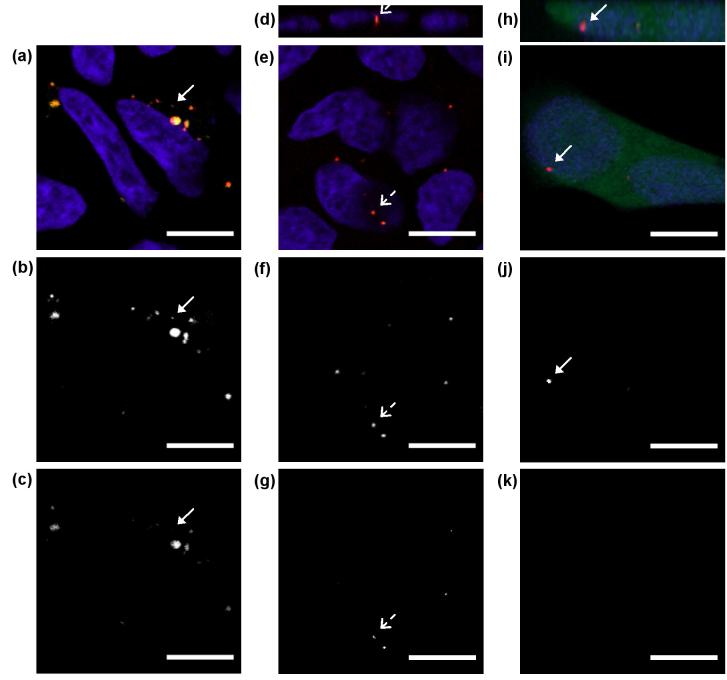
QD-FRET nanocomplexes were transfected into HEK293 cells and imaged by confocal microscopy to study their intracellular distribution at 24, 48, and 72 h post-transfection. At 24 h post-transfection, many nanocomplexes remained intact (yellow/orange punctuated spots) within the cytoplasm and localized around the nucleus (Figure 4a), which is similar to previous reports[15], suggesting that the trafficking behavior of the nanocomplexes was not affected by QD or Cy5 labeling. Intact nanocomplexes exhibited both QD and FRET-mediated Cy5 signal in Figure 4b and 4c, which show each respective detection channel individually for clarity. The onset of DNA release (arrow, Figure 4a-c) was detected at the time when the released plasmid only showed QD signal (red in Figure 4a and Figure 4b) and not Cy5 (Figure 4c). The sensitivity and digital nature of QD-FRET permitted the immediate detection of changes in nanocomplex state intracellularly. At 48 h post-transfection, most nanocomplexes have unpacked and the released plasmid (red only) accumulated in the perinuclear space (Figure 4e-g). Some nancomplexes appeared to be intact and located within the nucleus (broken arrow, Figure 4d-g), providing the tantalizing evidence that intact nanocomplexes may cross the nuclear membrane[10, 18]. In Figure 4h-k, a live HEK293 cell, expressing green fluorescent protein (GFP) at 72 h post-transfection, contained a released QD-labeled plasmid within the nucleus (arrow). The observation of live GFP-positive cells at this time point indicated that the QD-labeled plasmids partially retained their bioactivity.
Fig. 4.
Intracellular distribution of QD-FRET nanocomplexes. (a)-(c) HEK293 cells at 24 h post-transfection. Most nanocomplexes remained intact (Cy5, yellow/orange) and localized around the nucleus (blue). The onset of QD-labeled DNA release (red only in panel a) was detected at this time point. Only QD signal was detected (arrow, panel b) and not Cy5 (arrow, panel c) for the released plasmid. Grayscale images for each individual channel for 605QD and Cy5 are shown in the bottom two rows, respectively. (d)-(g) At 48 h post-transfection, most nanocomplexes have unpacked and released DNA (red only) in the perinuclear region. Note the intact nanocomplex (broken arrow) found within the nucleus in corresponding panels d-g. Panel d is a z-section showing the nuclear localization of the nanocomplex. (h)-(k) At 72 h post-transfection, a live HEK293 cell expressing GFP due to plasmid released from nanocomplexes. QD-labeled plasmid (arrow) was observed within the nucleus in corresponding panels h-j. Panel h is a z-section showing the nuclear localization of the QD-labeled plasmid. No signal was detected in the Cy5 channel (panel k). Scale bar: 10 μm.
