Abstract
Reports of frequent loss of heterozygosity (LOH) of markers on human chromosome 7q in malignant myeloid disorders as well as breast, prostate, ovarian, colon, head and neck, gastric, pancreatic, and renal cell carcinomas suggest the presence of a tumor suppressor gene (TSG). Functional assays have demonstrated that the introduction of an intact copy of human chromosome 7 (hchr7) can restore senescence to immortalized human fibroblast cell lines having LOH of markers within 7q31-q32 and can inhibit the tumorigenic phenotype of a murine squamous cell carcinoma cell line. To facilitate the cloning of the putative TSG, we have constructed a high-resolution physical map of this region of hchr7, specifically that encompassing the markers D7S522 and D7S677 within 7q31.1-q31.2. By using a lower resolution yeast artificial chromosome-based map as a starting framework, we established complete clone coverage of the implicated critical region in bacterial-artificial chromosomes (BACs) and P1-derived artificial chromosomes (PACs). The resulting BAC/PAC-based contig map has provided suitable clones for the systematic sequencing of the entire interval. In addition, we have already identified 29 clusters of overlapping expressed-sequence tags (ESTs) and 4 known genes contained within these clones. Together, the physical map reported here coupled with the evolving sequence and gene maps should hasten the identification of the putative TSG residing within this region of hchr7.
Keywords: tumor suppressor gene, human chromosome 7, physical mapping, expressed-sequence tag, bacterial artificial chromosome
Introduction
Frequent loss of heterozygosity (LOH) of a specific chromosomal marker(s) is considered an indication of a closely linked tumor suppressor gene (TSG) [1]. The association of specific interstitial deletions within chromosome 7q and a variety of human neoplasias is now well-established [2]. Specifically, frequent LOH of markers located within 7q31.1-q31.2 has been observed in breast [3,4], prostate [5,6], ovarian [7,8], colon [9], head and neck [9], gastric [10,11], pancreatic [12], and renal cell carcinomas [13] as well as in malignant myeloid disorders [14]. This collection of molecular cytogenetic data in conjunction with numerous classical cytogenetics studies showing frequent deletions and alterations within chromosome 7q in a variety of human neoplasias [15] suggests the presence of a TSG.
The results of functional complementation studies also indicate the presence of a TSG on chromosome 7q. Specifically, microcell-mediated chromosome-transfer studies have shown that the introduction of an intact copy of human chromosome 7 (hchr7) can restore senescence to human fibroblast cell lines known to have LOH within 7q31-q32 [16]. We have shown that the introduction of a single hchr7 inhibits the tumorigenicity of a mouse squamous cell carcinoma cell line [17] and a highly malignant human prostate carcinoma cell line (PC3) (J.C.Z. and E.D.G., unpublished data). Furthermore, reversion of the resulting suppressed cell lines back to a more tumorigenic state is associated with deletions in 7q31.1-q31.2. Finally, Matsuda and coworkers demonstrated that hchr7 can suppress tumorigenicity of a choriocarcinoma cell line [18]. Taken together, these findings strongly suggest that a TSG resides in human chromosome 7q31.1-q31.2.
To accelerate the isolation of this putative TSG, we sought to construct a highly detailed physical map of this well-defined region of hchr7. As part of the ongoing Human Genome Project, our laboratory has been heavily engaged in the mapping and sequencing of hchr7; for example, we recently reported the completion of a yeast artificial chromosome (YAC)-based physical map of the entire chromosome [19]. The latter provides information about the relative positions of numerous sequence-tagged sites (STSs) across hchr7. These mapped STSs provide a powerful framework for assembling more detailed maps, such as higher-resolution physical maps consisting of bacterial artificial chromosomes (BACs) [20,21] and P1-derived artificial chromosomes (PACs) [22].
The availability of complete BAC/PAC coverage of the hchr7 region believed to harbor the TSG would be valuable for several purposes. First, the use of large-insert bacterial clones would allow the construction of a highly accurate physical map of the region, providing both ready access to the DNA in an experimentally convenient form and the ability to position sequences of interest at high resolution. The latter effort could include the more precise localization of the numerous expressed-sequence tags (ESTs) being coarsely mapped throughout the human genome [23,24] (http://www.ncbi.nlm.nih.gov/genemap). Specifically, polymerase chain reaction (PCR) assays can be developed for ESTs (or EST clusters) mapping to the general vicinity of 7q31-q32 and used to test the available BACs/PACs, thereby establishing whether the corresponding sequences truly reside within the critical region of interest. Second, a properly constructed BAC/PAC contig map can be used for the systematic sequencing of the region. The resulting sequence data would provide information about additional ESTs and insight as to the position and structure of the corresponding genes. Finally, the available mapped BACs/PACs can be harnessed with appropriate mammalian selectable markers [25] and introduced into appropriate cell lines for functional complementation studies to test for the resence of the TSG.
