Abstract
Cyclooxygenase is a key enzyme in prostanoid biosynthesis. Mammalian species have two cyclooxygenases, constitutively expressed cyclooxygenase-1 (Cox-1) and inducible cyclooxygenase-2 (Cox-2). Cox-1 and/or Cox-2 have been also identified in other vertebrates, including fish. We identified a second zebrafish Cox-2 gene orthologue, Cox-2b. All of the functionally important amino acids for cyclooxygenase enzymes are conserved in Cox-2b. The 3′ untranslated region of the Cox-2b message contains AU rich elements characteristic of regulation at the level of mRNA stability. Constitutive tissue expression patterns for Cox-2a and Cox-2b are distinct, but overlap. Both Cox-2a and Cox-2b expression are inducible in the kidney when fish are exposed to tetradecanoylphorbol acetate. Like Cox-2a, Cox-2b protein, expressed in COS cells is functionally active. Thus, the zebrafish genome contains two functional, inducible Cox-2 genes. Database searching demonstrates that some fish genomes contain multiple Cox-1 or Cox-2 cyclooxygenase genes, suggesting alternate duplication and retention of this gene.
Keywords: Danio rerio, prostaglandins, gene duplication, teleost genome
Introduction
The cyclooxygenase (Cox) enzymes catalyze the conversion of arachidonic acid to prostaglandin H2, the common precursor for the various prostanoids (the prostaglandins, prostacyclins and thromboxanes). Prostanoids play important roles in a wide range of biological processes that include platelet aggregation, reproduction, thermoregulation, wound healing, water balance, glomerular filtration and hemostasis. One class of the most commonly used pharmacologic agents, the non-steroidal anti-inflammatory drugs or NSAIDs, exert their pharmacological effects by inhibition of cyclooxygenase enzyme activity. The effects of NSAIDs suggest a role for cyclooxygenase products in a wide range of pathophysiological conditions that include chronic and acute inflammation, neurodegenerative diseases, cardiovascular disease and a number of different cancers [1].
Cyclooxygenase genes have been identified in all vertebrates investigated, including mammals, birds, teleosts, and cartilaginous fishes. In contrast, cyclooxygenases have not been identified in unicellular organisms, insects or plants. Mammalian species have two cyclooxygenase genes, constitutively expressed cyclooxygenase-1 (Cox-1) and inducible cyclooxygenase-2 (Cox-2). The Cox-1 and Cox-2 proteins share conserved amino acid sequences (approximately 60%), similar three-dimensional structures, and identical reactions in prostanoid synthesis [2–4].
In fish species, cyclooxygenase orthologues have been cloned from rainbow trout [5], brook trout [6], and zebrafish [7]. Knockdown of Cox-1 in zebrafish, using antisense morpholino oligonucleotides, results in gastrulation arrest and defects in vascular tube formation during development. In addition, PGE2 produced by Cox-1 is essential for gastrulation movement in the zebrafish [8]. In contrast, knockdown in the zebrafish of the single Cox-2 described previously does not cause a developmental phenotype [7, 9].
Genome sequencing projects have revealed sequences of entire genomes for a number of organisms. Sequence analysis can identify potential orthologous genes in different species, and conserved syntenic regions can help to define chromosomal evolution. The last common ancestor of fish and mammals is likely to be the origin of the vertebrate lineage. Sequence comparisons between such distantly related organisms are informative, and understanding of genome evolution provides leads to understanding the function of genes [10]. In this study, we identified a second zebrafish Cox-2 orthologue, by searching the publicly available genome database. We have characterized this gene, which we refer to a Cox-2b, as a second inducible and functional Cox-2 homologue in the zebrafish, produced from a gene distinct from the zebrafish Cox-2 gene first identified by Grosser et al. [7]. Database search also revealed two potential Cox-1 genes in some fish species and two potential Cox-2 genes in other fish, suggesting that duplication of alternative chromosomal regions during teleost evolution has resulted in differential retention of cyclooxygenase genes.
