Abstract
Aniridia is a panocular human eye malformation caused by heterozygous null mutations within PAX6, a paired-box transcription factor, or cytogenetic deletions of chromosome 11p13 that encompass PAX6. Chromosomal rearrangements also have been described that disrupt 11p13 but spare the PAX6 transcription unit in two families with aniridia. These presumably cause a loss of gene expression, by removing positive cis regulatory elements or juxtaposing negative DNA sequences. We report two submicroscopic de novo deletions of 11p13 that cause aniridia but are located >11 kb from the 3′ end of PAX6. The clinical manifestations are indistinguishable from cases with chain-terminating mutations in the coding region. Using human × mouse retinoblastoma somatic cell hybrids, we show that PAX6 is transcribed only from the normal allele but not from the deleted chromosome 11 homolog. Our findings suggest that remote 3′ regulatory elements are required for initiation of PAX6 expression.
The PAX6 transcription factor is highly conserved among metazoans and controls critical steps during eye morphogenesis (1). Human PAX6 mutations are associated with a range of eye abnormalities, including anophthalmia, aniridia, various anterior segment defects, and isolated foveal hypoplasia (2–8). In general, heterozygosity for null alleles causes aniridia, a progressive disorder consisting of severe iris hypoplasia, cataracts, glaucoma, foveal hypoplasia, and nystagmus, whereas heterozygosity for missense alleles produces milder phenotypes (9). Aniridia is most simply explained by a 50% reduction in PAX6 activity at the cellular level, inasmuch as most mutations are predicted to truncate the polypeptide prematurely and may trigger nonsense-mediated mRNA decay. Cases with point mutations and cytogenetic deletions (WAGR syndrome, MIM194072) have similar eye findings, and a clear dosage effect has been established among different PAX6 genotypes (6, 10, 11). However, a dominant-negative mechanism has also been suggested for some aniridia alleles on the basis of cDNA overexpression data in vitro and in transgenic mice (12, 13), and an allelic inactivation model has been proposed to explain the semidominant phenotypes of most Pax genes (14).
Although PAX6 is expressed abundantly in the developing eye, forebrain, ventral spinal cord, and endocrine pancreas (15–18), heterozygous phenotypes are limited to the eye. Cis regulatory elements have been defined at the 5′ end of mouse Pax6. They extend over 25 kb, act upon three different promoters, include specific enhancers for retina and lens, and are evolutionarily conserved (19–21). At the same time, a paracentric inversion and a reciprocal chromosome translocation with breakpoints 85–124 kb from the 3′ end of human PAX6 are correlated with aniridia in different pedigrees (2, 22). The relationship between the 5′ enhancer elements and 3′ PAX6 rearrangements is unknown. In this report, we characterize two mutations as regulatory alleles, show that 3′ sequences are essential for PAX6 expression, and establish their primacy in controlling PAX6 transcription.
Materials and Methods
Cell Culture.
The human embryonic retinal cell line Ad12 (23) was kindly provided by René Bernards (Netherlands Cancer Institute, Amsterdam). The mouse retinoblastoma cell line 661 was developed from the retinal tumor of a transgenic mouse expressing SV40 large T antigen under the control of the human interphotoreceptor retinoid-binding protein (IRBP) promoter (24). 661TGr was derived from 661 by ethyl methanesulfonate mutagenesis (400 μg/ml EMS for 18 h) and selection in 5 μg/ml 6-thioguanine (25). Lymphoblastoid cell lines were established by Epstein–Barr virus transformation of peripheral white blood cells (26). Cells were fused by low-speed cocentrifugation for 3 min in serum-free media containing 40% (wt/vol) polyethylene glycol (PEG 1000; Baker). Hypoxanthine phosphoribosyl transferase-positive (HPRT+) hybrid clones were selected in DMEM with 10% (vol/vol) FCS, plus hypoxanthine-aminopterin-thymidine (HAT) and 1 μM ouabain, which kill the mouse and human parental cells, respectively.
Genomic DNA Analysis.
