Abstract
Myosin VI moves toward the pointed (minus) end of actin filaments, the reverse direction of other myosin classes. The myosin VI structure demonstrates that a unique insert at the end of the motor repositions its lever arm and is at least in part responsible for the reversal of directionality. However, it has been proposed that there must be additional modifications within the motor that contribute to its large step size and to the reversal of directionality. To ascertain the inherent directionality of the motor core, we attached the myosin V lever arm to myosin VI, with and without the unique insert. If the insert was maintained, the motor moved toward the minus end of actin filaments, but if removed, movement was redirected toward the plus end. Single-molecule studies revealed that further adaptations within the motor increase the magnitude and variability of the plus-end directed converter movements, and unexpectedly provide the source of the highly variable myosin VI step size. Thus, the unique insert is necessary and sufficient to reverse an inherently plus-end directed myosin.
Keywords: lever arm, motility, unconventional myosin, reverse direction
The myosin superfamily is composed of at least 20 classes of molecular motors that move along actin filaments (1). Actin binding drives conformational changes in the myosin motor that leads to sequential release of the hydrolysis products of ATP (Pi followed by ADP). These conformational changes in the motor are amplified by a “lever arm,” which is an extended α-helix composed of consensus calmodulin (CaM) and/or CaM-like light chain binding sites (IQ motifs). This lever arm is attached to a region of the motor known as the converter, which amplifies and directs (converts) the movements resulting from the actin-coupled changes in the motor conformation. This results in a swinging of the lever arm, roughly parallel to the actin filament (2). This swing is known as the myosin powerstroke, which begins when myosin binds to actin with Pi and ADP bound at the active site (prepowerstroke), and ends when the hydrolysis products have been released (rigor). Rigor positions of the lever arms of class VI and V myosins are schematized in Fig. 1 A and B, respectively.
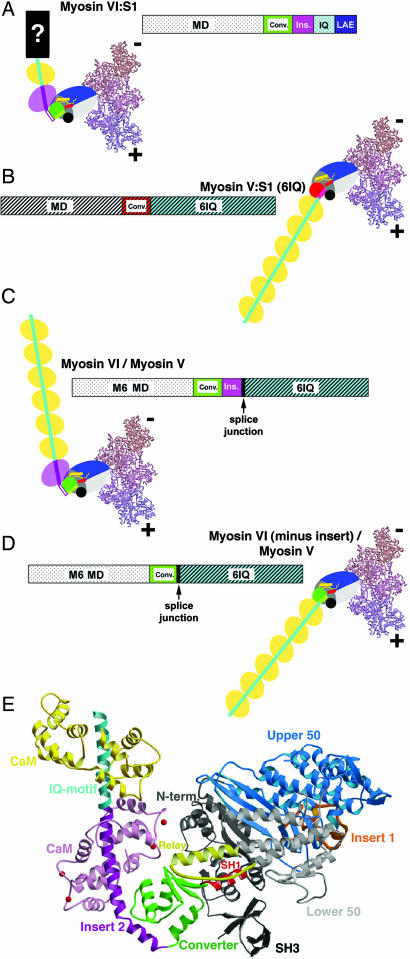
Fig. 1.
Myosin constructs. (A–D) Schematics of wild-type and chimeric myosin constructs. Both bar representation and cartoons of the wild-type myosin VI and myosin V, as well as the chimeric proteins are depicted. Wild-type myosin VI:S1 and myosin V:S1 constructs (single-headed constructs truncated proximal to their respective coiled coil dimerization domains) are depicted in A and B. The bar diagrams indicate the position of the respective motor domains, converters, insert 2 (myosin VI only), and the CaM-binding IQ motifs for myosin VI and myosin V. In the cartoons, the motor domains are shown as solid colored (blue, gray, green/red) large ovals, to which the lever arms are attached. (The small yellow and red cylinders within each motor domain represent the relay helix and SH1 helix, respectively; see E.) The lever arms (IQ motifs with bound CaMs) are depicted by the sequential yellow ovals, representing individual CaM molecules. Note that, as shown in A, for the wild-type myosin VI:S1, the lever arm consists of one IQ-bound CaM that is extended by a region of unknown structure (7). This is denoted by a “?” in the cartoon and labeled LAE (lever arm extension) in the bar diagram. For the cartoons, the converter subdomain of each motor (shown in detail in Fig. 2) is represented by a green circle in myosin VI (A, C, and D) and by a red circle in myosin V (B). This is consistent with the coloring of the myosin VI and myosin V converters shown in Fig. 2. The unique myosin VI insert (insert 2) and bound CaM are in purple (A and C), followed by the IQ-bound CaMs in yellow. In the chimeras (C and D), the myosin V lever arms are fused onto the myosin VI converter with (C) or without (D) the myosin VI-specific insert (insert 2). The junction for each of the chimeras is indicated in the bar diagrams. The splice junction is at Lys-809 from myosin VI (followed by Ala-764 from myosin V) in C (insert 2 maintained). The splice junction is at Lys-771 from myosin VI (followed by Ala-764 from myosin V) in D (insert 2 removed). In all cartoons (A–D), the motor domains are docked onto two actin monomers positioned as in the Holmes model of the actin filament (10). The positioning is based on the high-resolution structures of myosin VI and myosin V (9, 11) and cryo-EM reconstructions of actin-myosin rigor complexes (3, 10). (E) Ribbon diagram of the myosin VI motor with insert 2, the IQ motif and associated CaMs. The structural elements of the myosin VI motor are indicated in different colors, maintaining the same color-coding as in A–D. The subdomains of the motor, which move relative to each other in different states of the motor cycle, are: upper 50 kDa subdomain (blue); lower 50 kDa subdomain (light gray); N-terminal subdomain (dark gray/black); and the converter (green). Within the motor, the relay (dark yellow) and SH1-helix (red) are elements that determine the positioning of the converter, and insert 1 (orange) is involved in altering nucleotide binding (9). The unique myosin VI insert (insert 2) and bound CaM are in purple (red balls indicating Ca2+ ions), followed by the IQ-motif (cyan) and bound CaM (yellow).