In a separate experiment to quantify the bioactivity level of QD-labeled plasmids, HEK293 cells were transfected with Lipofectamine, and the transfection efficiency was evaluated by flow cytometry. For unlabeled plasmid, >90% of cells were GFP-positive, whereas for QD-labeled plasmid, <10% of cells were GFP-positive. The decreased transfection efficiency is possibly due to the combination of random incorporation of biotin at critical coding sites within the plasmid and the potential of QDs to sterically hinder transcription factor binding or transcription in general. Despite these issues, the main intent of QD-labeling for FRET is to allow the direct study of complexation and trafficking behaviors of nanocomplexes intracellularly.
Conclusion
This report demonstrated that QD-FRET provides a highly sensitive and quantitative tool to study the structural composition and dynamic behavior of polymeric DNA nanocomplexes intracellularly. The high signal-to-noise ratio of QD-FRET allows characterization of single nanocomplexes, which may be performed to capture the immediate onset of dissociation and transient states during DNA release, against various intracellular microenvironments. QD-FRET also provides a convenient method to follow the intracellular trafficking of polymeric DNA nanocomplexes over time and digitally monitor their unpacking behavior with conventional or confocal microscopy. The development of this sensitive and quantitative QD-FRET technique will facilitate the optimization of gene carrier characteristics such as molecular weight, charge density, and chemical composition. It is also expected that QD-FRET will find interesting applications in studying the intracellular transport of natural and synthetic nanostructures.
Acknowledgment
This work is supported by NIH EB002849, NSF DBI-0352407, and the Whitaker Foundation.
References
- [1].Roy K, Mao HQ, Huang SK, Leong KW. Oral gene delivery with chitosan--DNA nanoparticles generates immunologic protection in a murine model of peanut allergy. Nat Med. 1999;5(4):387–391. doi: 10.1038/7385. [DOI] [PubMed] [Google Scholar]
- [2].Salem AK, Searson PC, Leong KW. Multifunctional nanorods for gene delivery. Nat Mater. 2003;2(10):668–671. doi: 10.1038/nmat974. [DOI] [PubMed] [Google Scholar]
- [3].Putnam D, Zelikin AN, Izumrudov VA, Langer R. Polyhistidine-PEG:DNA nanocomposites for gene delivery. Biomaterials. 2003;24(24):4425–4433. doi: 10.1016/s0142-9612(03)00341-7. [DOI] [PubMed] [Google Scholar]
- [4].Luo D, Han E, Belcheva N, Saltzman WM. A self-assembled, modular DNA delivery system mediated by silica nanoparticles. J Control Release. 2004;95(2):333–341. doi: 10.1016/j.jconrel.2003.11.019. [DOI] [PubMed] [Google Scholar]
- [5].Mao HQ, Roy K, Troung-Le VL, Janes KA, Lin KY, Wang Y, August JT, Leong KW. Chitosan-DNA nanoparticles as gene carriers: synthesis, characterization and transfection efficiency. J Control Release. 2001;70(3):399–421. doi: 10.1016/s0168-3659(00)00361-8. [DOI] [PubMed] [Google Scholar]
- [6].Niidome T, Huang L. Gene therapy progress and prospects: nonviral vectors. Gene Ther. 2002;9(24):1647–1652. doi: 10.1038/sj.gt.3301923. [DOI] [PubMed] [Google Scholar]