Pulse-field gel electrophoresis (PFGE) was performed with a hexagonal rig (CHEF Mapper; Bio-Rad). Southern analysis of conventional and pulsed-field agarose gels was performed using probes p32.1 (27), pFix2RS1 (5), pC1DBRI (an 0.5-kb EcoRI subclone derived from the distal breakpoint of case 1; accession no. AF301379), and P1NS (the 3′ terminal NcoI-SfiI fragment of a 58.5-kb human P1 clone spanning PAX6; accession no. AF301378).
Inverse PCR and Sequence Analysis.
Long-range inverse PCR was performed as described (28) on restricted genomic DNA that was ligated at high dilution (<100 μg/ml), using divergent primer pairs and a mixture of Taq and Pwo polymerases (EXPAND; Roche). Primers were designed on the basis of DNA sequence from phage (5) and P1 subclones. For case 1, we amplified the 3.0-kb EcoRI junction fragment (Fig. 1a). For case 2, we amplified 1.9-kb XbaI and 11.9-kb ApaI junction fragments. DNA fragments corresponding to the telomeric deletion breakpoints were obtained by screening a genomic phage library (case 1) or performing a secondary inverse PCR on normal DNA (case 2). The sequence of all four breakpoints is available from GenBank (accession nos. AF301378–81). The telomeric breakpoints of cases 1 and 2 are also represented in the public sequence database as human PAC clones (GenBank accession nos. AL135932 and AL122014, respectively).
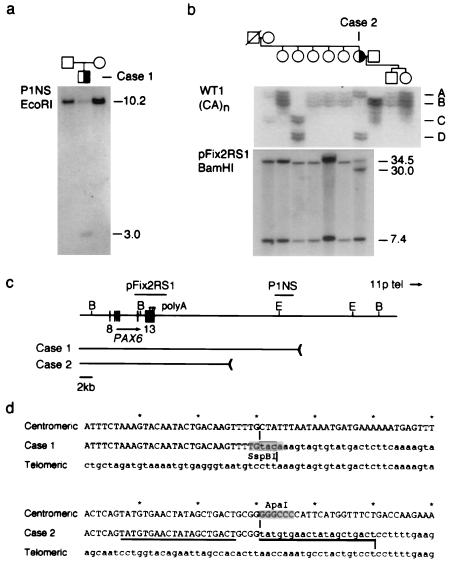
Figure 1.
Identification of 3′ PAX6 rearrangements in sporadic aniridia. (a) Southern analysis of case 1. Probe P1NS detects a novel 3.0-kb EcoRI fragment in the affected child, but not in his parents, and a 10.2-kb EcoRI fragment in normal DNA. (b) Haplotype and Southern analysis of case 2. (Upper) Segregation of the WT1 (CA)n repeat polymorphism. Four alleles are distinguished. Each allele is represented by an ensemble of fragments that differ from the mean length by 1–2 repeat units; these arise by replication slippage during PCR amplification. Case 2 is heterozygous (AD) at WT1. (Lower) Probe pFix2RS1 detects 34.5-kb and 7.4-kb BamHI fragments in normal DNA, and an additional 30-kb BamHI fragment was in the affected female, who is heterozygous. The mutation arose on a paternal chromosome 11 and is linked to WT1 allele D. (c) Restriction map showing PAX6 exons 8–13, polyadenylation sites (arrowheads), hybridization probes, and the centromeric deletion breakpoints in these two cases. The breakpoints are located 22.1 kb and 11.6 kb from the 3′ end of PAX6. E, EcoRI; B, BamHI. Some of the EcoRI sites have been omitted. (d) DNA sequence spanning the centromeric (uppercase) and telomeric (lowercase) breakpoints of cases 1 and 2 in normal DNA and the deletion junctions. The junction points are indicated by vertical lines and the 20-bp duplication in case 2 is underlined. The rearrangement in case 1 creates an SspBI restriction site at the junction.
Haplotype Analysis.