Here we report the construction of a complete BAC/PAC-based physical map of the hchr7 critical region harboring the putative TSG. Information about the rapidly evolving sequence and gene map of the region is also provided. Together, these studies are facilitating the identification and evaluation of candidates for this TSG.
Materials and Methods
Polymerase Chain Reaction
PCR assays were performed in a total volume of 10 µland contained either 0.1 µg of genomic DNA, purified by a standard phenol-chloroform extraction protocol [26], or BAC DNA, prepared by a modified alkaline lysis protocol [25], 60 pmol of each primer, 200 µmol/L dNTPs (dATP, dCTP, dGTP, dTTP), 1X buffer (10 mmol/L Tris-HCl [pH 8.3], 1.5 mmol/L MgCl2, 50 mmol/L KCl, and 0.001% gelatin), and 0.5 units of AmpliTaq polymerase. (The latter three components were purchased from Perkin-Elmer, Norwalk, CT). Thermal cycling consisted of 35 cycles of 20 seconds at 94°C, 30 seconds at 55°C, and 15 seconds at 72°C in a 9600 Thermocycler (Perkin-Elmer). PCR products were electrophoresed in 2.5% MetaPhor (FMC Bioproducts, Rockland, ME) agarose gels at 3.5 V/cm2 in TBE buffer [89 mmol/L Tris-borate, 89 mmol/L boric acid, 2 mmol/L ethylenetetraamineacetic acid (pH 7.5)] containing 0.5 mg/ml ethidium bromide and imaged with UV light.
LOH Determination
LOH analyses of tumor DNA samples were performed as previously described [4,5,7,9]. LOH was defined as the complete loss or a greater than 70% reduction in the signal intensity for 1 allele.
BAC/PAC Isolation
BACs were isolated from the Research Genetics (http://www.resgen.com) and Genome Systems (http://www.genomesystems.com) human BAC libraries by PCR screening according to the suppliers' instructions. PACs were isolated from the Roswell Park Cancer Institute (Buffalo, NY) human PAC library (http://bacpac.med.buffalo.edu) by hybridization-based screening. Following isolation, clones were colony purified and tested for the presence of sequence-tagged sites (STSs) by PCR.
Generation of BAC Insert-End Sequences
Insert-end sequences from selected BACs were obtained as follows. BAC DNA was purified using an Autogen 740 Automated Nucleic Acid System (Integrated Separation Systems, Princeton, NJ, and concentrated to 200 ng/µl using a Microcon-100 column (Amicon, Bedford, MA). Fluorescent DNA sequencing was done with -40M13F and -28M13R primers (Clontech, Palo Alto, CA) and BigDye terminators (Perkin-Elmer). The 20-µl sequencing reaction contained 11 µl of purified BAC DNA (at 200 ng/µl), 1 µl of primer (at 10 µM), and 8 µl of BigDye reaction mixture. Thermal cycling was performed as suggested by the manufacturer. The products from each reaction were then purified on a Centrisep column (Princeton Separations, Adelphia, NJ), dried, resuspended in 2 µl of loading buffer, and analyzed on an automated fluorescent sequencing instrument (Applied Biosystems 377, Norwalk, CT). Repetitive elements in the resulting sequences were then masked using the program RepeatMasker (http://ftp.genome.washington.edu/cgi-bin/RepeatMasker), and primers suitable for PCR were designed with MacVector software (Oxford Molecular, Madison, WI).
Detection of Matching ESTs in Genomic Sequence Data
Prefinished and finished BAC/PAC sequences were imported from the Washington University Genome Sequencing Center (ftp://www.genome.wustl.edu / pub / gsc1 /sequence/st.louis/human). After the masking of repetitive sequences with RepeatMasker, PowerBLAST [27] analysis was done, and the results retrieved in hypertext (HTML) format. The resulting reports were manually browsed to identify matching ESTs as well as known genes.
Results
Previous studies [4,5,7,9] indicated that the critical region on hchr7 harboring the putative TSG was defined by the flanking markers D7S522 and D7S677 (Figure 1). In an effort to refine this interval, we analyzed a representative set of tumors from a variety of epithelial sources (breast, prostate, ovary, and colon) that we had previously characterized for LOH in this region [4,5,7,9]. Specifically, 14 specimens with a small area of LOH comprising the markers D7S522 and/or D7S677 were analyzed for 3 STSs (sWSS843, sWSS844, and sWSS850) that both map to the region [19] and amplify polymorphic segments. One of these STSs, sWSS850, failed to show LOH in some of the tumors, indicating that this marker resided outside the interval of interest. These results reduce the critical region by approximately 200 kb from the telomeric end (Figure 1).
