Abstract
Protein crystallization is important for determining protein structures by X-ray diffraction. Nanoliter-sized plugs —aqueous droplets surrounded by a fluorinated carrier fluid —have been applied to the screening of protein crystallization conditions. Preformed arrays of plugs in capillary cartridges enable sparse matrix screening. Crystals grown in plugs inside a microcapillary may be analyzed by in situ X-ray diffraction. Screening using plugs, which are easily formed in PDMS microfluidic channels, is simple and economical, and minimizes consumption of the protein. This approach also has the potential to improve our understanding of the fundamentals of protein crystallization, such as the effect of mixing on the nucleation of crystals.
Introduction
This paper reviews recent applications of nanoliter plugs in protein crystallization and the resulting improvements in our understanding of the crystallization process. Macro-molecular crystals are important in structural biology, both for determining the three-dimensional structures and functions of molecules and complexes [1,2••,3–6], and for understanding ultrafast macromolecular dynamics [7•,8]. Protein crystallization is carried out by combining and confining a protein solution with crystallization reagents. The crystallization of many proteins is still difficult [9] and, because of a lack of fundamental understanding, the optimal conditions for protein crystallization cannot be predicted a priori [10–12]. Mixing of solutions, phase behavior, and nucleation and growth of crystals need to be thoroughly understood in order to make protein crystallization a more rational process. In the mean time, a large number of screens are still required to find the best crystallization conditions for a protein [1].
The protein supply is usually limited, especially for eukaryotic proteins. The 0.5–1.0 μl volume of concentrated protein solution per trial, used in traditional crystallization methods, makes screening large numbers of precipitants difficult for such proteins. It is also labor intensive to manually set up a large number of screens, and to harvest and mount the crystals. New tools for protein crystallization are required and are being developed [12–17], to both improve our understanding of and accelerate protein crystallization.
Tools are needed to improve screening of crystallization conditions and to understand crystallization
In traditional methods of protein crystallization, the protein and reagents are deposited into crystallization wells by pipetting [1]. The volume and composition of the mixture may then be further controlled through equilibration by vapor diffusion or dialysis. By confining the mixture of protein and precipitant to a smaller volume, the consumption of the protein can be reduced. Robotic systems have been developed to automate crystallization processes and to dispense solutions into nanoliter volumes (much smaller than traditional pipetting methods) [13,14,18–20,21•,22]. In robotic systems, nanoliter volumes of reagents are dispensed into microwells or microchambers. Thousands of trials can be easily set up per day. These systems are finding important applications in high-throughput protein crystallization centers [19,20,23] and in the pharmaceutical industry. However, the upfront equipment cost of these systems is higher than what many individual laboratories can justify. Lower cost, smaller systems are needed for such laboratories, which often work on difficult but scientifically very important macromolecular targets.
Microfluidics provides a method for combining and confining crystallization reagents in volumes as small as or smaller than those dispensed by robotic systems [24,25]. Microfluidic chambers combined with elastomeric PDMS [poly(dimethylsiloxane)] valves have been developed, and used for protein crystallization [15] and for studying the crystallization phase diagrams of proteins [26••]. In these systems, crystallization requires several actively controlled microfluidic valves for each trial, necessitating the use of specialized microfluidic chips that are still rather expensive. The permeability of PDMS to water and organic molecules, which is problematic for crystallization experiments, is being addressed by the development of new fluorinated elastomeric materials [27].
These new crystallization techniques have not yet provided a complete solution to the crystallization problem. Further development of both techniques and knowledge is required, and is bound to occur. For example, liquid droplets of picoliter to microliter volumes that contain the mixture of protein and precipitants can be levitated [28] to provide a clean and simple environment for protein crystallization [12]. This approach has not yet been widely adopted for the crystallization of proteins, but it has already provided valuable scientific insights into the mechanism of nucleation of protein crystals. Below, we describe a promising approach that may complement the existing methods — crystallization of proteins in nanoliter plugs generated in microfluidic systems.