MATERIALS and METHODS
Cloning of the zebrafish Cox-2b coding region
Three zebrafish genomic sequences with high homology to the mouse Cox-2 amino acid sequence were identified by BLAST search (tblastn) of the Ensemble zebrafish genome. Two of these sequences, Cox-1 and Cox-2, have previously been reported by Grosser et al. [7]. Two zebrafish expressed sequence tag (EST) clones (GenBank accession numbers CF997612 and BQ450716) with high similarity to the third zebrafish genomic Cox sequence were obtained and sequenced.
RT-PCR detection of zebrafish COX mRNA
Total RNA samples from a number of zebrafish tissues were isolated with the Trizol reagent (Invitrogen). cDNA was reverse-transcribed with AMV RT XL (Takara). To amplify each cyclooxygenase, the following primer sets were used; zCOX1F3 (5′-ATCTGAAACCCTACACATCCTTCGC-3′) and zCOX1R1 (5′-AGACGTTTTGCTAAAGTTCGCCGTG-3′) for Cox-1, zCOX2aF3 (5′-TACTCATCCTTTGAGGAGATGACAG-3′) and zCOX2aR1 (5′-GACCTTTTACAGCTCTGAACTCCGC) for Cox-2a, zCOX2bF2 (5′-TTTCACAACAGCCCTGAACC-3′) and zCOX2bR2 (5′-GTTGAAGGACTCAACCAAGC-3′) for Cox-2b, zGAPDHF1 (5′-CATTGAGAAGGCCTCAGCTC-3′) and zGAPDHR1 (5′-ACGGACACATCAGCGACTGG-3′) for zebrafish GAPDH.
Induction of Cox-2 genes with TPA in adult zebrafish
TPA (12-O-tetradecanoylphorbol-13-acetate, Sigma) was added to the tank water at a final concentration of 50 ng/ml. After 1 hour, the fishes were dissected, and RNA samples were isolated.
Cyclooxygenase activity
The zebrafish cyclooxygenase expression vectors were constructed by cloning open reading frame sequences into pcDNA3.1 (Invitrogen). The open reading frame sequences were amplified using the following primer sets; zCOX1F1 (5′-TGTGTTCAAAACTACAATGAGAGAG-3′) and zCOX1R1 for Cox-1, zCOX2aF1 (5′-AACACTTCTGGAATGAATAAACTGG-3′) and zCOX2aR1 for Cox-2a, zCOX2bF1 (5′-CACGCCACTGGAATGAAAAGTTCGG-3′) and zCOX2bR1 (5′-TACACTAATCAGAGCTCAGATGTCC-3′) for Cox-2b. For PCR templates, the following EST clones were used; AW342787 for Cox-1, BC056736 for Cox-2a, CF997612 for Cox-2b. Each expression vector was transiently transfected with Lipofectamine 2000 (Invitrogen) into COS-1 cells. Cyclooxygenase activity in microsomal fraction was measured with COX Activity Assay kit (Cayman) according to the manufacturer’s instructions.
Database search
The genomic sequences for cyclooxygenases of the puffer fish Takifugu rubripes and Tetraodon nigroviridis were obtained using BLAST search (tblastn) of Ensemble fugu (assembly 4) and tetraodon genome (assembly 7), respectively. The deduced protein sequences of rainbow trout cyclooxygenases were obtained by BLAST search (tblastn) of The Institute of Genomic Research (TIGR) Oncorhynchus mykiss gene index. The Cox gene sequences for medaka (Oryzias latipes) were obtained from the medaka genome sequencing project. This data has been provided freely by the National Institute of Genetics and the University of Tokyo for use in this publication only.
RESULTS
Blast search of the genome databases of zebrafish (Danio rerio) using the mouse Cox-2 sequence identified three highly homologous sequences. Two of the mammalian Cox-related sequences are identical to zebrafish Cox-1 (Chr. 5) and Cox-2 (Chr. 2) cDNA sequences reported previously [7]. The third Cox-related sequence, not previously reported, is present on Chr. 20 (GenBank accession number DQ494791). Zebrafish expressed sequence tag (EST) clones related to this third Cox zebrafish genomic sequence were identified in the Washington University Genome Resources EST database.