Figure 1.
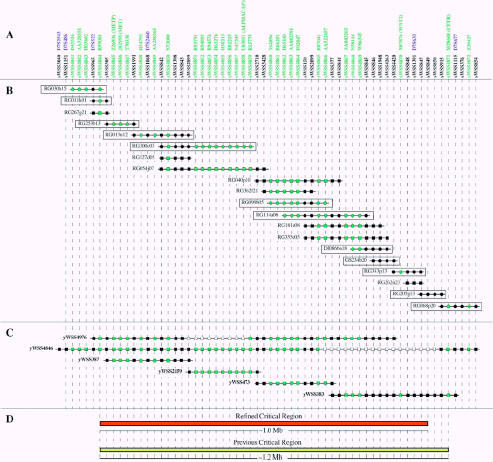
Clone-based physical map of the region of human chromosome 7q31.1-q31.2 containing the putative TSG (oriented with the centromere leftward and the 7q telomere rightward). (A) The deduced positions of the indicated STSs are depicted (STS names begin with the prefix sWSS). Relevant information about the STSs and their corresponding PCR assays is available in GenBank (see http://www.ncbi.nlm.nih.gov/dbSTS). STSs are depicted in an equidistant fashion from one another. Listed above many of the STSs are the corresponding D7S numbers (in the case of genetic markers; shown in blue) and GenBank accession numbers (in the case of genes and ESTs; shown in green). For the genes/ESTs, the indicated GenBank accession number corresponds to the sequence from which the STS was derived. (See Table 2 for additional GenBank records corresponding to the genes and ESTs). In the case of a known gene, the STS was developed from the sequence of the first exon (with the name of the gene provided in parenthesis). An additional STS was also developed from the promoter region of MET (‘METP’). (B) The relative positions of BACs and PACs, indicated as horizontal lines, are indicated. Clones are named starting with the following prefixes, which reflect their library of origin: RG (Research Genetics human BAC library), GS (Genome Systems human BAC library), and DJ (Roswell Park Cancer Institute human PAC library). A square indicates that the STS was verified in that clone by PCR testing, whereas a circle indicates that the STS was verified by both PCR testing and identification of the STS within sequence generated from that clone. Note that this figure depicts a representative subset of the total BACs/PACs known to map to this region, with a larger contig constructed by restriction enzyme digest-based fingerprint analysis [28] as part of a collaborative effort with the Washington University Genome Sequencing Center. A map of the latter contig is available as an electronic supplement to this paper (see http://genome.nhgri.nih.gov/CHR7/Neoplasia99). The clones selected for sequencing by the Washington University Genome Sequencing Center (see http://genome.wustl.edu/gsc) are boxed. (C) A small, representative set of YACs spanning the region. (See Bouffard et al. [19] for additional details about the complete YAC contig map.) Solid and open squares indicate if the STS was present or not, respectively, in that YAC by PCR testing. (D) The previous and refined critical regions containing the putative TSG are indicated (additional details in text), with the approximate sizes indicated as deduced from the BAC/PAC sequence data.
Our YAC-based STS-content map of hchr7 [19] (http://www.nhgri.nih.gov/DIR/GTB/CHR7) provided a valuable starting point for mapping the interval containing the putative TSG. Specifically, the defined critical region resides on a single YAC contig [contig O (sWSS23)], with a minimal set of the YACs spanning this interval shown in Figure 1. We assembled a complete bacterial clone-based contig map encompassing this critical region. Specifically, positive BACs and/or PACs were isolated for each of the 30 STSs previously localized on our YAC contig map. An additional 32 STSs corresponding to known genes and ESTs were also mapped relative to these BACs/PACs. These included several STSs (sWSS4859, sWSS4864, and sWSS4872) derived from EST clusters positioned on the genomewide transcript map [23,24] (http://www.ncbi.nlm.nih.gov/genemap). Based on the resulting STS content of the clones, two nascent BAC/PAC contigs were assembled. Two new STSs (sWSS4263 and sWSS4420) were then developed from the insert-ends of clones located at the ends of the two nascent contigs, and these were in turn used to isolate additional clones. This process resulted in the merger of the two smaller contigs to form the single contig shown in Figure 1. Note that the depicted BAC/PAC contig spans approximately 1.4 Mb and contains 62 STSs, thereby providing an average STS spacing of approximately 23 kb.
In parallel with our STS mapping efforts, the clones shown in Figure 1 (as well as a larger set of BACs/PACs isolated by hybridization-based screening) were analyzed at the Washington University Genome Sequencing Center (http://genome.wustl.edu/gsc) by a restriction enzyme digest-based fingerprint strategy [28]. The resulting BAC/PAC contig (available at http://genome.nhgri.nih.gov/CHR7/Neoplasia99) is fully consistent with that depicted in Figure 1 and serves to verify the accuracy of our assembled map.