Whereas, in general, myosin motors move toward the barbed (plus) end of an actin filament, myosin VI was the first superfamily member shown to traffic toward the pointed (minus) end of the actin filament (3). This allows it to fulfill a number of unique cell biological functions (4, 5). In addition to reverse directionality, myosin VI has a number of additional unusual features. Like myosin V, dimers of myosin VI are capable of taking multiple steps (processive movement) on an actin filament without detachment (6). These steps are similar in size to those of myosin V, even though the lever arm of myosin VI contains only one IQ motif (CaM-binding site), whereas that of myosin V contains six (Fig. 1 A and B). As also noted in Fig. 1A, it appears that the lever arm of myosin VI is extended by a somewhat flexible structure of unknown nature that is distal to the IQ motif (7). This lever arm extension precedes the region responsible for dimerization (6). There is also a second calmodulin bound to one of two inserts (insert 2) that are both unique to class VI myosins. The calmodulin bound to insert 2 is structural and contains four tightly bound calcium ions (8, 9). Based on the high-resolution structure of the myosin VI motor (Fig. 1E), this CaM and insert 2 appear to be integral components of the myosin VI converter (9).
In the initial description of reverse directionality of myosin VI, it was postulated that the unique insert at the end of the motor domain (insert 2) was likely a component of a redesigned converter that allows repositioning of the lever arm and reversal of directionality (3). The recently published structure of myosin VI is consistent with this speculation, and reveals that the insert wraps around the converter and binds a calmodulin that interacts with the converter. The result is a ≈120° repositioning of the myosin VI lever arm. However, it was also noted that the large movement size of the crystallized myosin VI construct could not be explained unless myosin VI initiates its movement (powerstroke) on actin in a state (prepowerstroke state) unlike that previously seen for plus-end directed myosins (9). This observation raises the possibility that part of the mechanism of directionality reversal is within the motor domain itself, and that the repositioning of the lever arm simply amplifies the magnitude of the reverse movement. This would be consistent with a previous study on chimeras between myosin V and myosin VI that appeared to demonstrate that it is the core of the myosin VI motor, and not the unique insert, that is responsible for reverse directionality (12). This conclusion of Homma et al. (12) was based on the inability of chimeras that either removed the insert from myosin VI and added myosin V lever arms or added the insert to myosin V with the myosin VI lever arms to reverse directionality. Additionally, attempts to add the myosin VI converter plus insert 2 to myosin V failed to alter the direction of movement. We have analyzed the chimeras that were created in the study of Homma et al. (12) in light of the structural data now available. Based on this, we postulate why their experiments may not have unambiguously addressed the role of the unique insert. Accordingly, we have used the structural information to design new chimeras to definitively evaluate the role of the insert in the myosin VI converter in the determination of motor directionality.
Results
Structural Insights into Chimera Design.