PCR genotyping of family 2 was performed using primers flanking a polymorphic (CA)n repeat in the 3′ untranslated region of the Wilms' tumor locus WT1 (29), which is 700 kb centromeric to PAX6. The wild-type chromosome 11 was identified by PCR, using primers spanning the HV translocation breakpoint at D11S495 (30), HVnormfor (5′-CACACGGTGAAGCTAACCTATAGG-3′), and HVnormrev (5′-CAAGCTGTAGTATGCATCTC-3′), standard reaction conditions [10 mM Tris⋅HCl (pH 8.3), 1.5 mM MgCl2, 50 mM KCl, 0.2 mM dNTPs, 20 pmol of each primer, and 1 unit Taq polymerase in a 50-μl volume], and parameters (2 min at 94°C, followed by 40 cycles of 30 s of denaturation at 94°C, 30 s of annealing at 60°C, and 60 s of extension at 72°C, followed by 5 min at 72°C). The mutant chromosome 11 in case 2 was identified similarly, using primers immediately flanking the deletion breakpoints, BPprox1for 5′-GTCTGCCATTCTCAACAAGGGG-3′ and BPforrev 5′-GAGGAACTGAGAAGTATTGCC-3′, and an annealing temperature of 65°C.
RNA Analysis.
To detect PAX6 transcripts, total RNA (2.5 μg) from somatic cell hybrids and control cell lines was reverse transcribed in 50 μl for 1 h at 42°C (SuperscriptII; GIBCO/BRL), using standard conditions and a primer (5′-TTTTTTTTTTTTTTACTGTAATCTT-3′) complementary to the human PAX6 sequence around the stop codon. First-strand synthesis was terminated by incubating the samples at 70°C for 15 min, and the RNA was removed by treating the reactions with 2 units of RNaseH for 20 min at 37°C. PCR amplification was performed for 40 cycles in 0.2-ml thin-walled tubes, using an MJ thermocycler (MJ Research, Watertown, MA), under the conditions described above, with 1–2 μl cDNA/50-μl reaction. Cycles consisted of 30 s of denaturation at 94°C, 30 s of annealing, and 60 s of extension at 72°C, with an initial denaturation step of 2 min and a final extension step of 5 min. Human transcripts were specifically amplified using primers SCHup2 (5′-CTACCAACCAATTCCACAA-3′) from exon 10 and SCHdown1 (5′-CTTGAACTGGAACTGACACA-3′) from exon 13, and an annealing temperature of 62°C. Mouse and human transcripts were coamplified using primers 103ax (5′-TCCTTCACRTCAGGCTCCATGTTGGGC-3′) from exon 11 and 11hu (5′-CCGGGAACTTGAACTGGAAC-3′) from exon 13, and an annealing temperature of 56°C. The same first-strand cDNA products were used for both PCRs. PAX6 polyadenylation sites were mapped using a 3′RACE procedure (31) and total RNA from human fetal eyes (17–20 weeks). PCR products were cloned, sequenced, and compared with genomic DNA. We recovered 2, 7, and 11 RACE clones corresponding to polyA sites 573 bp, 800 bp, and 967 bp, respectively, past the start of exon13.
Results
DNA rearrangements telomeric to PAX6 were discovered in two unrelated patients by Southern analysis (Fig. 1). Both cases are sporadic and have typical clinical features of aniridia. They are otherwise healthy and have apparently normal karyotypes. We detected a novel 3-kb EcoRI fragment in case 1, by using probe P1NS, which is located 19.4 kb beyond the 3′ end of PAX6, and a novel 30-kb BamHI fragment in case 2, using probe pFix2RS1, which spans terminal exon 13. These rearrangements were also apparent when we used other restriction enzymes (not shown), but no further PAX6 alteration was found in either patient by Southern or single-strand conformation (SSCP) analysis (5). The novel fragments were not present in parents or siblings. WT1 haplotype analysis showed that the mutation in case 2 arose on the paternal chromosome 11 (Fig. 1b). To characterize the rearrangements precisely in relation to PAX6, the novel fragments were amplified using inverse PCRs (28) and compared with normal genomic DNA (Fig. 1d). PAX6 uses three major polyadenylation sites in fetal eye tissue located 573 bp, 800 bp, and 967 bp past the start of exon 13 (not shown). The breakpoints in cases 1 and 2 were mapped 22.1 kb and 11.6 kb past the third polyA site, which defines the 3′ end of the PAX6 transcription unit (Fig. 1c). Both rearrangements are interstitial deletions, inasmuch as DNA probes immediately beyond these breakpoints gave normal hybridization patterns in Southern blots, and because the novel sequences recovered by inverse PCR map to chromosome 11p (not shown). The deletion endpoints do not share extensive homology. A short tandem duplication (20 bp) of DNA from the centromeric side was observed at the junction in case 2, suggesting that replication slippage or break-induced replication accompanied this deletion (Fig. 1d).