- [7].Zhang XQ, Wang XL, Zhang PC, Liu ZL, Zhuo RX, Mao HQ, Leong KW. Galactosylated ternary DNA/polyphosphoramidate nanoparticles mediate high gene transfection efficiency in hepatocytes. J Control Release. 2005;102(3):749–763. doi: 10.1016/j.jconrel.2004.10.024. [DOI] [PubMed] [Google Scholar]
- [8].Schaffer DV, Fidelman NA, Dan N, Lauffenburger DA. Vector unpacking as a potential barrier for receptor-mediated polyplex gene delivery. Biotechnol Bioeng. 2000;67(5):598–606. doi: 10.1002/(sici)1097-0290(20000305)67:5<598::aid-bit10>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- [9].Kiang T, Wen J, Lim HW, Leong KW. The effect of the degree of chitosan deacetylation on the efficiency of gene transfection. Biomaterials. 2004;25(22):5293–5301. doi: 10.1016/j.biomaterials.2003.12.036. [DOI] [PubMed] [Google Scholar]
- [10].Huang M, Fong CW, Khor E, Lim LY. Transfection efficiency of chitosan vectors: effect of polymer molecular weight and degree of deacetylation. J Control Release. 2005;106(3):391–406. doi: 10.1016/j.jconrel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- [11].Wiethoff CM, Middaugh CR. Barriers to nonviral gene delivery. J Pharm Sci. 2003;92(2):203–217. doi: 10.1002/jps.10286. [DOI] [PubMed] [Google Scholar]
- [12].Pouton CW, Seymour LW. Key issues in non-viral gene delivery. Adv Drug Deliv Rev. 2001;46(13):187–203. doi: 10.1016/s0169-409x(00)00133-2. [DOI] [PubMed] [Google Scholar]
- [13].Lechardeur D, Lukacs GL. Intracellular barriers to non-viral gene transfer. Curr Gene Ther. 2002;2(2):183–194. doi: 10.2174/1566523024605609. [DOI] [PubMed] [Google Scholar]
- [14].Lucas B, Van Rompaey E, Remaut K, Sanders N, De Smedt SC, Demeester J. On the biological activity of anti-ICAM-1 oligonucleotides complexed to non-viral carriers. J Control Release. 2004;96(1):207–219. doi: 10.1016/j.jconrel.2003.12.028. [DOI] [PubMed] [Google Scholar]
- [15].Kiang T, Bright C, Cheung CY, Stayton PS, Hoffman AS, Leong KW. Formulation of chitosan-DNA nanoparticles with poly(propyl acrylic acid) enhances gene expression. J Biomater Sci Polym Ed. 2004;15(11):1405–1421. doi: 10.1163/1568562042368112. [DOI] [PubMed] [Google Scholar]
- [16].Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc Natl Acad Sci U S A. 2003;100(7):3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Panyam J, Labhasetwar V. Dynamics of endocytosis and exocytosis of poly(D,L-lactide-co-glycolide) nanoparticles in vascular smooth muscle cells. Pharm Res. 2003;20(2):212–220. doi: 10.1023/a:1022219003551. [DOI] [PubMed] [Google Scholar]
- [18].Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc Natl Acad Sci U S A. 1999;96(9):5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lucas B, Remaut K, Sanders NN, Braeckmans K, De Smedt SC, Demeester J. Studying the intracellular dissociation of polymer-oligonucleotide complexes by dual color fluorescence fluctuation spectroscopy and confocal imaging. Biochemistry. 2005;44(29):9905–9912. doi: 10.1021/bi0476883. [DOI] [PubMed] [Google Scholar]
- [20].Lucas B, Remaut K, Sanders NN, Braeckmans K, De Smedt SC, Demeester J. Towards a better understanding of the dissociation behavior of liposome-oligonucleotide complexes in the cytosol of cells. J Control Release. 2005;103(2):435–450. doi: 10.1016/j.jconrel.2004.12.017. [DOI] [PubMed] [Google Scholar]