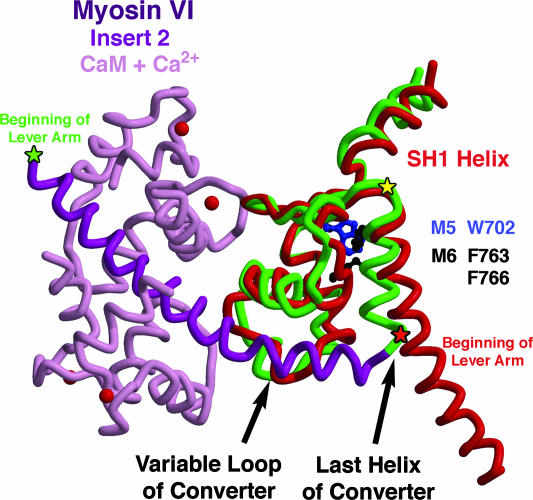
Our attempts to exchange the entire converter of myosin VI with that of either myosin II (chicken smooth) or myosin V (chicken) resulted in constructs with either no or poor movement and low enzymatic activity, making directionality determination impossible (data not shown). We did not attempt to duplicate the chimera of Homma et al. (12) that added insert 2 from myosin VI into the myosin V converter (junction at Gly-761 of myosin VI and Ala-753 of myosin V, marked by the yellow star in Fig. 2), as it is clear from the structure of myosin VI that this would not lead to a reversal of direction due to two structural mismatches. The first is a steric clash between W702 of the myosin V converter and F763 of the last helix of the myosin VI converter (shown in Fig. 2), which would have prevented the last helix to be oriented as found in myosin VI. Secondly, to be positioned properly, the insert requires specific interactions with the variable loop of the converter (depicted in Fig. 2), which only the myosin VI converter provides. Unable to wrap around the converter, the insert in the context of a myosin V/myosin VI chimera likely would simply have lengthened the lever arm of a plus-end motor, consistent with the observations of Homma et al. (12).
Fig. 2.
The converter regions of myosin VI and myosin V. The converter subdomain of the myosin motor immediately follows the SH1 helix and precedes the lever arm. The myosin VI converter (green) is extended by insert 2 (dark purple) plus a CaM (light purple; with Ca2+ ions as red balls), which function to redirect the lever arm helix (beginning at green star; Lys-809). For comparison, the SH1 helix and converter of myosin V (red) are overlaid. To create the myosin VI/myosin V chimeras for this study, the myosin V lever arm was fused onto the myosin VI converter either following insert 2 (green star), or at the end of the last helix of the converter preceding the insert (red star; Lys-771 of myosin VI). The former should position the beginning of the lever arm as in myosin VI, whereas the latter should position it as in myosin V, as schematized in Fig. 1 C and D, respectively. Note that the chimeras of Homma et al. (12) that involved either removal of insert 2 from myosin VI or insertion of insert 2 into myosin V were joined at the position of the yellow star (Gly-761), which is the beginning of the last helix of the converter, as discussed in the text.
The one type of chimera that would seem simple to design would entail removal of the unique insert of myosin VI and simply extending the lever arm in the manner of a plus-end-directed myosin motor, similar to what is shown in the cartoon of Fig. 1D. Although the earlier study of Homma et al. (12) did create a chimera in which insert 2 from myosin VI was removed and the myosin V lever arm was added to the myosin VI converter, the junction between the myosin VI and myosin V sequences created a structural incompatibility. The junction was again at the position marked by the yellow star in Fig. 2 (Gly-761 in myosin VI), which likely detached the last helix of the converter from the myosin VI motor due to the loss of myosin VI-specific interactions (F763 and F766, shown in Fig. 2). Not surprisingly, their construct displayed a 6-fold slowing of velocity, but inexplicably was reported to move toward the minus end of the actin filament, like wild-type myosin VI. We also constructed similar chimeras before the solving of the myosin VI structure, but observed either no movement, or movement so slow that directionality could not be unambiguously assigned (data not shown).
Based on these considerations, we created redesigned chimeras of myosin VI with myosin V lever arms, either with insert 2 maintained (Fig. 1C), or without insert 2 (Fig. 1D). First we sought to avoid the problems associated with removing insert 2 from the myosin VI converter (Fig. 1D) that were created in the earlier chimeras (12) by moving the myosin VI/myosin V splice junction to the end of the last helix of the converter. This insures that the last helix of the converter remains coupled to the myosin VI converter and positions the six IQ-containing myosin V lever arm as in a plus-end motor when insert 2 is removed (compare Fig. 1D and Fig. 1B). Thus, the chimera that deleted the insert 2 (Fig. 1D) was made at the position of the red star in Fig. 2 (end of the last helix of the myosin VI converter). This entailed joining myosin VI residue K771 to myosin V residue A764. To construct the chimera that maintained insert 2 (Fig. 1C), the first IQ of myosin V replaced the IQ of myosin VI and was followed by the rest of the myosin V lever arm. Fusion took place at the position of the green star (K809 of myosin VI) in Fig. 2.
The Unique Insert Alters Directionality.