Sequence comparisons of the predicted zebrafish cyclooxygenase proteins
Sequence analysis of these previously uncharacterized, Cox-related ESTs reveals a 1818 bp open reading frame. The deduced amino acid sequence (606 a.a.) was determined from the ORF and is compared in Fig. 1a with zCox-1 (597 a.a.) and the “zCox-2” cDNA described by Grosser et al. [7], which we now term Cox-2a (601 a.a.). For optimal alignment of the zebrafish Cox-2 related sequences, it is necessary to insert a seven amino acid deletion that follows amino acid residue 152 in the Cox-2a sequence.
Figure 1.

(a) Comparison of the deduced amino acid sequences of the Cox-1, Cox-2a and Cox-2b cyclooxygenases from the zebrafish. Conserved amino acids of the cyclooxygenases are shaded. Functionally important amino acids are indicated by asterisks; the active site of Cox (tyrosine-385, histidine-388 and serine-530), the substrate binding site (arginine-120), the N-glycosylation site (asparagines-68, -144 and -410), and the sites crucial for peroxidase activity (glutamine-203 and histidine-207). The two domains that define the haem-binding sites are identified by the black boxes. The amino acids which define conformational differences of the substrate binding channels between Cox-1 and Cox-2 in the mammalian enzymes are indicated by a “+” (arginine-513 and valine-523 in mammalian Cox-2 proteins). (b) The sequences of the AU-rich elements present in the murine Cox-2, zebrafish Cox-1a, Cox-2a and Cox-2b 3′ untranslated regions. Shaw-Kamen AUUUA pentamer motifs are underlined.
All the important structural and functional domains implicated in cyclooxygenase enzyme function are conserved in the new putative cyclooxygenase enzyme, which we term Cox-2b. Like the mammalian cyclooxygenases and the zebrafish Cox-1 and Cox-2a cyclooxygenases previously described, Cox-2b contains an active site tyrosine (Tyr-385), proximal and distal haem-binding histidines (His-207 and His-388), the aspirin acetylation site (Ser-388), potential N-glycosylation sites and a haem-binding domain. The two haem-binding sites are identified by the black boxes in Fig. 1a. The zebrafish Cox-2b sequence is more closely related to mouse Cox-2 (70.5% identity) than to mouse Cox-1 (61.0%), suggesting this gene is a second zebrafish orthologue of the mammalian Cox-2 gene. Zebrafish Cox-2b contains an 18 amino acid C-terminal insertion that is conserved in mammalian Cox-2 proteins (and in zebrafish Cox-2a) and absent in the Cox-1 protein. In contrast, the N-terminal hydrophobic amino acid insertion characteristic of Cox-1 proteins [11] is absent in zebrafish Cox-2b (Fig. 1a).
Amino acids at positions 513 and 523 are postulated to determine the differences in flexibility for the substrate channels of mammalian Cox-1 and Cox-2, and to be responsible for the difference in the specificity of Cox-2 specific inhibitors [reviewed in 12]. Mammalian Cox-1 enzymes have His-513 and Ile-523; while mammalian Cox-2 enzymes have Arg-513 and Val 523 at these residues. Grosser et al. [7] report that both Cox-1 and Cox-2 have Arg and Val in position 513 and 523. The Arg and Val residues at these positions are conserved between Cox-2a and Cox-2b (Fig. 1a).