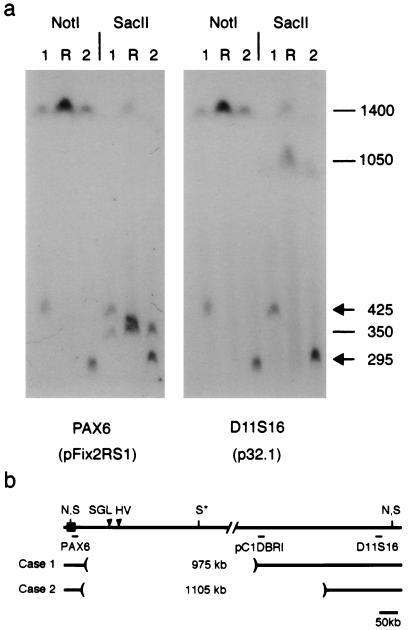
To measure the size of the deletions, genomic DNA was digested with NotI or SacII, separated by pulsed-field gel electrophoresis, and analyzed by Southern blotting (Fig. 2). In normal DNA, a 1,400-kb NotI fragment extends from PAX6 toward the telomere and terminates beyond D11S16 (2, 22, 32). This fragment was reduced to 425 kb and 295 kb, respectively, in cases 1 and 2. Identical NotI hybridization patterns were observed with pFix2RS1 (PAX6) from the centromeric side and p32.1 (D11S16) from the telomeric side, as expected for probes flanking an internal deletion. However, the 295-kb NotI fragment did not hybridize with a probe (pC1DBRI) derived from the distal breakpoint of case 1 (data not shown). The deletion in case 2 thus encompasses this site. The SacII data provide further support for this configuration. The internal SacII restriction site is deleted in both patients, creating fragments that are similar in size to the novel NotI fragments and that hybridize with probes p32.1 and pFix2RS1. The deletions are 975 kb (case 1) and 1105 kb (case 2) and are nested (Fig. 2b). They span the breakpoints of the two large-scale chromosomal rearrangements known to cause aniridia in other pedigrees (22, 33, 34).
Figure 2.
Physical mapping of 3′ PAX6 deletions. (a) Pulsed-field gel of lymphoblastoid genomic DNA from cases 1 and 2 and a normal reference individual (R) hybridized with probes pFix2RS1 and p32.1. These probes detect one 1400-kb NotI (N) fragment and two SacII (S) fragments, which are 350 kb (centromeric) and 1050 kb (telomeric). The internal SacII (S*) site is partially cleaved (22, 32). The second SacII fragment in the reference (R) lane reflects a likely sequence polymorphism. (b) Restriction map showing hybridization probes and deletion breakpoints relative to PAX6. Probe pC1PBRI detects an altered NotI fragment in case 1 but not case 2 (data not shown). The deletions encompass breakpoints of the HV reciprocal translocation and the SGL paracentric inversion (arrowheads), which are located, respectively, 124 kb and 85–100 kb from the 3′ end of PAX6 (22, 33, 34).