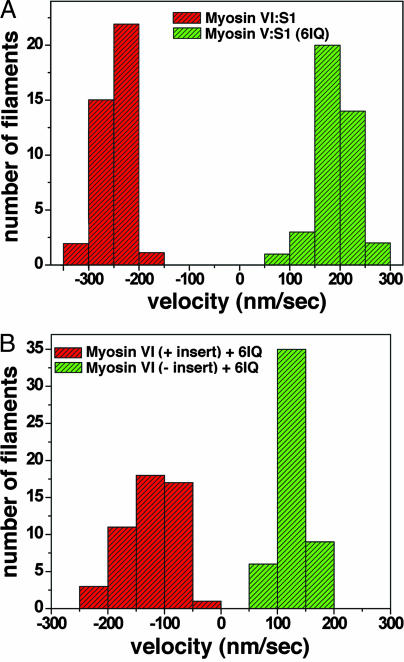
The directional movement of monomeric constructs is summarized in the histogram in Fig. 3 A [see supporting information (SI) Movies 1–4]. The wild-type myosin V single-headed construct (S1) moved actin filaments with the gelsolin (plus) end trailing, and thus with the myosin motor directing movement toward the plus end (average velocity of 190 ± 37 nm/s). For the myosin VI wild-type S1, the opposite movement (average velocity of 245 ± 30 nm/s) was observed (Fig. 3A), with the gelsolin (plus) end of the filament leading (minus-end directed movement). For the chimera depicted in Fig. 1C, the results summarized in Fig. 3B demonstrate that replacement of the IQ-CaM of myosin VI with the six IQ motif-lever arm of myosin V (Myosin VI + 6IQ) maintained minus-end movement, but the velocity of actin filament translocation was slowed (129 ± 27 nm/s) as compared with the wild type (one IQ-lever arm plus extension). This difference is explained mostly by the fact that the kinetics are altered in the chimera, which has a maximal actin-activated ATPase activity of 3.1 ± 0.14 s−1, as opposed to 5.3 ± 0.21 s−1 for the wild type (Myosin VI:S1+extension) at 20°C. Although it is unclear what effect strain may have on the kinetics and motility, the difference in kinetics suggests that the two constructs have similar unitary displacements.
Fig. 3.
Histograms of directionality and velocities of actin filament sliding by wild type and chimeric myosin VI and myosin V motors. (A) Plotted are the sliding velocities of individual actin filaments that contained labeled gelsolin. The direction of movement was determined based on gelsolin labeling of the plus-end of the actin filament. Filaments moved by the wild-type myosin VI:S1 (Fig. 1A) are represented by red bars, and green bars are used for the wild-type myosin V:S1 (Fig. 1B). (B) Filaments moved by the chimera between the myosin VI motor and myosin V lever arm in which insert 2 was retained (Fig. 1C) are represented by red bars, whereas those moved by the chimera in which insert 2 was removed (Fig. 1D) are shown as green bars. Minus velocity signifies minus end-directed actin movement and positive velocity signifies plus-end directed movement.
Removal of the insert from myosin VI and addition of the myosin V lever arm (Myosin VI-insert + 6IQ; depicted in Fig. 1D) resulted in reversal of directionality (Fig. 3B) and a further slowing of the maximal actin-activated ATPase activity (2.3±.08 s−1) and motility (121 ± 42 nm/s). Given the kinetic differences, the unitary step size is likely similar (or greater) to that of the wild-type myosin VI and the other chimera.
Movements of the Myosin VI Converter Lead to the Variable Step Sizes of the Dimer.
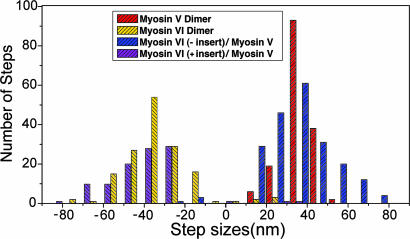
To gain further insight into the movements of the myosin VI converter, in the absence of both insert 2 and the lever arm extension, we extended the myosin V sequence of the two myosin VI / myosin V (6IQ) chimeras through most of the native coiled coil to create two-headed (dimeric) chimeras. We first examined the directionality of the dimers in the actin gliding assays, as was done for the single-headed constructs. The dimeric construct without insert 2 moved toward the plus-end of the actin filaments and the dimer with the insert maintained moved to the minus-end of actin filaments, as had their single-headed counterparts. We then used the single molecule technique known as FIONA (13, 14) to measure the step sizes of single dimeric molecules moving on a stationary actin filament. The centroid of movement was monitored by binding a Cy3-conjugated antibody (anti-FLAG) to the C terminus of the chimera, following the coiled coil. As shown in the histogram of Fig. 4, the average step size of the myosin VI (minus insert)/myosin V chimera was 36.0 nm (±14.7 nm), which is similar to the published average values for two-headed wild-type myosin V (13, 15). For the myosin VI (plus insert)/myosin V chimera, the average step size was 39.7 nm (±15.6 nm). Thus, not only is the rotation of the myosin VI converter plus-end directed, but the amplitude of the rotation (powerstroke) must be at least as large or larger than that of the myosin V converter.
Fig. 4.
Comparison of step sizes of dimeric constructs. Plotted are the distributions of the sizes of individual steps taken by single molecules moving along a stationary actin filament for wild-type myosin VI (yellow) and myosin V (red) dimers, as well as the myosin VI (minus insert)/myosin V chimeric dimer (blue) or the myosin VI (plus insert)/myosin V chimeric dimer (purple). Step sizes were determined by using the FIONA technique (13, 14), but in this case, the centroid of motion was measured by attaching a Cy3-labeled antibody to the C terminus of the individual molecules. Positive steps denote movements toward the plus end of the actin filament, whereas negative steps indicate movements toward the minus end of the filament.