Crystallizing proteins in nanoliter plugs is a promising approach
Plugs — aqueous droplets confined in microfluidic channels and surrounded by an immiscible carrier fluid [29] —provide an attractive platform for protein crystallization on the nanoliter scale. Formation of droplets in micro-channels has been widely studied [30–32], and plugs have been used in emulsions [33,34], kinetic studies [35] and chemical synthesis [36,37]. Plugs form spontaneously in microfluidic channels, without the need for valves, for example, when continuous aqueous streams of proteins and precipitants are combined and injected into the flow of a carrier fluid. Each plug containing protein and precipitant is equivalent to one crystallization trial. Thousands of crystallization trials in plugs can be set up rapidly within a microfluidic channel, each consuming only a few nanoliters of the protein solution. Plugs can be flowed out of the microchannels and into a microcapillary. After the flow is stopped, plugs are incubated and crystal formation is monitored (Figure 1).
Figure 1.
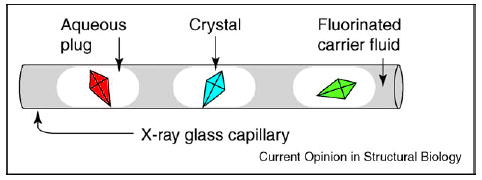
Schematic illustration of protein crystals in aqueous plugs surrounded by fluorinated carrier fluid inside a glass capillary.
Precipitation commonly takes place as solutions of the protein and precipitants are combined. This is a potential problem for microfluidic methods, as precipitates tend to adhere to the walls of the microchannels and perturb or even obstruct the flow. For plug-based methods, precipitation is not a significant problem. Plugs are surrounded by the fluorinated carrier fluid, which separates the contents of plugs from the walls of the microchannels, and enables the reliable transport of plugs containing solids [36]. This feature is especially important for the screening of crystallization conditions at high supersaturations and it has been used to understand the effect of mixing on the nucleation of protein crystals (described below). In addition, crystals formed in plugs appear to be less likely to adhere to the walls, simplifying the manipulation and extraction of crystals.
Preformed cartridges of plugs facilitate sparse matrix screening
The results of a sparse matrix screen are used to determine the best reagents for crystal formation. Sparse matrix screens are carried out by combining a protein with a large number of precipitants, each containing different crystallization reagents. To implement such an approach in plugs, one may use a cartridge — a preformed array of plugs containing different precipitants stored in a capillary [38••]. In simple cartridges, plugs are separated by the fluorinated carrier fluid (Figure 2a). Alternatively, three-phase fluid systems may be used in more robust cartridges, in which the aqueous plugs of the array are separated both by the fluorinated carrier fluid and by gas bubbles [38••]. To perform a screen, the array and the protein solution are flowed simultaneously into two separate channels of a microfluidic T-junction (Figure 2b). When the array of plugs merges with a stream of the protein solution (Figure 2a), a new array of larger plugs forms. Each plug of this newly formed array contains a mixture of the precipitant and the protein. The array of these plugs flows into a receiving capillary. Then the flow is stopped and protein crystallization takes place.
Figure 2.
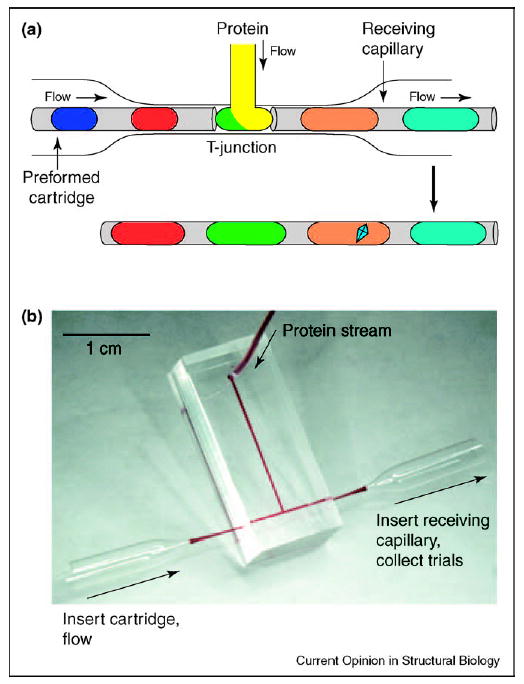
Sparse matrix screening is performed using preformed cartridges and a microfluidic T-junction. (a) A preformed cartridge of plugs containing different precipitants merges with a stream of protein solution at a T-junction (top). The resulting array of plugs is transferred into a receiving capillary, the flow is stopped, the array is incubated and the optimal precipitant yields a protein crystal (bottom). (b) Photograph of a T-junction microfluidic device. Two funnel-shaped glass capillaries are used to couple the cartridge to the inlet and the receiving capillary to the outlet of the microfluidic channel.