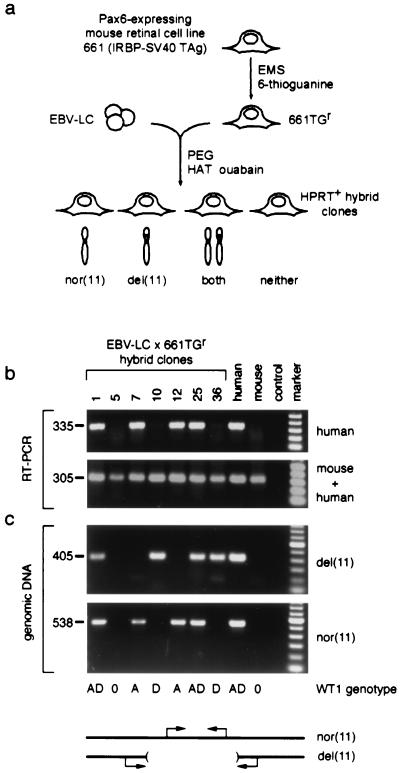
The simplest explanation for these combined mapping data would be if PAX6 expression requires a positive regulatory element located >124 kb from its 3′ end, past the most distal translocation breakpoint (HV in Fig. 2b). To test this model and determine if the 3′ deletions affect PAX6 transcription, we used a somatic cell hybrid strategy (35) to segregate mutant and wild-type chromosome 11 homologs. Hybrid clones were selected by fusing EBV-transformed lymphoblastoid cells from case 2 to 661TGr, an HPRT-deficient mouse retinoblastoma cell line with a high level of endogenous Pax6 expression (Fig. 3a). Clones retaining mutant and wild-type chromosomes 11 were identified by allele-specific PCR (Fig. 3c) and WT1 genotype analysis (Fig. 1b). Among 37 hybrid clones, 4 retained only the wild-type chromosome 11 and 8 retained only the deleted chromosome 11, whereas 21 clones retained both homologs and 4 retained neither. We then tested PAX6 expression, using a human-specific reverse transcriptase (RT) PCR assay. Although expression of mouse Pax6 was maintained in all clones, only hybrids retaining a normal chromosome 11 (n = 25) expressed human PAX6 (Fig. 3b and data not shown). The cellular environment of the mouse somatic cell hybrids is thus sufficient to activate and maintain transcription of the human PAX6 gene. However, PAX6 was essentially silent in all eight hybrids retaining only the mutated copy of chromosome 11, even though the gene itself is intact and the RT-PCR is extremely sensitive. The correlation between the PAX6 genotype and expression is absolute (for 12 clones retaining one homolog, for Fisher's exact test, P < 0.002, df = 1), and the difference in expression is striking.
Figure 3.
Human PAX6 is not expressed from the deleted chromosome 11 in somatic cell hybrids. (a) Strategy used to make hybrid cell lines. EBV-lymphoblastoid cells from aniridia case 2 (EBV-LC) were fused to 661TGr, an HPRT-deficient mouse retinoblastoma cell line. Hybrid clones (n = 37) were selected in HAT + ouabain and classified on the basis of human chromosome 11 content. In the absence of selective pressure for human autosomes, both homologs were free to segregate. (b) RNA analysis of representative hybrids. Human PAX6 and mouse Pax6 expression is demonstrated by parallel RT-PCRs, using conditions that amplify human transcripts only or transcripts from both species (see Materials and Methods). Hybrid clones 1, 7, 12, and 25 retain the nor(11) and express PAX6, whereas clones 10 and 36 retain only the del(11) and do not express PAX6. Similar results were obtained for all 37 clones. Reactions performed on RNA from Ad12 (human) and 661TGr (mouse) retinal cells, and a no-RT control, are shown for comparison. The size of each PCR product is indicated in base pairs. (c) DNA analysis of representative hybrids. The chromosome 11 content is demonstrated by diagnostic genomic PCRs. An amplicon within the deletion identifies nor(11), and an amplicon spanning the breakpoint junction identifies del(11). PCRs performed on genomic DNA from case 2 (human) and 661TGr (mouse) cells, and a no-template control reaction, are shown for comparison. WT1 genotype data for the hybrid cell lines are indicated below the panels; the D allele is linked to the deletion. marker, 100-bp ladder (Roche); nor(11), normal; del(11), deleted.