Constitutive and induced expression of the zebrafish Cox genes
Mammalian Cox-2 mRNAs contain AU-rich elements (AREs) in their 3′ untranslated regions (3′UTR). The presence of AREs is a common characteristic of immediate-early genes and is implicated in post-transcriptional regulation of mRNA stability [reviewed in 13, 14]. AREs of mammalian Cox-2 genes contain multiple, often overlapping, copies of the pentameric AUUUA motif originally described by Shaw and Kamen [15]. As shown in Fig. 1b, the proximal 3′UTR sequences of Cox-2a and Cox-2b also possess 5 and 7 AUUUA sequences, respectively (Fig.1b). In contrast, no AREs are present in the Cox-1 message. The Cox-2b 3′UTR contains two clusters of three overlapping ARE sequences; the murine Cox-2 3′UTR contains a similar set of three overlapping ARE sequences. In contrast, the Cox-2a 3′UTR contains only a single overlapping ARE doublet. The presence of AREs in their 3′UTR sequence suggests that Cox-2a and Cox-2b might both be inducible genes.
Constitutive expression of the three zebrafish Cox-2 genes, Cox-1, Cox-2a and Cox-2b, was examined by RT-PCR in adult zebrafish organs (Fig. 2a). Cox-2b transcripts are present in many tissues, and are particularly high in the gill, heart and ovary. Intermediate constitutive Cox-2b message levels are present in kidney, gut and testis, with lower transcript levels observed in the brain and eye. Constitutive Cox-2b expression is not detected in liver. The Cox-1 and Cox-2a expression patterns are similar to those previously reported by Grosser et al. [7]. Cox-1 message is present in all tissues, albeit weakly in brain. The most robust constitutive Cox-2a signals are observed in the gills, gut and testis, followed by heart and brain, with relatively low expression levels in the eye, liver and kidney. Thus the constitutive expression patterns of the two Cox-2 related zebrafish genes are different, but overlap in the various organs.
Figure 2.
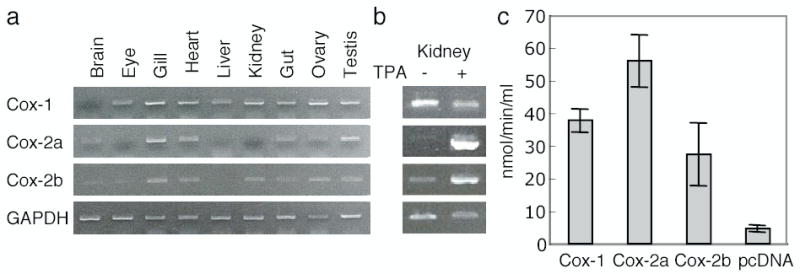
(a), Constitutive Cox-1, Cox-2a and Cox-2b mRNA expression in adult zebrafish organs. Total RNA (100 ng) from each adult organ was reverse-transcribed and amplified with specific primer sets for Cox-1, Cox-2a or Cox-2b. GAPDH was also amplified as a control. (b), Induction of cyclooxygenase expression by TPA in zebrafish tissues. Fish were treated with TPA (50 ng/ml) in the tank water for 1 hour. Total RNA from kidney was reverse-transcribed and amplified to detect the individual cyclooxygenase isoforms and the GAPDH control. (c) Enzymatic activity of the cyclooxygenases produced from plasmids expressing the zebrafish cyclooxygenase isoforms. COS cells were transfected with Cox-1, Cox-2a, Cox-2b or control pcDNA expression vectors. Two days after transfection, cell lysates were prepared, microsome fractions were isolated, and peroxidase enzyme activity was measured. Data are the averages ± S.D. from triplicate assays.
The mouse Cox-2 gene was originally identified in our laboratory as a gene induced by TPA in 3T3 fibroblasts [2]. To examine whether the Cox-2b gene is inducible, zebrafish Cox transcripts were analyzed after challenge of fish in vivo with TPA (Fig. 2b). Constitutive expression of all three cyclooxygenases is observed in kidney, although Cox-2a transcripts are quite low. When the fish are treated with TPA, both the Cox-2a and Cox-2b message levels increase substantially in this organ. In contrast, the kidney Cox-1 transcript level is, if anything, reduced following TPA treatment.