Gradients generated over arrays of plugs are used for optimization of crystallization conditions
Sparse matrix screening provides a set of initial crystallization hits that suggest the best crystallization reagents [2••]. The next step is to identify optimal concentrations of each reagent (and additives) that yield the highest quality crystals. To identify these concentrations, stock solutions of reagents may be diluted to different concentrations and combined with the protein solution in different ratios. Such optimization gradient screens may be performed by forming plugs. The dilutions are performed [17,35] by simultaneously flowing — into a stream of the carrier fluid — the protein, the dilution buffer and the solutions of reagents, and by varying the relative flow rates of these streams (as illustrated in Figure 3 for a single reagent). Plugs with a higher concentration of the reagent form at a higher flow rate of the reagent and a lower flow rate of the dilution buffer (Figure 3b). The concentration of the reagent is reduced as the flow rate of the reagent is decreased and the flow rate of the buffer is increased by the same amount (Figure 3c); this preserves the total flow rate of the aqueous streams and maintains the size of plugs. The concentration of the protein may be adjusted in the same manner. The array of plugs representing such an optimization screen is flowed into a capillary, the flow is stopped and the plugs are incubated. Crystal formation, precipitation, liquid–liquid phase-separated states and clear plugs may be visualized and analyzed. Visualization inside plugs, especially at the rounded edges of plugs, is facilitated by matching the refractive index of the fluorinated carrier fluid with that of the aqueous crystallization solutions.
Figure 3.
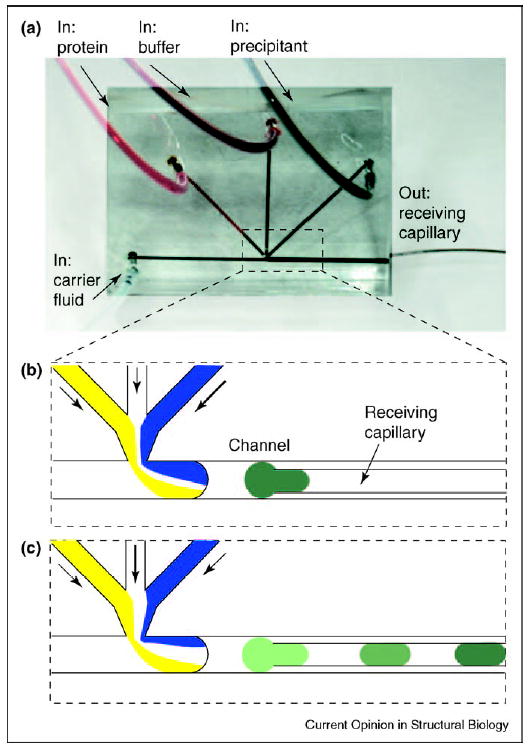
Optimization screens are performed by varying the flow rates of aqueous streams of crystallization reagents. (a) Microphotograph showing the microfluidic device for generating aqueous plugs. There are three inlets for aqueous streams and one inlet for carrier fluid. The receiving capillary is inserted into the outlet channel. The entire chip is approximately 1 × 1 inch. The background of the image was brightened in Photoshop for clarity. (b) When the flow rate of the precipitant (blue) is high, the concentration of the precipitant in the plug is also high. (c) As the flow rate of the precipitant is decreased and the flow rate of the buffer is increased, the concentration of the precipitant decreases accordingly.