A small number of BACs/PACs (roughly 8–9 clones) provide a minimal tiling path across the hchr7 critical region containing the putative TSG. Specific clones from the contig have been selected (Table 1) and are actively being sequenced at the Washington University Genome Sequencing Center (http://genome.wustl.edu/gsc). Preliminary analyses of the resulting data are revealing the presence of additional, previously unknown gene sequences. Specifically, in addition to the 3 EST clusters already localized on the genome-wide transcript map [23,24] (http://www.ncbi.nlm.nih.gov/genemap), we have identified 26 additional matching EST clusters within the sequence generated from this region (Table 2). The corresponding genes represent viable candidates for the TSG.
Table 1.
BACs/PACs Being Sequenced by the Washington University Genome Sequencing Center.
| BAC/PAC | Insert Size (kb)* | Status | GenBank Accession No.† |
| RG030h15 | 151 | Finished | AC002066 |
| RG011k01 | 157 | Prefinished‡ | AC006159 |
| RG253b13 | 171 | Finished | AC002080 |
| RG013n12 | 184 | Finished | AC004416 |
| RG300c03 | 150 | Finished | AC002543 |
| RG099b05 | 130 | Prefinished‡ | AC005063 |
| RG114a06 | 188 | Finished | AC002542 |
| DJ0866n18 | 101 | Finished | AC003987 |
| RG234b20 | 157 | Prefinished‡ | |
| RG343p13 | 155 | Finished | AC002465 |
| RG205g13 | 82 | Finished | AC003045 |
| RG068p20 | 150 | Finished | AC000111 |
Derived from sequence data.
Sequencing in progress, with preliminary data already available.
Table 2.
Summary of ESTs and Known Genes Identified in Genomic Sequence from the hchr7 TSG Critical Region.
| BAC/PAC* | Strand† | GenBank No.‡ | Source§ | Gene Name |
| RG030h11 | - | D45516 | Lung | |
| - | AA528351 | Ewing sarcoma | ||
| - | H03662 | Placenta | ||
| + | R99005 (3′), R99779 (5′) | Liver/spleen | ||
| RG253b13 | + | Z26936 | MET (Promoter) | |
| + | J02958 | MET | ||
| + | M37520 | MET H | ||
| - | T70330 (3′), T70414 (′5) | Liver/spleen | ||
| RG013n12 | + | H54792 | Liver/spleen | |
| + | AA280265 | Germ. B Cell | ||
| RG300c03 | - | N73000 | Melanocyte | |
| - | R93791 | Liver/spleen | ||
| - | R94893 | Liver/spleen | ||
| + | R94575 (3′), R94574 (5′) | Liver/spleen | ||
| - | H63276 | Liver/spleen | ||
| - | H38213 | Liver/spleen | ||
| - | R02106 (3′), R02219 (5′) | Liver/spleen | ||
| - | N47349 | Multiple sclerosis | ||
| + | U03851 | ALPHACAP | ||
| RG099b05 | - | T62896 | Liver | |
| - | R66201 | Placenta | ||
| RG114a06 | + | H65143 | Olfactory | |
| + | AA325774 | Cerebellum | ||
| + | F02047 (3′), F05800 (5′) | Multiple sclerosis | ||
| + | R07041 (3′), R07091 (5′) | Liver/spleen | ||
| + | AA522457 | Prostate | ||
| + | AA043204 (3′), AA043203 (5′) | Uterus | ||
| DJ0866n18 | + | N58116 | Liver/spleen | |
| + | W86346 (3′), W86345 (5′) | Liver/spleen | ||
| RG343p13 | + | X07876 | WNT2 | |
| RG068p20 | + | M28668 | CFTR |
Some ESTs/genes are present in more than one clone. For simplicity, this is not indicated here (see Figure 1 for the complete EST/gene content of each BAC/PAC).
Arbitrarily assigned strand containing that sequence: (+) sense and (-) antisense.
GenBank accession numbers for ESTs and known genes (see http://www.ncbi.nlm.nih.gov/Entrez/nucleotide.html). In some cases, two accession numbers are provided, reflecting 3′ and 5′ sequence reads (as indicated).
Tissue or cell line from which the mRNA used for cDNA library construction was derived.