Thus, as summarized in Fig. 4, the average step size for both of the dimeric chimeras was similar to that of myosin V (somewhat larger in the case of the dimeric chimera with insert 2 maintained). However, the distribution of step sizes was much broader, and appeared more like the distribution that had been previously published for dimers of myosin VI (14, 16–18). To verify this finding, we used the FIONA technique to track the centroid of a Cy3-labeled antibody bound to the C terminus of dimers of wild-type myosin V or myosin VI under the same assay conditions used for the dimeric chimeras. Indeed, as shown in Fig. 4, the distribution of step sizes was broader than for myosin V (36.0 ± 7.2) for both the myosin VI (minus insert)/myosin V chimera (36.0 ± 14.7 nm) and myosin VI (plus insert)/myosin V chimera (39.7 ± 15.6 nm). In both cases, the broadness of the distribution was similar to that of wild-type myosin VI (35.5 ± 12.4 nm). Also in both cases a small number of backward (opposite direction) steps were observed as for wild-type myosin VI.
Discussion
The data clearly indicate that insert 2 is the origin of reverse directionality in myosin VI. The use of the myosin V lever arm, which is unlikely to have multiple conformations, as has been suggested for the myosin VI lever arm plus extension (7), provides an easily interpreted amplification of the movements of the myosin VI converter. With insert 2 removed, myosin VI is a plus-end directed motor, and thus the core of the motor is not involved in directionality reversal. However, it was somewhat unexpected (discussed below) that the movements of the chimeras would be similar in velocity, and likely in unitary displacements. It was also unexpected that the removal of the native myosin VI IQ-CaM would have an impact on the kinetics of the motor. This finding suggests that CaM–converter and CaM–CaM interactions may influence the conformation of the converter and kinetics of the myosin VI motor in some yet-to-be-determined manner.
As we previously pointed out (9), the large movement associated with the myosin VI construct truncated after its IQ-bound calmodulin (7) is impossible to reconcile with the prepowerstroke structure that has been characterized for other myosins and the rigor-like structure of myosin VI (beginning and end of powerstroke, respectively), because it predicts a much smaller powerstroke than measured (2.5 nm predicted vs. 11 nm measured). It also would predict that the end of the lever arm would be positioned close to the actin filament in the prepowerstroke state, which could interfere with processive movement in a two-headed motor. These points are illustrated in Fig. 5. This prompted us to postulate two fundamentally different mechanisms by which myosin VI movements could be accomplished: that myosin VI either has a unique prepowerstroke state in which the converter positioning is unlike that of other myosins, or that the converter may uncouple from the motor domain in the prepowerstroke state (essentially disrupting the normal prepowerstroke positioning of the converter), leading to a plus end-biased positioning. The latter mechanism (uncoupling) would predict that the movement of a construct with the insert removed would be much smaller than with the insert maintained, and minus-end directed movement might even be maintained with insert 2 removed. Because we see similar velocities of movement (and step sizes) with or without the insert, but in opposite directions, we now exclude the uncoupling model and favor a model in which myosin VI has a unique converter position in its prepowerstroke state. Based on the step size data summarized in Fig. 4, this unique converter position generates a powerstroke that must be similar in amplitude to that of myosin V. In this model, the unique prepowerstroke state does not alter the directionality of the motor, but merely alters the amplitude of the lever arm swing. This may explain why earlier attempts (12) to use the myosin VI converter plus insert to reverse the direction of myosin V were not successful. The motor domain of myosin V simply may not accommodate the unique myosin VI converter conformation. Further structural studies of the myosin VI prepowerstroke state will be required to provide additional insights into the mechanism of myosin VI movement, and to allow the design of chimeras in which the entire converter is exchanged.
Fig. 5.
Comparison of the rigor (A) and hypothetical prepowerstroke (B) states of myosin VI. The structures are oriented as in Fig. 1, as if they were docked onto an actin filament, which is represented by an arrow with polarity noted. The first part of the myosin VI-specific insert (insert 2) is in purple and a blue line represents the axis for the rest of the insert 2 helix that binds Ca2+–CaM. The converter is in green with its last helix in bright pink. (A) The published rigor-like structure (9). (B) A model of the prepowerstroke state of myosin VI in which the converter (including insert 2) from the myosin VI rigor structure (9) is positioned in the scallop myosin II prepowerstroke state structure (19). Note how this would position the insert 2-helix perpendicular to the actin filament. This would result in a small movement at the end of the helix (marked by red stars), in disagreement with the large movements that have been measured (7). Ideally, a novel myosin VI prepowerstroke state would orient the insert 2-helix parallel to the actin filament, directed toward the plus end of the filament, as indicated by the green line with a green star at the end. This positioning would correctly predict the 11-nm stroke size of the myosin VI:S1 (7). How this might be accomplished will require additional structural information.