Transfer of water can be controlled in plugs to facilitate crystallization
Evaporation occurs readily on the nanoliter scale. When a crystallization trial undergoes uncontrolled evaporation, the trial may be lost. Crystallizing proteins in PDMS microfluidic channels is difficult because PDMS is permeable to water and volatile or hydrophobic reagents. Controlling evaporation requires careful control of humidity and minimizing differences in osmotic pressure between adjacent crystallization wells. Crystallization in glass capillaries is preferred because glass is essentially impermeable and does not allow uncontrolled evaporation to occur. Plugs confined in glass capillaries, surrounded by water-impermeable fluorinated carrier fluid, do not suffer from evaporation and may be used in microbatch-style crystallization trials.
In crystallization by vapor diffusion, controlled loss of water from a trial results in increasing supersaturation, and can enhance both nucleation and growth of the protein crystal. To implement vapor diffusion crystallization in plugs inside glass capillaries [39••], controlled loss of water from a crystallization plug can be achieved by the transfer of water through a water-permeable fluorinated fluid to a plug of higher osmotic pressure (Figure 4). The rate of transfer is determined by the degree of permeability of the carrier fluid and by the distance between plugs. The amount of water transferred depends on the difference in osmotic pressure and on the relative size of the two plugs. To generate crystallization plugs that alternate with plugs of higher osmotic pressure, a cross-shaped microfluidic channel is employed. Aqueous streams are injected from opposite directions into the flow of the carrier fluid and an array of alternating plugs results when the flow rates are in the appropriate range, as determined by the Capillary number, a dimensionless parameter that describes the relative importance of viscous forces and surface tension acting at an interface [39••,40].
Figure 4.
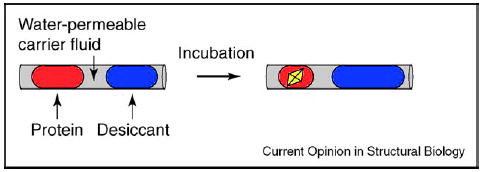
Schematic drawing showing the process of water transfer from a plug containing a protein and a precipitant to a plug containing a desiccant solution of higher osmotic pressure. The carrier fluid is water permeable. After incubation, the plug containing protein and precipitant will be concentrated, and could yield protein crystals.
Crystallization in plugs inside microcapillaries is well suited to on-chip, in situ X-ray diffraction analysis
X-ray diffraction is used both to evaluate the quality of crystals and to determine the crystal structure of proteins. To minimize radiation damage in the X-ray beam, crystals are usually cryoprotected for diffraction. Freezing and mounting a crystal for diffraction can be labor intensive and could be destructive. In situ diffraction has been proposed to avoid manipulation of fragile crystals [41,42]. An in situ diffraction test under crystallization conditions is certainly attractive for determining the intrinsic quality of crystals, before potential damage by handling and freezing [43,44]. Previously, data sets and structures were obtained from crystals grown in gels inside capillaries and then cryogenically frozen directly in the capillary [42–44]. Freezing a crystal together with the capillary and the mother liquor could be less desirable than freezing an isolated crystal, due to the slower rate of heat transfer through the capillary and the higher combined heat capacity of the capillary, mother liquor and crystal. These two factors lead to slower cooling of the crystal in the capillary and are presumably more likely to lead to damage of the crystal by the formation of crystalline ice. On the other hand, modern synchrotrons allow effective structural determination of crystals smaller than 50 μm [45–47]. Presumably, a 50 μm crystal (surrounded by an ~200 μm × 200 μm × 200 μm volume of the mother liquor and located inside a 200 μm X-ray capillary with thin 10 μm walls) could be cooled and frozen as rapidly as a larger 500 μm × 500 μm × 500 μm isolated crystal, which have been frozen successfully for structural studies.