The zebrafish Cox-2b gene, like the Cox-1 and Cox-2a genes, encodes a functional cyclooxygenase
Sequence analysis, mRNA expression and inducibility all suggest that Cox-2b, like Cox-2a, is a functional orthologue of mammalian Cox-2. To analyze Cox-2b function, we expressed the protein and measured its enzymatic activity. The ORFs of the Cox-1, Cox-2a, and Cox-2b cDNAs were subcloned into a mammalian expression vector and expressed in COS cells. Cyclooxygenase is a bifunctional enzyme exhibiting both cyclooxygenase and peroxidase activity. Lysates were prepared from transfected cells and assayed for peroxidase activity characteristic of the cyclooxygenases (Fig. 2c). Like Cox-1 and Cox-2a, Cox-2b encodes peroxidase activity, demonstrating that the zebrafish has three, rather than two, functional cyclooxygenase genes.
Structure of the zebrafish Cox-2 genes
The genomic structure of the zebrafish cyclooxygenase genes was determined by comparing their cDNA sequences with genomic database sequences. Maps of the murine and zebrafish cyclooxygenase gene structures are shown in Fig. 3. The exon/intron structures of zebrafish cyclooxygenases are similar to those of the mouse cyclooxygenase genes, although intron lengths vary substantially. The exon/intron structures of zebrafish Cox-1 and mouse Cox-1 are identical; 11 exons and 10 introns. Like murine Cox-2, zebrafish Cox-2b has one less intron and one less exon than the Cox-1 genes; both murine Cox-2 and zebrafish Cox-2b have 10 exons and nine introns. In contrast, in zebrafish Cox-2a, the intron separating exons four and five is missing; exons four and five form a continuous coding exon sequence in zCox-2a. Thus, in comparison to murine Cox-2 and zebrafish Cox-2b, Cox-2a has one less exon and one less intron. The “fused” exon four of the Cox-2a gene, encoding sequences homologous to exons four and five of murine Cox-2 and zebrafish Cox-2b, is 21 nucleotides shorter in length than the sum of the nucleotides present in the murine Cox-2 or zebrafish Cox-2b exons four and five (305 nucleotides versus 326 nucleotides). This difference in exon length accounts for the seven amino acid deletion observed in the zebrafish Cox-2a protein, when compared to murine Cox-1, Cox-2, and the zebrafish Cox-1 or Cox-2b proteins (Fig. 1a).
Figure 3.
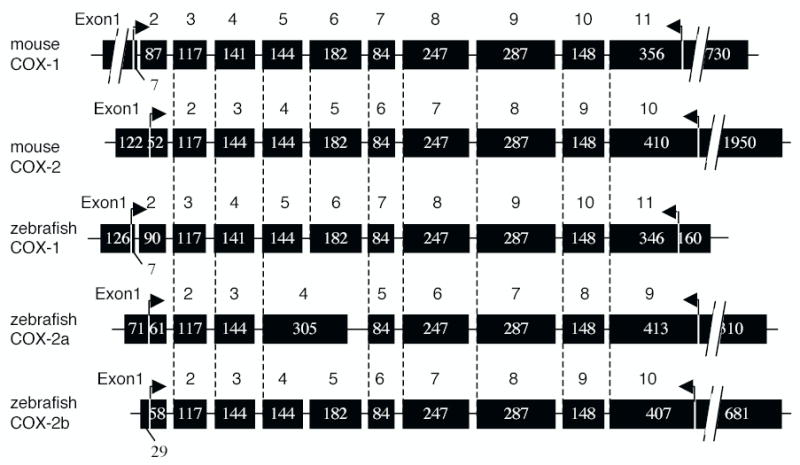
Structures of the mouse and zebrafish Cox genes. The number and length of the exons of the zebrafish Cox-1, Cox-2a and Cox-2b genes are compared with exons of the mouse Cox-1 and Cox-2 genes. The numbers in the boxes indicate the numbers of nucleotides in each exon. The arrows above the genes indicate the coding regions.
The mouse and human Cox-1 genes are approximately 22–25 kb in length, whereas Cox-2 genes are 8 kb [reviewed in 11]. In zebrafish, the Cox-1 gene is also greater than 20 kb in length. In contrast, both zebrafish Cox-2 genes, Cox-2a and Cox-2b, are relatively small; 7.3 and 5.5 kb in length respectively.