The step size histogram of Fig. 4 reveals that not only is the large size of the myosin VI step primarily a function of the movements of the converter, but so is the variability of the step size. We previously speculated that the step size variability might come from either a transient uncoupling of insert 2 (16) and/or from flexibility within the lever arm extension (7). Note that not only do we observe steps larger in size than any seen for myosin V, but back-steps are also seen (Fig. 4), as is characteristic for wild-type myosin VI. Because both insert 2 and the lever arm extension have been removed and replaced by the myosin V lever arm in one of the chimeric dimers (dimer based on monomer shown in Fig. 1D), the large and variable step size must come primarily from variable conformations and/or structural states involving positioning of the converter. This, in turn, leads to a variable positioning of the lever arm during hand-over-hand processive stepping, which we have directly observed (14). Ultimately, this creates a variable biasing of the lead head attachment site on the actin filament. How this converter movement is achieved is unclear. It does, however, imply that the lever arm extension may not be extensible, as we previously have speculated solely on the basis of the variable step size (7, 16, 20). This more easily explains the fact that the step size is not load dependent below the stall force (20). The structure of the lever arm extension remains unresolved, as is the reason for myosin VI adopting such a divergent lever arm design.
The implication of our results is that the myosin VI insert (insert 2) and converter are not portable elements that can reverse the directionality of other classes of myosin. They are specifically adapted to optimize minus-end directed actin movement of the myosin VI motor. In the myosin VI context, the unique insert reverses the motor core's intrinsic plus-end directed and highly variable movements.
Materials and Methods
Design of the Chimeras.
Myosin V lever arms were attached to the myosin VI converter with or without insert 2. The use of the long (six CaM-containing) myosin V lever arms was to allow a large amplification of the motions of the myosin VI converter, without the possible complications in interpretation that could arise if we used the myosin VI lever arm plus extension. The lever arm extension of myosin VI appears to be of variable length and possess an undetermined structure (7). The structure and function of the myosin V lever arm is well understood compared with the myosin VI lever arm plus extension (15).
To create the myosin VI:S1 construct (Fig. 1A), the porcine myosin VI cDNA was truncated at amino acid 917, as described (6). This truncation is at the end of the lever arm extension and precedes the dimerization domain of myosin VI. The myosin V:S1 (Fig. 1B) was formed as described (15) by truncating the chicken myosin V heavy chain at the beginning of the coiled coil, preserving the entire 6-IQ-containing myosin V lever arm. To create the myosin VI/V chimera in which the myosin V lever arm replaced the myosin VI lever arm plus extension (Fig. 1C), myosin VI was truncated after insert 2 at residue K809 and joined to the myosin V lever arm at myosin V residue A764. The myosin V (6IQ) lever arm ended at K909. To form the myosin VI/V chimera with insert 2 removed (Fig. 1D), myosin VI residue K771 was joined to myosin V residue A764 through the end of the 6-IQ lever arm (K909). All constructs had a GFP followed by a Flag tag (encoding GDYKDDDDK) appended to the C terminus. The GFP provided an antibody-binding site for motility assays, whereas the FLAG tag was used to facilitate purification.
The wild-type myosin V and myosin VI dimers were those described in refs. 15 and 16, respectively. The only difference was the removal of GFP + FLAG at the C terminus, which was replaced by the FLAG epitope alone (following the coiled coil). The chimeric dimers simply extended the myosin V sequence of the monomers (Fig. 1 C and D) through the coiled coil found in our wild-type myosin V dimer (15).
Expression and ATPase Assays.
The constructs described above were used to create recombinant baculovirus for expression in SF9 cells, as described (21). The myosin-expressing viruses were coinfected with a virus encoding chicken calmodulin, and the expressed calmodulin-containing myosin molecules were purified as described (21). ATPase assays (three to four separate preparations for each construct) were performed as described (22).
In Vitro Motility Assay.
All directionality assays with actin–myosin have used actin filaments that have one end of the filament differentially labeled (3, 12, 23). Although this provides reliable determination if the filaments are used immediately after being produced, over time, breaking and reannealing of filaments can lead to mismarked filaments, complicating directionality determination. For this study, we labeled the ends of the actin filament with a single, heavily Cy3-labeled N-terminal fragment of a gelsolin molecule (a plus-end actin capping protein), to unambiguously determine actin filament polarity. We then determined their velocity and direction of movement in a standard actin-gliding motility assay (24). In this assay, actin filaments are translocated by myosin motors that are held stationary on the surface.
The in vitro motility assay was performed following a previously described protocol (25) with modification. First, a sample chamber was prepared as described (26). Myosin was attached to a nitrocellulose-coated coverslip with anti-GFP (Sigma-Aldrich, St. Louis, MO) antibodies. A solution (20 mM Hepes/2 mM NaCl/25 mM KCl/1 mM EGTA/10 mM DTT/6.25 mM CaM) with 4 mg/ml casein was used to wash out unbound antibody and myosin. Actin filaments were visualized by using Alexa Fluor 647-labeled phalloidin, with Cy3-labeled gelsolin at the barbed (plus) end. Images were taken as described (27), and an oxygen scavenger system was used (13). An HQ615/130M emission filter (Chroma, Rockingham, VT) and CCD camera (MicroMax, Photometrics, Tucson, AZ) were used for fluorescence acquisition. The center of F-actin was calculated by weighting the position by signal intensity, allowing the speed to be calculated by the center's displacement divided by elapsed time.