Discussion
As an initial step toward the isolation of the putative TSG residing within human chromosome 7q31.1-q31.2, we have constructed a high-resolution physical map of the critical region established by LOH studies. This interval was initially defined by the flanking markers D7S522 and D7S677 [4,5,7,9]; however, based on analyses with additional polymorphic markers, we have slightly reduced the size of this region (Figure 1). The refined critical region (now flanked by D7S522 and sWSS850) is fully contained within a single YAC contig [19], with the STSs previously positioned along this contig serving as the starting point for the development of a bacterial clone-based physical map. Indeed, the availability of STSs mapped, on average, every 79 kb across the region [19] greatly accelerated clone isolation and map construction.
The resulting BAC/PAC contig (Figure 1) provides highly redundant coverage of the region as well as a set of clones suitable for systematic sequencing. Of note, one of the areas where clone coverage is particularly poor (sWSS2710 to sWSS3428) is covered by a BAC (RG099b05) that is thought to contain a chimeric insert (i.e., two segments of DNA that are not contiguous in the human genome). The latter was discovered when new STSs were developed from sequence generated from this clone, and some (but not all) of these STSs mapped to hchr7. Interestingly, the hchr7 DNA within and flanking RG099b05 contains a notably large amount of repetitive sequences. Such repeats may facilitate the formation of chimeric clones by recombination-based mechanisms. Furthermore, a common chromosomal fragile site (Fra7G) has recently been localized to this general region of hchr7 [14,29]. These various observations-poor clone coverage, a chimeric BAC, repeat-rich sequences, frequent neoplasia-associated LOH for markers from this region [2–15], and the likely presence of a nearby fragile site-may all be related. Of note, the recent discovery of a TSG (FHIT) within the most prominent fragile site of the human genome [30,31] suggests that TSGs may be located in areas of chromosomal fragility.
The availability of well-mapped bacterial clones covering the putative TSG critical region has catalyzed a couple of important areas of investigation. First, the entire interval is actively being sequenced (see Table 1), providing a wealth of sequence data for use in additional studies. Our initial analyses of this data have included both the identification of matching gene and EST sequences (Table 2) as well as the prediction of genes and their corresponding structures by various computer programs (GRAIL [32], GeneScan [33]; data not shown). To date, our evaluations of the matching ESTs and predicted new genes have not uncovered a particularly attractive candidate gene, although more detailed studies are ongoing (e.g., generation of full-length cDNA sequences). In total, 4 known genes have been identified (Table 2). Three of these are unlikely to be the TSG: 1) CFTR, the gene mutated in cystic fibrosis that encodes an AMP-regulated chloride channel [34]; 2 ) MET, which encodes an oncogene with a variety of known functions in tumorigenesis [35]; and 3 ) WNT2, which encodes a mitogenic factor that is up-regulated in breast cancer [36]. In contrast, the fourth known gene (ALPHACAP) encodes the F-actin capping protein alpha and represents a viable candidate. It is a member of a family of regulating proteins that are involved in the capping of the actin filament after elongation is completed. It is also known to release the actin filament allowing cytoskeletal reorganization as a result of mitogenic stimuli [37,38]. Interestingly, it has been suggested that members of the actin family may play a role in the regulation of angiogenesis by interacting with angiogenins [38]. Based on our studies showing that the repressed hybrid cell lines obtained by microcell fusion retain the proliferation rate of the parental cell lines [17], it seems unlikely that the TSG encodes a cell-cycle regulator; rather, it is more likely that the TSG product functions within a pathway such as cell-host interaction (e.g., angiogenesis, matrix remodeling). The latter notion makes ALPHACAP a good candidate for further evaluation.
A second area of investigation accelerated by the reported clone-based physical map involves the use of functional complementation assays with the characterized BACs/PACs. Specifically, clones of interest are being retrofitted with suitable mammalian selectable markers [25,39] and introduced into tumorigenic cell lines. Repression of tumor formation with specific BACs/PACs may provide additional clues about the location and functionality of the putative TSG.
In summary, the studies reported here illustrate how the fruits of the Human Genome Project can be used to accelerate research in human oncology. Specifically, an approximately 1-Mb segment of hchr7 presumed to harbor an important TSG has been isolated in bacterial clones, allowing the sequencing of the entire interval to commence and providing valuable informational and experimental tools that will facilitate the study of genes residing in the region.
Acknowledgements
We thank the Washington University Genome Sequencing Center for their BAC/PAC sequencing efforts and their unwavering commitment to the rapid and open release of the resulting sequence data. In particular, we acknowledge Drs. John McPherson and Marco Marra for their collaborative assistance in the construction of BAC/PAC-based physical maps of human chromosome 7. We also thank Nicole Dietrich for technical assistance.