Evolutionary relationships among fish Cox genes
Jarving et al. [16] report that database searches suggest the puffer fish fugu (Takifugu rubripes) genome sequence contains three cyclooxygenase related sequences. In contrast to zebrafish, two of the fugu cyclooxygenase genes resemble mammalian Cox-1, while the third fugu gene more closely resembles Cox-2. Our identification of a second Cox-2 gene in the zebrafish prompted us to examine the evolution of the cyclooxygenase genes in the teleost lineage, using existing sequence data. Homology searches were performed on publicly available fish genome databases. Blast searches of the genome database for the fresh water puffer fish Tetraodon nigroviridis, using known Cox sequences, also yields three cyclooxygenase-like sequences. Although these genomic sequences have some gaps, sequence similarities and the distribution of 3′UTR AUUUA sequences indicate that Tetraodon has two Cox-1 genes and one Cox-2 gene (the Cox-1 genes are on chromosomes 4 and 12; Cox-2 is on chromosome 1). Genome database searches for the Japanese killifish, medaka (Oryzias latipes), reveal that this genome also contains two Cox-1 and one Cox-2 like sequences (the Cox-1 sequences are on scaffolds 84 and 283; the Cox-2 gene is on scaffold 230). In contrast, we found a second Cox-2 like gene in the rainbow trout Oncorhynchus mykiss. Individual Cox-1 and Cox-2 orthologues have been reported and cloned previously in this species [5]. Searches of EST databases revealed that rainbow trout also have a third cyclooxygenase sequence. Because the predicted amino acid sequence for this third Cox species in the rainbow trout shows a 59.3% identity with mouse Cox-1 and 69.4% with Cox-2, and because the mRNA of this previously undescribed Cox-related sequence also contains 3′UTR AUUUA sequences, we suggest that this gene is a putative second Cox-2 in this genome. These predicted sequences and the known cyclooxygenases from fish species and ancestral corals have been used, in Fig. 4, to construct a phylogenetic tree. The results indicate that teleosts often possess three cyclooxygenase genes; some species have two Cox-1 genes and a single Cox-2 gene, while other species have a single Cox-1 gene and two Cox-2 genes. However, to date only this study in the zebrafish has demonstrated expression and function for three cyclooxygenases, encoded by three distinct genes, in a single species.
Figure 4.
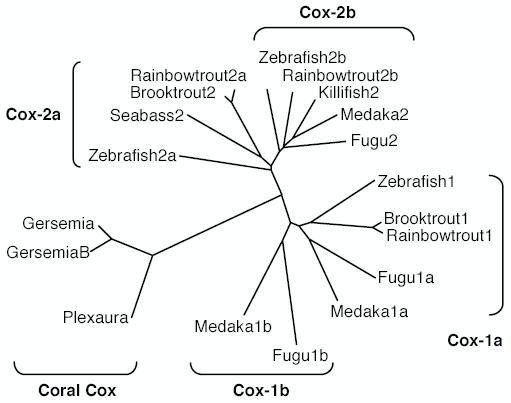
Unrooted phylogenetic distance dendrogram of fish and ancestral coral cyclooxygenases. The tree was constructed by the neighbor-joining method, using the CLUSTAL program.
Discussion
In this study we identified a third functional cyclooxygenase gene, Cox-2b, in the zebrafish. To our knowledge, expression of two distinct inducible, enzymatically active Cox-2 isoforms has not previously been described in any species.