Expression and Labeling of Gelsolin Fragments for Motility Assays.
DNA encoding human gelsolin residues 1–406 in the expression vector pMW172 was the generous gift of Alan Weeds (Medical Research Council Laboratory for Molecular Biology, Cambridge, U.K.). This construct corresponds to segments 1–3 of cytoplasmic gelsolin. The protein was expressed in Escherichia coli strain BL21 (DE3) and purified as described by Way et al. (28). Fluorescent labeling of this N-terminal fragment of gelsolin was carried out in 20 mM Hepes/1 mM DTT/1 mM NaN3, pH 7.5, with an ≈10-fold molar excess of monofunctional amino-reactive Cy3 (GE Healthcare, Piscataway, NJ) at 4°C for 3.5 h, in the dark. The labeled protein was separated from excess free dye by dialysis. Dye incorporation was measured spectrophotometrically, and the dye-to-protein ratio was determined to be 9.6:1.
Preparation of Doubly Labeled Actin Filaments.
Doubly labeled F-actin was prepared by adding 13 nM of Cy3-labeled gelsolin with 2 mM g-actin in F-buffer (300 mM KCl/10 mM MgCl2/40 mM Hepes, pH 7.1), and incubated for 10 min. Then, 9.9 mM phalloidin labeled with Alexa Fluor 647 (Invitrogen, Carlsbad, CA) was added in F-buffer at a ratio of 1:5 (phalloidin/actin). Polymerization was allowed to proceed overnight.
Single-Molecule Motility Assay.
For single-molecule motility studies, we applied the previously described technique termed FIONA (fluorescence imaging with one nanometer accuracy) that can track the position of a single fluorophore with ≈1.5-nm resolution (13). The assay was modified to track the centroid of movement of a processive dimer. To do so, FLAG-tagged dimers were labeled with Cy3 via anti-FLAG Cy3 conjugate antibody with three to six Cy3 molecules per antibody. Unlabeled FLAG-tagged myosin dimers were incubated with monoclonal anti-FLAG Cy3 conjugate antibody (Sigma-Aldrich) at the ratio of 1:2 overnight. The single molecule motility assay was performed as described (13, 14).
We have used a dynamical Student's t test approach to find the best global fit to a given data set by optimizing two parameters, the “probability threshold”, p, and the “group”, g, that are not known a priori for the particular motility recording. In a standard t test, the probability threshold or the so-called p value is an indicator for the statistical significance of the quantity 〈x1〉 − 〈x2〉 of the two populations under consideration with mean values 〈x1〉 and 〈x2〉. In our case, the distributions are neighboring clusters of position data. The lower the threshold the greater the significance of 〈x1〉 − 〈x2〉 and typically values of p < 0.05 are suggestive of a highly probable difference in the mean values. In our algorithm, we generate a sequence of kinetic events with the data for each pair of parameters p and g and calculate the reduced χ2 χr2 for that sequence. Here, p varies between 0.01 and 0.05, and g ranges from 2 to 10. The parameters that result in minimizing χr2 are used to create the best fit to the data.
Supplementary Material
Acknowledgments
We thank Xiaoyan Liu for help in protein purification, Dan Safer for his assistance with expression and labeling of gelsolin fragments, and Ben Blehm and Evan Graves for helpful discussions. This work was supported by grants from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (to H.L.S. and P.R.S.), the National Institute of General Medicine Sciences (to P.R.S.), and the Centre National de la Recherche Scientifique (to A.H.).
Abbreviation
- CaM
calmodulin.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610066104/DC1.