Abbreviations
- BAC
bacterial artificial chromosome
- EST
expressed-sequence tag
- hchr
human chromosome
- LOH
loss of heterozygosity
- PAC
P1-derived artificial chromosome
- STS
sequence-tagged site
- TSG
tumor suppressor gene
- YAC
yeast artificial chromosome
References
- 1.Gupta PK, Sahota A, Boyadjiev SA, Bye S, Shao C, O'Neill JP, Hunter TC, Albertini RJ, Stambrook PJ, Tischfield JA. High frequency in vivo loss of heterozygosity is primarily a consequence of mitotic recombination. Cancer Res. 1997;57:1188–1193. [PubMed] [Google Scholar]
- 2.Zenklusen JC, Conti CJ. Cytogenetic, molecular and functional evidence for novel tumor suppressor genes on the long arm of human chromosome 7. Mol Carcinog. 1996;15:167–175. doi: 10.1002/(SICI)1098-2744(199603)15:3<167::AID-MC2>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 3.Bieche I, Champeme MH, Matifas F, Hacene K, Callahan R, Lidereau R. Loss of heterozygosity on chromosome 7q and aggressive primary breast cancer. Lancet. 1992;339:139–143. doi: 10.1016/0140-6736(92)90208-k. [DOI] [PubMed] [Google Scholar]
- 4.Zenklusen JC, Bieche I, Lidereau R, Conti CJ. (C-A)n microsatellite repeat D7S522 is the most commonly deleted region in human primary breast cancer. Proc Natl Acad Sci USA. 1994;91:12155–12158. doi: 10.1073/pnas.91.25.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zenklusen JC, Thompson JC, Troncoso P, Kagan J, Conti CJ. Loss of heterozygosity in human primary prostate carcinomas: A possible tumor suppressor gene at 7q31.1. Cancer Res. 1994;54:6370–6373. [PubMed] [Google Scholar]
- 6.Oakahashi S, Shan AL, Ritland SR, Delacey KA, Bostwick DG, Lieber MM, Thibodeau SN, Jenkins RB. Frequent loss of heterozygosity at 7q31.1 in primary prostate cancer is associated with tumor aggressiveness and progression. Cancer Res. 1995;55:4114–4119. [PubMed] [Google Scholar]
- 7.Zenklusen JC, Weitzel JN, Ball HG, Conti CJ. Allelic loss at 7q31.1 in human primary ovarian carcinomas suggests the existence of a tumor suppressor gene. Oncogene. 1995;11:359–363. [PubMed] [Google Scholar]
- 8.Koike M, Takeuchi S, Yokota J, Park S, Hatta Y, Miller CW, Tsuruoka N, Koeffler HP. Frequent loss of heterozygosity in the region of the D7S523 locus in advanced ovarian cancer. Genes Chromosomes Cancer. 1997;19:1–5. doi: 10.1002/(sici)1098-2264(199705)19:1<1::aid-gcc1>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Zenklusen JC, Thompson JC, Klein-Szant AJ, Conti CJ. Frequent loss of heterozygosity in human primary squamous cell and colon carcinomas at 7q31.1: Evidence for a broad range tumor suppressor gene. Cancer Res. 1995;55:1347–1350. [PubMed] [Google Scholar]
- 10.Nishizuka S, Tamura G, Terashima M, Satodate R. Commonly deleted region on the long arm of chromosome 7 in differentiated adenocarcinoma of the stomach. Br J Cancer. 1997;76:1567–1571. doi: 10.1038/bjc.1997.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kuniyasu H, Yasui W, Yokozaki H, Akagi M, Akama Y, Kitahara K, Fujii K, Tahara E. Frequent loss of heterozygosity of the long arm of chromosome 7 is closely associated with progression of human gastric carcinomas. Int J Cancer. 1994;59:597–600. doi: 10.1002/ijc.2910590504. [DOI] [PubMed] [Google Scholar]
- 12.Achille A, Biasi MO, Zamboni G, Bogina G, Magalini AR, Pederzoli P, Perucho M, Scarpa A. Chromosome 7q allelic losses in pancreatic carcinoma. Cancer Res. 1996;56:3808–3813. [PubMed] [Google Scholar]
- 13.Shridhar V, Sun QC, Miller OJ, Kalemkerian GP, Petros J, Smith DI. Loss of heterozygosity on the long arm of human chromosome 7 in sporadic renal cell carcinomas. Oncogene. 1997;15:2727–2733. doi: 10.1038/sj.onc.1201448. [DOI] [PubMed] [Google Scholar]