- [21].Remaut K, Lucas B, Braeckmans K, Sanders NN, Demeester J, De Smedt SC. Protection of oligonucleotides against nucleases by pegylated and non-pegylated liposomes as studied by fluorescence correlation spectroscopy. J Control Release. 2005;110(1):212–226. doi: 10.1016/j.jconrel.2005.09.048. [DOI] [PubMed] [Google Scholar]
- [22].Itaka K, Harada A, Nakamura K, Kawaguchi H, Kataoka K. Evaluation by fluorescence resonance energy transfer of the stability of nonviral gene delivery vectors under physiological conditions. Biomacromolecules. 2002;3(4):841–845. doi: 10.1021/bm025527d. [DOI] [PubMed] [Google Scholar]
- [23].Kong HJ, Liu J, Riddle K, Matsumoto T, Leach K, Mooney DJ. Non-viral gene delivery regulated by stiffness of cell adhesion substrates. Nat Mater. 2005;4(6):460–464. doi: 10.1038/nmat1392. [DOI] [PubMed] [Google Scholar]
- [24].Lakowicz JR. Principles of Fluorescence Spectroscopy. Kluwer Academic/Plenum; 1999. [Google Scholar]
- [25].Zhang CY, Yeh HC, Kuroki MT, Wang TH. Single-quantum-dot-based DNA nanosensor. Nat Mater. 2005;4(11):826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- [26].Medintz IL, Clapp AR, Mattoussi H, Goldman ER, Fisher B, Mauro JM. Self-assembled nanoscale biosensors based on quantum dot FRET donors. Nat Mater. 2003;2(9):630–638. doi: 10.1038/nmat961. [DOI] [PubMed] [Google Scholar]
- [27].Clapp AR, Medintz IL, Mauro JM, Fisher BR, Bawendi MG, Mattoussi H. Fluorescence resonance energy transfer between quantum dot donors and dye-labeled protein acceptors. J Am Chem Soc. 2004;126(1):301–310. doi: 10.1021/ja037088b. [DOI] [PubMed] [Google Scholar]
- [28].Hohng S, Ha T. Single-molecule quantum-dot fluorescence resonance energy transfer. Chemphyschem. 2005;6(5):956–960. doi: 10.1002/cphc.200400557. [DOI] [PubMed] [Google Scholar]
- [29].Bruchez M, Jr., Moronne M, Gin P, Weiss S, Alivisatos AP. Semiconductor nanocrystals as fluorescent biological labels. Science. 1998;281(5385):2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- [30].Chan WC, Nie S. Quantum dot bioconjugates for ultrasensitive nonisotopic detection. Science. 1998;281(5385):2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- [31].Dubertret B, Skourides P, Norris DJ, Noireaux V, Brivanlou AH, Libchaber A. In vivo imaging of quantum dots encapsulated in phospholipid micelles. Science. 2002;298(5599):1759–1762. doi: 10.1126/science.1077194. [DOI] [PubMed] [Google Scholar]
- [32].Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- [33].Wang TH, Peng Y, Zhang C, Wong PK, Ho CM. Single-molecule tracing on a fluidic microchip for quantitative detection of low-abundance nucleic acids. J Am Chem Soc. 2005;127(15):5354–5359. doi: 10.1021/ja042642i. [DOI] [PubMed] [Google Scholar]
- [34].Danielsen S, Strand S, de Lange Davies C, Stokke BT. Glycosaminoglycan destabilization of DNA-chitosan polyplexes for gene delivery depends on chitosan chain length and GAG properties. Biochim Biophys Acta. 2005;1721(13):44–54. doi: 10.1016/j.bbagen.2004.10.011. [DOI] [PubMed] [Google Scholar]
- [35].Ho YP, Kung MC, Yang S, Wang TH. Multiplexed hybridization detection with multicolor colocalization of quantum dot nanoprobes. Nano Lett. 2005;5(9):1693–1697. doi: 10.1021/nl050888v. [DOI] [PubMed] [Google Scholar]