Both Cox-2a and Cox-2b messenger RNAs contains ARE sequences in their 3′UTRs. In contrast, like the mammalian Cox-1 genes, zebrafish Cox-1 has no ARE sequences in its mRNA. AREs are divided generally into three categories; the ARE region of mammalian Cox-2 is classified as class II, in which mRNAs contain multiple AUUUA pentamers, often with some overlap [13, 14]. AREs act as potent mRNA destabilizing sequences. The mouse Cox-2 ARE has three overlapping and four separated AUUUA motifs in close proximity. Cox-2b has 6 overlapping and 1 separated ARE motifs, while Cox-2a has 2 overlapping and 3 separated ARE motifs. It will be instructive to compare the mRNA destabilizing effects of the zebrafish Cox-2a and Cox-2b 3′UTRs with that of human and or mouse Cox-2 3′UTRs. Such a comparison might help to elucidate which of the two zebrafish Cox-2 genes is more closely related functionally to the single mammalian Cox-2 gene.
Exon-intron junctions of cyclooxygenases are highly conserved in all mammalian species studied. Cox-1 genes contain an extra intron (intron 1) compared with Cox-2 genes. Comparison of the zebrafish Cox-2b cDNA sequence with genomic database sequences indicates that the exon-intron structure of the zebrafish Cox-2b gene is identical to mammalian Cox-2, with 10 exons and 9 introns. In contrast, zebrafish Cox-2a lacks one intron when compared both to mammalian Cox-2 genes and to zebrafish Cox-2b. Thus comparison of gene structures also suggests that zebrafish Cox-2a is more divergent than is zebrafish Cox-2b from mammalian Cox-2 genes.
By database search of genomic and EST sequences, we also detected three cyclooxygenase genes in other teleosts; the puffer fishes Takifugu rubripes and Tetraodon nigroviridis, the Japanese killifish Oryzias latipes, and the rainbow trout Oncorhynchus mykiss. Among these species, both puffer fish species and killifish have two putative Cox-1 genes and one putative Cox-2 gene. In contrast, like the zebrafish, the rainbow trout has two Cox-2 genes. For many gene families, fish have more members than do tetrapods. The frequency of two paralogous genes in fish species corresponding to one ortholog in tetrapods has suggested that a whole-genome duplication occurred in the teleost lineage after its divergence from the tetrapod lineage [17–20]. Recently, Woods et al. [21] showed that different duplicated genes have been lost in zebrafish and Tetraodon, although similar overall numbers of duplicated genes may have been retained in these two species. The differences in retention of the Cox-2 and Cox-1 gene pairs respectively in zebrafish and Tetraodon are consistent with the conclusions of Woods et al [21]. The Cox-1 and Cox-2 genes are likely to have been duplicated after the divergence of teleosts from tetrapods; subsequently puffer fishes and killifish retained two Cox-1 genes, while zebrafish and rainbow trout retained two Cox-2 genes.
Expression of the single Cox-2 genes in mammals is induced by wide variety of signals, including growth factors, cytokines, and neuronal depolarization, as well as in many pathophysiological conditions such as neuronal degeneration, inflammation and cancer [22]. It will be of great interest to compare the regulation of the two Cox-2 genes present in zebrafish and rainbow trout, to determine whether induction of the two genes shows substantial overlap in response to alternative ligands, or demonstrates differential induction responses to alternative stimuli.
Acknowledgments
We thank Art Catapang for technical assistance, and Naveen K. Jain for helpful discussions and comments. This work was supported by NIH Grant RO1 CA123055 (HRH) and Exploration-Hypothesis Development Award from the Department of Defense CMLRP (KJPG).