When X-ray-quality glass capillaries are used to carry out crystallization in plugs, such in situ diffraction can be easily performed without freezing. High-quality diffraction patterns have been obtained using this approach on a model protein (Figure 5) [38••]. It remains to be seen whether problems with radiation damage can be reduced, and whether complete data sets and corresponding electron densities can be obtained from such crystals under non-cryogenic conditions. It also remains to be seen if the freezing of crystals in plugs inside capillaries can be carried out effectively. In the mean time, crystals grown in plugs inside capillaries can be easily extracted by flowing them out and catching them with a cryoloop for subsequent traditional flash-freezing.
Figure 5.
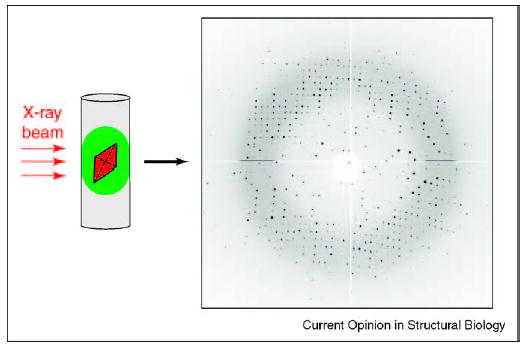
In situ diffraction of a thaumatin crystal at 1.8 Å resolution. A crystal was grown inside a plug in a capillary. The capillary was mounted on a goniometer and the crystal, while still in the plug, was subjected to the X-ray beam to obtain the diffraction pattern. The diffraction pattern is comparable to those obtained from crystals grown using conventional methods. The thaumatin crystal was ~100 × 100 μm and data were collected at Argonne National Laboratory’s BioCARS beamline sector 14.
New science: crystallization in plugs clarifies the effect of mixing on protein crystallization
Plugs provide additional degrees of control to enhance our scientific understanding of protein crystallization. Inconsistency of mixing of protein and precipitants has been proposed to play a role in the irreproducibility of the nucleation of protein crystals in batch crystallization [48]. Thus, the effect of mixing on the nucleation of protein crystals was studied in plugs (Figure 6a) [49•]. Mixing in plugs can be carefully controlled. Chaotic mixing may be induced in plugs flowing through winding channels and the rate of mixing can be controlled by varying the velocity of flow [29,50]. Chaotic mixing is caused by repeated stretching and folding of the fluid, similar to the folding and stretching performed by a baker kneading dough (baker’s transformation) [51]. As a plug of the protein and precipitant travels through a straight section of the microchannel, the interface between the two solutions stretches. As the plug flows through a turn, it undergoes reorientation, which is equivalent to folding. This folding and stretching of the fluid in the plug leads to an exponential increase in the area of the interface between the solutions of the protein and precipitant (Figure 6a). At this interface, the concentrated solutions of the protein and precipitant come into contact, giving rise to high supersaturation and leading to nucleation. If mixing is chaotic but slow, this interface is sufficiently long-lived to give rise to excessive nucleation (Figure 6b), and the formation of precipitates or microcrystalline showers. For fast chaotic mixing, the lifetime of the interface is shorter and only a few nucleation events take place, leading to a few well-formed crystals (Figure 6c). Mixing could emerge as an additional tool with which to control crystallization.
Figure 6.
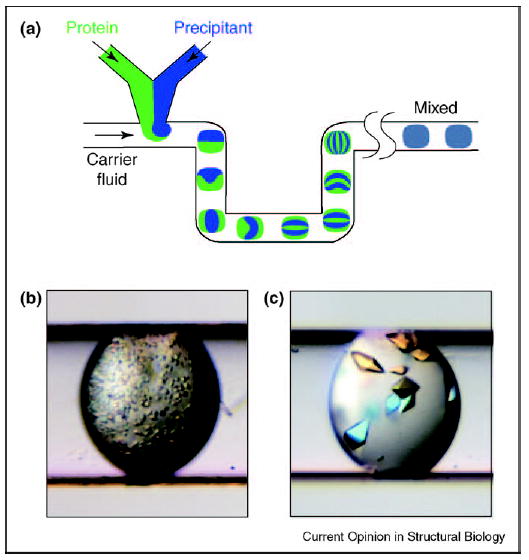
Control of mixing can lead to control of nucleation in protein crystallization. (a) Schematic illustration of the plug-based microfluidic setup for studying the effect of mixing on the nucleation of protein crystals. Mixing by chaotic advection is illustrated. Microphotographs showing an example of the effect of (b) slow chaotic mixing and (c) fast chaotic mixing on protein crystallization under otherwise identical conditions. The diameter of the receiving capillary is 200 μm.