Discussion
Our data prove that 3′ deletions prevent PAX6 expression in cis. The tight correlation among the somatic cell hybrids between genotype and expression is inconsistent with an allelic inactivation model, which predicts a random inactivation of PAX6 alleles among the hybrid clones (14). Together with the clinical, structural, and 3′ RACE data, these results strongly suggest that the deletions affect PAX6 transcription rather than mRNA stability. There is no evidence for a remote untranslated 3′ exon and no need to invoke a second independent aniridia gene within 11p13. Our results also provide further support for haploinsufficiency as the basis of aniridia rather than a dominant-negative mechanism. Finally, these findings are comparable to earlier results obtained using somatic cell hybrids for deletions of the β-globin locus control region (LCR) in δβγo-thalassemia (36). The on-off effect of the 3′ deletion upon PAX6 transcription is consistent with clinical findings in these two patients, whose disease differs from that of patients with documented partial loss-of-function alleles (9) and suggests the workings of an LCR or an array of critical enhancers, which could influence the transcriptional activity of adjoining chromatin regions in an all-or-none fashion (37, 38). This hypothesis is supported by the convergence of mapping data on the 3′ side of PAX6, including experiments in which a human YAC transgene spanning PAX6 was able to rescue the Small eye mouse phenotype if the 3′ flanking DNA was intact (11). Alternatively, the deletions may disrupt a specific insulator or boundary element that normally shields the PAX6 transcription unit (39). The discontinuous nature of our results is less consistent with gene silencing via nonspecific position effects (40, 41) or the fractional loss of tissue-specific enhancer elements, which has been suggested to explain the milder phenotypes of translocation cases for genetic diseases such as holoprosencephaly (SHH) or Saethre–Chotzen syndrome (TWIST; refs. 42 and 43) and remote 3′ deletions altering the expression of Bmp5 in mice and dpp in Drosophila (44, 45). All known eyeless mutations in Drosophila Pax6 are regulatory alleles (46). We have functionally characterized cis regulatory alleles of human PAX6. The molecular identity and properties of the 3′ element(s) should be particularly interesting.
Acknowledgments
We thank the patients and their families for participating in the study; René Bernards, Muayyad Al-Ubaidi, and Anand Swaroop for cell lines; David Kurnit for loaning PFGE equipment; Hua Li and David Law for technical assistance; Richard Maas for help in the initial stages of this project; Gail Bruns, Manfred Gessler, and Tom Wilson for valuable advice; and David Burke, David Ginsburg, John Moran, and Diane Robins for critically reading the manuscript. This work was supported by grants from the National Institutes of Health and the Howard Hughes Medical Institute to T.G.