- 14.Neuman WL, Rubin CM, Rios RB, Larson RA, LeBeau MM, Rowley JD, Vardiman JW, Schwartz JL, Farber RA. Chromosomal loss and deletion are the most common mechanisms for loss of heterozygosity from chromosomes 5 and 7 in malignant myeloid disorders. Blood. 1992;79:1501–1510. [PubMed] [Google Scholar]
- 15.Mitelman F, Mertens F, Johansson B. Breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat Genet. 1997;15:417–474A. doi: 10.1038/ng0497supp-417. [DOI] [PubMed] [Google Scholar]
- 16.Ogata T, Ayusawa D, Namba M, Takahashi E, Oshimura M, Oishi M. Chromosome 7 suppresses indefinite division of nontumorigenic immortalized human fibroblast cell lines KMST-6 and SUSM-1. Mol Cell Biol. 1993;13:6036–6043. doi: 10.1128/mcb.13.10.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zenklusen JC, Oshimura M, Barrett JC, Conti CJ. Inhibition of tumorigenicity of a murine squamous cell carcinoma (SCC) cell line by a putative tumor suppressor gene on human chromosome 7. Oncogene. 1994;9:2817–2825. [PubMed] [Google Scholar]
- 18.Matsuda T, Sasaki M, Kato H, Yamada H, Cohen M, Barrett JC, Oshimura M, Wake N. Human chromosome 7 carries a putative tumor suppressor gene(s) involved in choriocarcinoma. Oncogene. 1997;15:2773–2781. doi: 10.1038/sj.onc.1201461. [DOI] [PubMed] [Google Scholar]
- 19.Bouffard GG, Idol JR, Braden VV, Iyer LM, Cunningham AF, Weintraub LA, Touchman JW, Mohr-Tidwell RM, Peluso DC, Fulton RS, Ueltzen MS, Weissenbach J, Magness CL, Green ED. A physical map of human chromosome 7: An integrated YAC contig map with average STS spacing of 79 kb. Genome Res. 1997;7:673–692. doi: 10.1101/gr.7.7.673. [DOI] [PubMed] [Google Scholar]
- 20.Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y, Simon M. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birren B, Mancino V, Shizuya H. Bacterial Artificial Chromosomes. In: Birren B, Green ED, Klapholz S, Myers RM, Riethman H, Roskams J, editors. Genome Analysis. A Llaboratory Manual (Volume 3: Cloning Systems) Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1997. pp. 241–296. [Google Scholar]
- 22.Ioannou PA, Amemiya CT, Garnes J, Kroisel PM, Shizuya H, Chen C, Batzer MA, de Jong PJ. A new bacteriophage P1-derived vector for the propagation of large human DNA fragments. Nat Genet. 1994;6:84–89. doi: 10.1038/ng0194-84. [DOI] [PubMed] [Google Scholar]
- 23.Schuler GD, Boguski MS, Stewart EA, Stein LD, Gyapay G, Rice K, White RE, Rodriguez-Tome P, Aggarwal A, Bajorek E, Bentolila S, Birren BB, Butler A, Castle AB, Chiannilkulchai N, Chu A, Clee C, Cowles S, Day PJR, Dibling T, Drouot N, Dunham I, Duprat S, East C, Edwards C, Fan J-B, Fang N, Fizames C, Garret C, Green L, Hadley D, Harris M, Harrison P, Brady S, Hicks A, Holloway E, Hui L, Hussain S, Louis-Dit-Sully C, Ma J, MacGilvery A, Mader C, Maratukulam A, Matise TC, McKusick KB, Morissette J, Mungall A, Muselet D, Nusbaum HC, Page DC, Peck A, Perkins S, Piercy M, Qin F, Quackenbush J, Ranby S, Reif T, Rozen S, Sanders C, She X, Silva J, Slonim DK, Soderlund C, Sun W-L, Tabar P, Thangarajah T, Vega-Czarny N, Vollrath D, Voyticky S, Wilmer T, Wu X, Adams MD, Auffray C, Walter NAR, Brandon R, Dehejia A, Goodfellow PN, Houglatte R, Hudson JR, Jr, Ide SE, Iorio KR, Lee WY, Seki N, Nagase T, Ishikawa K, Nomura N, Phillips C, Polymeropoulos MH, Sandusky M, Schmitt K, Berry R, Swanson K, Torres R, Venter JC, Sikela JM, Beckmann JS, Weissenbach J, Myers RM, Cox DR, James MR, Bentley D, Deloukas P, Lander ES, Hudson TJ. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 24.Deloukas P, Schuler GD, Gyapay G, Beasley EM, Soderlund C, Rodriguez-Tome P, Hui L, Matise TC, McKusick KB, Beckmann JS, Bentolila S, Bihoreau M-T, Birren BB, Browne J, Butler A, Castle AB, Chiannikulchai N, Clee C, Day PJR, Dehejia A, Dibling T, Drouot N, Duprat S, Fizames C, Fox S, Gelling S, Green L, Harrison P, Hocking R, Holloway E, Hunt S, Keil S, Lijnzaad P, Louis-Dit-Sully C, Ma J, Mendis A, Miller J, Morissette J, Muselet D, Nusbaum HC, Peck A, Rozen S, Simon D, Slonim DK, Staples R, Stein LD, Stewart EA, Suchard MA, Thangarajah T, Vega-Czarny N, Webber C, Wu X, Auffray C, Nomura N, Sikela JM, Polymeropoulos MH, James MR, Lander ES, Hudson TJ, Myers RM, Cox DR, Weissenbach J, Boguski MS, Bentley DR. A physical map of 30,000 human genes. Science. 1998;282:744–746. doi: 10.1126/science.282.5389.744. [DOI] [PubMed] [Google Scholar]