References
- 1.Herschman HR. In: Historical aspects of COX-2: Cloning and characterization of the cDNA, protein and gene, in COX-2 blockade in cancer prevention and therapy. Harris RE, editor. Jumana Press; Clifton, NJ: 2003. pp. 13–32. [Google Scholar]
- 2.Kujubu DA, Fletcher BS, Varnum BC, Lim RW, Herschman HR. TIS10, a phorbor ester tumor promoter-inducible mRNA from Swiss 3T3 cells, encodes a novel prostaglandin synthase/cyclooxygenase homologue. J Biol Chem. 1991;266:12866–12872. [PubMed] [Google Scholar]
- 3.O’Banion MK, Sadowski HB, Winn V, Young DA. A serum-and glucocorticoid-regulated 4-kilobase mRNA encodes a cyclooxygenase-related protein. J Biol Chem. 1991;266:12866–12872. [PubMed] [Google Scholar]
- 4.Xie W, Chipman JG, Robertson DL, Erikson RL, Simmons DL. Expression of mitogen-responsive gene encoding prostaglandin synthase/cyclooxygenase homologue. Proc Natl Acad Sci USA. 1991;88:2692–2696. doi: 10.1073/pnas.88.7.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou J, Neumann NF, Holland JW, Belosevic M, Cunningham C, Secombes CJ, Rowley AF. Fish macrophage express a cyclo-oxygenase-2 homologue after activation. Biochem J. 1999;340:153–159. [PMC free article] [PubMed] [Google Scholar]
- 6.Roberts SB, Langenau DM, Goetz FW. Cloning and characterization of prostaglanin endoperoxide synthase-1 and -2 from the brook trout ovary. Mol Cell Engocrinol. 2000;160:89–97. doi: 10.1016/s0303-7207(99)00252-x. [DOI] [PubMed] [Google Scholar]
- 7.Grosser T, Yusuff S, Cheskis E, Pack MA, Fitzgerald GA. Developmental expression of functional cyclooxygenases in zebrafish. Pro Natl Aca Sci. 2002;99:8418–8423. doi: 10.1073/pnas.112217799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cha YI, Kim SH, Sepich D, Buchanan FG, Solnica-Krezel S, Dubois R. Cyclooxygenase-1-derived PGE2 promotes cell motility via the G-protein-coupled EP4 receptor during vertebrate gastrulation. Gene Dev. 2006;20:77–86. doi: 10.1101/gad.1374506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cha YI, Kim SH, Solnica-Krezel S, DuBois RN. Cyclooxygenase-1 signaling is required for vascular tube formation during development. Dev Biol. 2005;282:274–283. doi: 10.1016/j.ydbio.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Crollius HR, Wessenbach J. Fish genomics and biology. Genome Res. 2005;15:1675–1682. doi: 10.1101/gr.3735805. [DOI] [PubMed] [Google Scholar]
- 11.Herschman HR, Talley JJ, Dubois R. Cyclooxygenase 2 (COX-2) as a target for therapy and noninvasive imaging. Mol Imaging and Biol. 2003;5:286–303. doi: 10.1016/j.mibio.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Flower RJ. The development of Cox2 inhibitors. Nature Rev Drug Discov. 2003;2:179–191. doi: 10.1038/nrd1034. [DOI] [PubMed] [Google Scholar]
- 13.Espel E. The role of the AU-rich elements of mRNAs in controlling translation. Sem Cell Dev Biol. 2005;16:59–67. doi: 10.1016/j.semcdb.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Dean JLE, Sully G, Clark AR, Saklatvala J. The involvement of AU-rich element-binding proteins in p38 mitogen-activated protein kinase pathway-mediated mRNA stabilisation. Cell Signal. 2004;16:1113–1121. doi: 10.1016/j.cellsig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 15.Shaw G, Kamen R. A conserved AU sequence from the 3′ untraslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 16.Jarving R, Jarving I, Kurg R, Brash AR, Samel N. On the evolutionary origin of cyclooxygenase (COX) isozymes. J Biol Chem. 2004;279:13624–13633. doi: 10.1074/jbc.M313258200. [DOI] [PubMed] [Google Scholar]
- 17.Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 18.Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene function. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 19.Taylor JS, Baasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaillon O, Aury J-M, Brunet F, Petit J-L, Stange-Thomann N, Maucell E, Bouneau L, Fischer C, Ozouf-Coxtaz C, Bernot A, Nicaud S, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- 21.Woods IG, Wilson C, Friedlander B, Chang P, Reyes DK, Nix R, Kelly PD, Chu F, Postlethwait JH, Talbot WS. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herschman HR. In: Regulation and function of prostaglandin synthase 2/cyclooxygenase, in The eicosanoids. Curtis-Prior P, editor. John Wiley & Sons, Ltd; 2004. [Google Scholar]