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF301378–AF301381).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240398797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240398797
References
- 1.Gehring W J, Ikeo K. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- 2.Ton C C, Hirvonen H, Miwa H, Weil M M, Monaghan P, Jordan T, van Heyningen V, Hastie N D, Meijers-Heijboer H, Drechsler M, et al. Cell. 1991;67:1059–1074. doi: 10.1016/0092-8674(91)90284-6. [DOI] [PubMed] [Google Scholar]
- 3.Hanson I, Jordan T, Van Heyningen V. In: Molecular Genetics of Inherited Eye Disorders. Wright A F, Jay B, editors. Switzerland: Harwood Academic; 1994. pp. 445–467. [Google Scholar]
- 4.Prosser J, van Heyningen V. Hum Mutat. 1998;11:93–108. doi: 10.1002/(SICI)1098-1004(1998)11:2<93::AID-HUMU1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 5.Glaser T, Walton D S, Maas R L. Nat Genet. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- 6.Glaser T, Jepeal L, Edwards J G, Young S R, Favor J, Maas R L. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- 7.Hanson I M, Fletcher J M, Jordan T, Brown A, Taylor D, Adams R J, Punnett H H, van Heyningen V. Nat Genet. 1994;6:168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- 8.Azuma N, Nishina S, Yanagisawa H, Okuyama T, Yamada M. Nat Genet. 1996;13:141–142. doi: 10.1038/ng0696-141. [DOI] [PubMed] [Google Scholar]
- 9.Hanson I, Churchill A, Love J, Axton R, Moore T, Clarke M, Meire F, van Heyningen V. Hum Mol Genet. 1999;8:165–172. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]
- 10.Hill R E, Favor J, Hogan B L, Ton C C, Saunders G F, Hanson I M, Prosser J, Jordan T, Hastie N D, van Heyningen V. Nature (London) 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- 11.Schedl A, Ross A, Lee M, Engelkamp D, Rashbass P, van Heyningen V, Hastie N D. Cell. 1996;86:71–82. doi: 10.1016/s0092-8674(00)80078-1. [DOI] [PubMed] [Google Scholar]
- 12.Singh S, Tang H K, Lee J Y, Saunders G F. J Biol Chem. 1998;273:21531–21541. doi: 10.1074/jbc.273.34.21531. [DOI] [PubMed] [Google Scholar]
- 13.Duncan M K, Cvekl A, Li X, Piatigorsky J. Invest Ophthalmol Vis Sci. 2000;41:464–473. [PubMed] [Google Scholar]
- 14.Nutt S L, Busslinger M. Biol Chem. 1999;380:601–611. doi: 10.1515/BC.1999.077. [DOI] [PubMed] [Google Scholar]
- 15.Walther C, Gruss P. Development (Cambridge, UK) 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 16.Grindley J C, Davidson D R, Hill R E. Development (Cambridge, UK) 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- 17.Ericson J, Morton S, Kawakami A, Roelink H, Jessell T M. Cell. 1996;87:661–673. doi: 10.1016/s0092-8674(00)81386-0. [DOI] [PubMed] [Google Scholar]
- 18.St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Nature (London) 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- 19.Williams S C, Altmann C R, Chow R L, Hemmati-Brivanlou A, Lang R A. Mech Dev. 1998;73:225–229. doi: 10.1016/s0925-4773(98)00057-4. [DOI] [PubMed] [Google Scholar]
- 20.Kammandel B, Chowdhury K, Stoykova A, Aparicio S, Brenner S, Gruss P. Dev Biol. 1999;205:79–97. doi: 10.1006/dbio.1998.9128. [DOI] [PubMed] [Google Scholar]
- 21.Xu P X, Zhang X, Heaney S, Yoon A, Michelson A M, Maas R L. Development (Cambridge, UK) 1999;126:383–395. doi: 10.1242/dev.126.2.383. [DOI] [PubMed] [Google Scholar]
- 22.Fantes J, Redeker B, Breen M, Boyle S, Brown J, Fletcher J, Jones S, Bickmore W, Fukushima Y, Mannens M, et al. Hum Mol Genet. 1995;4:415–422. doi: 10.1093/hmg/4.3.415. [DOI] [PubMed] [Google Scholar]