Conclusions
New tools to understand and to accelerate protein crystallization are needed and are being developed. The diversity of approaches being pursued is important, as it is conceivable that no ‘universal’ approach will emerge that is perfect for both fundamental studies and screening, performed both in industrialized high-throughput centers and in small individual laboratories. The precise control of fluid flow provided by plug-based crystallization has already been shown to be a useful tool for fundamental studies. The simplicity of this system makes it promising for screening protein crystallization conditions in ‘small laboratory’ settings. In addition, integrating this system into a high-throughput screening platform has the potential to reduce screening costs. Although it remains to be determined how such integration can be carried out, it is encouraging that this opportunity is being investigated by a technology development center of the Protein Structure Initiative [21•].
Plug-based protein crystallization is far from being mature and at least four questions need to be addressed. First, dynamic methods beyond vapor diffusion, such as seeding and addition of reagents, have been used to control crystallization [10,52,53]. What dynamic control methods are compatible with crystallization in plugs? Second, heterogeneous nucleation of protein crystals is well established [1]. Might manipulation of the surface chemistry of plugs [54] be important for the control of the nucleation and growth of protein crystals? Third, solutions of membrane proteins in detergents are characterized by low surface tension and high viscosity, complicating fluid handling. Will nanoliter plugs be applicable to the crystallization of membrane proteins? Finally, the miniaturization of crystallization trials from microliter to nanoliter volumes certainly affects the dynamics of the nucleation and growth of protein crystals by changing the surface-to-volume ratio, and the relative importance of convective and diffusive transport. What are the manifestations of these changes, how do we understand them quantitatively, and how do we use them to increase the success rate of crystallization and improve the diffraction quality of crystals? Crystallization in nanoliter plugs would benefit from answers to these questions. In addition, crystallization in plugs may provide some of the answers, further advancing our understanding of macromolecular crystallization.
Acknowledgments
The work described in this review has been supported by the National Institute for Biomedical Imaging and Bioengineering (R01 EB001903), by the National Institute of General Medical Sciences and National Center for Research Resources under the PSI-2 Specialized Center program (U54 GM074961), by the Beckman Young Investigator Program and by the DuPont Young Professor Award. We thank Keith Moffat, Chuan He and Phoebe Rice for their help in advancing the work described in this review. We thank our colleagues in the Ismagilov laboratory who have contributed to the work described here, and Peter Kuhn, Ray Stevens and Lance Stewart for helpful discussions.