References
- 1.Berg JS, Powell BC, Cheney RE. Mol Biol Cell. 2001;12:780–794. doi: 10.1091/mbc.12.4.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmes KC, Geeves MA. Philos Trans R Soc London B. 2000;355:419–431. doi: 10.1098/rstb.2000.0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wells AL, Lin AW, Chen LQ, Safer D, Cain SM, Hasson T, Carragher BO, Milligan RA, Sweeney HL. Nature. 1999;401:505–508. doi: 10.1038/46835. [DOI] [PubMed] [Google Scholar]
- 4.Buss F, Spudich G, Kendrick-Jones J. Annu Rev Cell Dev Biol. 2004;20:649–676. doi: 10.1146/annurev.cellbio.20.012103.094243. [DOI] [PubMed] [Google Scholar]
- 5.Frank DJ, Noguchi T, Miller KG. Curr Opin Cell Biol. 2004;16:189–194. doi: 10.1016/j.ceb.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Park H, Ramamurthy B, Travaglia M, Safer D, Chen L-Q, Franzini-Armstrong C, Selvin PR, Sweeney HL. Mol Cell. 2006;21:331–336. doi: 10.1016/j.molcel.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 7.Rock RS, Ramamurthy B, Dunn AR, Beccafico S, Morris CA, Spink B, Rami B, Franzini-Armstrong C, Spudich JA, Sweeney HL. Mol Cell. 2005;17:603–609. doi: 10.1016/j.molcel.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 8.Bahloul A, Chevreux G, Wells AL, Martin D, Nolt J, Yang Z, Chen LQ, Potier N, Van Dorsselaer A, Rosenfeld S, et al. Proc Natl Acad Sci USA. 2004;101:4787–4792. doi: 10.1073/pnas.0306892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ménétrey J, Bahloul A, Wells AL, Yengo CM, Morris CA, Sweeney HL, Houdusse A. Nature. 2005;435:779–785. doi: 10.1038/nature03592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes KC, Schroder RR, Sweeney HL, Houdusse A. Philos Trans R Soc London B. 2004;359:1819–1828. doi: 10.1098/rstb.2004.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coureux PD, Wells AL, Ménétrey J, Yengo CM, Morris CA, Sweeney HL, Houdusse A. Nature. 2003;425:419–423. doi: 10.1038/nature01927. [DOI] [PubMed] [Google Scholar]
- 12.Homma K, Yoshimura M, Saito J, Ikebe R, Ikebe M. Nature. 2001;412:831–834. doi: 10.1038/35090597. [DOI] [PubMed] [Google Scholar]
- 13.Yildiz A, Forkey JN, McKinney SN, Ha T, Goldman YE, Selvin PR. Science. 2003;300:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 14.Yildiz A, Park H, Safer D, Yang Z, Chen LQ, Selvin PR, Sweeney HL. J Biol Chem. 2004;279:37223–37226. doi: 10.1074/jbc.C400252200. [DOI] [PubMed] [Google Scholar]
- 15.Purcell TJ, Morris C, Spudich JA, Sweeney HL. Proc Natl Acad Sci USA. 2002;99:14159–14164. doi: 10.1073/pnas.182539599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rock RS, Rice SE, Wells AL, Purcell TJ, Spudich JA, Sweeney HL. Proc Natl Acad Sci USA. 2001;98:13655–13659. doi: 10.1073/pnas.191512398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishikawa S, Homma K, Komori Y, Iwaki M, Wazawa T, Hikikoshi Iwane A, Saito J, Ikebe R, Katayama E, Yanagida T, Ikebe M. Biochem Biophys Res Commun. 2002;290:311–317. doi: 10.1006/bbrc.2001.6142. [DOI] [PubMed] [Google Scholar]
- 18.Okten Z, Churchman LS, Rock RS, Spudich JA. Nat Struct Mol Biol. 2004;11:884–887. doi: 10.1038/nsmb815. [DOI] [PubMed] [Google Scholar]
- 19.Houdusse A, Szent-Györgyi A, Cohen C. Proc Natl Acad Sci USA. 2000;97:11238–11243. doi: 10.1073/pnas.200376897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altman D, Sweeney HL, Spudich JA. Cell. 2004;116:737–749. doi: 10.1016/s0092-8674(04)00211-9. [DOI] [PubMed] [Google Scholar]
- 21.Sweeney HL, Rosenfeld S, Brown F, Faust L, Smith J, Stein L, Sellers J. J Biol Chem. 1998;273:6262–6270. doi: 10.1074/jbc.273.11.6262. [DOI] [PubMed] [Google Scholar]
- 22.De La Cruz EM, Ostap EM, Sweeney HL. J Biol Chem. 2001;276:32373–32381. doi: 10.1074/jbc.M104136200. [DOI] [PubMed] [Google Scholar]
- 23.Tsiavaliaris G, Fujita-Becker S, Manstein DJ. Nature. 2004;427:558–561. doi: 10.1038/nature02303. [DOI] [PubMed] [Google Scholar]
- 24.Kron SJ, Spudich JA. Proc Natl Acad Sci USA. 1986;83:6272–6276. doi: 10.1073/pnas.83.17.6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sellers JR, Cuda G, Wang F, Homsher E. Methods Cell Biol. 1993;39:23–49. doi: 10.1016/s0091-679x(08)60159-4. [DOI] [PubMed] [Google Scholar]
- 26.Snyder GE, Sakamoto T, Hammer JA, III, Sellers JR, Selvin PR. Biophys J. 2004;87:1776–1783. doi: 10.1529/biophysj.103.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park H, Hanson GT, Duff SR, Selvin PR. J Microsc. 2004;216:199–205. doi: 10.1111/j.0022-2720.2004.01416.x. [DOI] [PubMed] [Google Scholar]
- 28.Way M, Gooch J, Pope B, Weeds AG. J Cell Biol. 1989;109:593–605. doi: 10.1083/jcb.109.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.