- 25.Mejia JE, Monaco AP. Retrofitting vectors for Escherichia coli-based artificial chromosomes (PACs and BACs) with markers for transfection studies. Genome Res. 1997;7:179–186. doi: 10.1101/gr.7.2.179. [DOI] [PubMed] [Google Scholar]
- 26.Moore DD. Preparation and analysis of DNA. In: Ausubel FM, Brent R, Kington RE, Moore DD, Seidman JG, Smith JA, Struhl S, editors. Current Protocols in Molecular Biology. Princeton, NJ: John Wiley & Sons; 1993. pp. 2.1.1–2.1.6. [Google Scholar]
- 27.Zhang J, Madden TL. PowerBLAST: a new network BLAST application for interactive or automated sequence analysis and annotation. Genome Res. 1997;7:649–656. doi: 10.1101/gr.7.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marra MA, Kucaba TA, Dietrich NL, Green ED, Brownstein B, Wilson RK, McDonald KM, Hillier LW, McPherson JD, Waterston RH. High throughput fingerprint analysis of large-insert clones. Genome Res. 1997;7:1072–1084. doi: 10.1101/gr.7.11.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang H, Qian J, Proffit J, Wilber K, Jenkins R, Smith DI. FRA7G extends over a broad region: coincidence of human endogenous retroviral sequences (HERV-H) and small polydispersed circular DNAs (spcDNA) and fragile sites. Oncogene. 1998;16:2311–2319. doi: 10.1038/sj.onc.1200202. [DOI] [PubMed] [Google Scholar]
- 30.Inoue H, Ishii H, Alder H, Snyder E, Druck T, Huebner K, Croce CM. Sequence of the FRA3B common fragile region: implications for the mechanism of FHIT deletion. Proc Natl Acad Sci USA. 1997;94:14584–14589. doi: 10.1073/pnas.94.26.14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huebner K, Hadaczek P, Siprashvili Z, Druck T, Croce CM. The FHIT gene, a multiple tumor suppressor gene encompassing the carcinogen sensitive chromosome fragile site, FRA3B. Biochim Biophys Acta. 1997;1332:M65–M70. doi: 10.1016/s0304-419x(97)00009-7. [DOI] [PubMed] [Google Scholar]
- 32.Xu Y, Einstein JR, Mural RJ, Shah M, Uberbacher EC. An improved system for exon recognition and gene modeling in human DNA sequences. Ismb. 1994;2:376–384. [PubMed] [Google Scholar]
- 33.Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 34.Seibert FS, Loo TW, Clarke DM, Riordan JR. Cystic fibrosis: channel, catalytic, and folding properties of the CFTR protein. J Bioenerg Biomembr. 1997;29:429–442. doi: 10.1023/a:1022478822214. [DOI] [PubMed] [Google Scholar]
- 35.Vande Woude GF, Jeffers M, Cortner J, Alvord G, Tsarfaty I, Resau J. Met-HGF/SF: Tumorigenesis, invasion and metastasis. Ciba Found Symp. 1997;212:119–132. doi: 10.1002/9780470515457.ch8. [DOI] [PubMed] [Google Scholar]
- 36.Dale TC, Weber-Hall SJ, Smith K, Huguet EL, Jayatilake H, Gusterson BA, Shuttleworth G, O'Hare M, Harris AL. Compartment switching of WNT-2 expression in human breast tumors. Cancer Res. 1996;56:4320–4323. [PubMed] [Google Scholar]
- 37.Maruta H. F-actin cappers. Gan To Kagaku Ryoho. 1997;24:1442–1447. [PubMed] [Google Scholar]
- 38.Strydom DJ. The Angiogenins. Cell Mol Life Sci. 1998;54:811–824. doi: 10.1007/s000180050210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim SY, Horrigan SK, Altenhofen JL, Arbieva ZH, Hoffman R, Westbrook CA. Modification of bacterial artificial chromosome clones using Cre recombinase: Introduction of selectable markers for expression in eukaryotic cells. Genome Res. 1998;8:404–412. doi: 10.1101/gr.8.4.404. [DOI] [PMC free article] [PubMed] [Google Scholar]