- 23.Vaessen R T, Houweling A, Israel A, Kourilsky P, van der Eb A J. EMBO J. 1986;5:335–341. doi: 10.1002/j.1460-2075.1986.tb04217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Al-Ubaidi M R, Font R L, Quiambao A B, Keener M J, Liou G I, Overbeek P A, Baehr W. J Cell Biol. 1992;119:1681–1687. doi: 10.1083/jcb.119.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsie A W, Brimer P A, Mitchell T J, Gosslee D G. Somatic Cell Genet. 1975;1:247–261. doi: 10.1007/BF01538449. [DOI] [PubMed] [Google Scholar]
- 26.Anderson M A, Gusella J F. In Vitro. 1984;20:856–858. doi: 10.1007/BF02619631. [DOI] [PubMed] [Google Scholar]
- 27.Feder J, Yen L, Wijsman E, Wang L, Wilkins L, Schroder J, Spurr N, Cann H, Blumenberg M, Cavalli-Sforza L L. Am J Hum Genet. 1985;37:635–649. [PMC free article] [PubMed] [Google Scholar]
- 28.Ochman H, Medhora M M, Garza D, Hartl D L. In: PCR Protocols: A Guide to Methods and Applications. Innis M, Gelfand D, Sninsky J, White T, editors. San Diego: Academic; 1990. pp. 219–227. [Google Scholar]
- 29.Haber D A, Buckler A J, Glaser T, Call K M, Pelletier J, Sohn R L, Douglass E C, Housman D E. Cell. 1990;61:1257–1269. doi: 10.1016/0092-8674(90)90690-g. [DOI] [PubMed] [Google Scholar]
- 30.Gessler M, Simola K O, Bruns G A. Science. 1989;244:1575–1578. doi: 10.1126/science.2544995. [DOI] [PubMed] [Google Scholar]
- 31.Frohman M A. PCR Protocols: A Guide to Methods and Applications. San Diego: Academic; 1990. pp. 28–38. [Google Scholar]
- 32.Gessler M, Bruns G A. Genomics. 1989;5:43–55. doi: 10.1016/0888-7543(89)90084-0. [DOI] [PubMed] [Google Scholar]
- 33.Simola K O, Knuutila S, Kaitila I, Pirkola A, Pohja P. Hum Genet. 1983;63:158–161. doi: 10.1007/BF00291536. [DOI] [PubMed] [Google Scholar]
- 34.Fukushima Y, Hoovers J, Mannens M, Wakui K, Ohashi H, Ohno T, Ueoka Y, Niikawa N. Hum Genet. 1993;91:205–209. doi: 10.1007/BF00218257. [DOI] [PubMed] [Google Scholar]
- 35.Weiss M C, Green H. Proc Natl Acad Sci USA. 1967;58:1104–1111. doi: 10.1073/pnas.58.3.1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forrester W C, Epner E, Driscoll M C, Enver T, Brice M, Papayannopoulou T, Groudine M. Genes Dev. 1990;4:1637–1649. doi: 10.1101/gad.4.10.1637. [DOI] [PubMed] [Google Scholar]
- 37.Bulger M, Groudine M. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 38.Engel J D, Tanimoto K. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]
- 39.Bell A C, Felsenfeld G. Curr Opin Genet Dev. 1999;9:191–198. doi: 10.1016/S0959-437X(99)80029-X. [DOI] [PubMed] [Google Scholar]
- 40.Bedell M A, Jenkins N A, Copeland N G. Nat Genet. 1996;12:229–232. doi: 10.1038/ng0396-229. [DOI] [PubMed] [Google Scholar]
- 41.Kleinjan D J, van Heyningen V. Hum Mol Genet. 1998;7:1611–1618. doi: 10.1093/hmg/7.10.1611. [DOI] [PubMed] [Google Scholar]
- 42.Belloni E, Muenke M, Roessler E, Traverso G, Siegel-Bartelt J, Frumkin A, Mitchell H F, Donis-Keller H, Helms C, Hing A V, et al. Nat Genet. 1996;14:353–356. doi: 10.1038/ng1196-353. [DOI] [PubMed] [Google Scholar]
- 43.Krebs I, Weis I, Hudler M, Rommens J M, Roth H, Scherer S W, Tsui L C, Fuchtbauer E M, Grzeschik K H, Tsuji K, et al. Hum Mol Genet. 1997;6:1079–1086. doi: 10.1093/hmg/6.7.1079. [DOI] [PubMed] [Google Scholar]
- 44.DiLeone R J, Russell L B, Kingsley D M. Genetics. 1998;148:401–408. doi: 10.1093/genetics/148.1.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masucci J D, Miltenberger R J, Hoffmann F M. Genes Dev. 1990;4:2011–2023. doi: 10.1101/gad.4.11.2011. [DOI] [PubMed] [Google Scholar]
- 46.Quiring R, Walldorf U, Kloter U, Gehring W J. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]