Footnotes
This review comes from a themed issue on Biophysical methods Edited by Wah Chiu and Keith Moffat
References
Papers of particular interest, published within the annual period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.McPherson A. Crystallization of Biological Macromolecules. edn 1. Woodbury, NY: Cold Spring Harbor Laboratory Press; 1999. [Google Scholar]
- 2.••.McPherson A. Introduction to protein crystallization. Methods. 2004;34:254–265. doi: 10.1016/j.ymeth.2004.03.019. A useful introduction to protein crystallization methods for those new to the field, and a review of recent developments. [DOI] [PubMed] [Google Scholar]
- 3.Berkovitch F, Nicolet Y, Wan JT, Jarrett JT, Drennan CL. Crystal structure of biotin synthase, an S-adenosylmethionine-dependent radical enzyme. Science. 2004;303:76–79. doi: 10.1126/science.1088493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass RB, Strop P, Barclay M, Rees DC. Crystal structure of Escherichia coli MscS, a voltage-modulated and mechanosensitive channel. Science. 2002;298:1582–1587. doi: 10.1126/science.1077945. [DOI] [PubMed] [Google Scholar]
- 5.Ke A, Doudna JA. Crystallization of RNA and RNA-protein complexes. Methods. 2004;34:408–414. doi: 10.1016/j.ymeth.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 6.Khademi S, O’Connell J, Remis J, Robles-Colmenares Y, Miericke LJW, Stroud RM. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.3.5 angstrom. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 7.•.Moffat K. Time-resolved biochemical crystallography: a mechanistic perspective. Chem Rev. 2001;101:1569–1581. doi: 10.1021/cr990039q. This study reviews crystallographic approaches to the time-resolved study of dynamic biochemical processes. [DOI] [PubMed] [Google Scholar]
- 8.Schotte F, Lim MH, Jackson TA, Smirnov AV, Soman J, Olson JS, Phillips GN, Wulff M, Anfinrud PA. Watching a protein as it functions with 150-ps time-resolved X-ray crystallography. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 9.Chayen NE. Tackling the bottleneck of protein crystallization in the post-genomic era. Trends Biotechnol. 2002;20:98. doi: 10.1016/s0167-7799(02)01916-9. [DOI] [PubMed] [Google Scholar]
- 10.Chayen NE. Turning protein crystallisation from an art into a science. Curr Opin Struct Biol. 2004;14:577–583. doi: 10.1016/j.sbi.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Gliko O, Neumaier N, Pan W, Haase I, Fischer M, Bacher A, Weinkauf S, Vekilov PG. A metastable prerequisite for the growth of lumazine synthase crystals. J Am Chem Soc. 2005;127:3433–3438. doi: 10.1021/ja043218k. [DOI] [PubMed] [Google Scholar]
- 12.Knezic D, Zaccaro J, Myerson AS. Nucleation induction time in levitated droplets. J Phys Chem B. 2004;108:10672–10677. [Google Scholar]
- 13.Kuhn P, Wilson K, Patch MG, Stevens RC. The genesis of high-throughput structure-based drug discovery using protein crystallography. Curr Opin Chem Biol. 2002;6:704–710. doi: 10.1016/s1367-5931(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 14.Stevens RC. High-throughput protein crystallization. Curr Opin Struct Biol. 2000;10:558–563. doi: 10.1016/s0959-440x(00)00131-7. [DOI] [PubMed] [Google Scholar]
- 15.Hansen CL, Skordalakes E, Berger JM, Quake SR. A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion. Proc Natl Acad Sci USA. 2002;99:16531–16536. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nollert P. Lipidic cubic phases as matrices for membrane protein crystallization. Methods. 2004;34:348–353. doi: 10.1016/j.ymeth.2004.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng B, Roach LS, Ismagilov RF. Screening of protein crystallization conditions on a microfluidic chip using nanoliter-size droplets. J Am Chem Soc. 2003;125:11170–11171. doi: 10.1021/ja037166v. [DOI] [PubMed] [Google Scholar]
- 18.Luft JR, Wolfley J, Jurisica I, Glasgow J, Fortier S, DeTitta GT. Macromolecular crystallization in a high throughput laboratory-the search phase. J Cryst Growth. 2001;232:591–595. [Google Scholar]
- 19.Rupp B. High-throughput crystallography at an affordable cost: The TB Structural Genomics Consortium Crystallization Facility. Acc Chem Res. 2003;36:173–181. doi: 10.1021/ar020021t. [DOI] [PubMed] [Google Scholar]
- 20.Adams MWW, Dailey HA, Delucas LJ, Luo M, Prestegard JH, Rose JP, Wang BC. The Southeast Collaboratory for Structural Genomics: a high-throughput gene to structure factory. Acc Chem Res. 2003;36:191–198. doi: 10.1021/ar0101382. [DOI] [PubMed] [Google Scholar]
- 21.•.Service R. Structural biology - Structural genomics, round 2. Science. 2005;307:1554–1557. doi: 10.1126/science.307.5715.1554. A recent editorial on the goals and the status of the Protein Structure Initiative and high-throughput protein crystallization efforts. [DOI] [PubMed] [Google Scholar]
- 22.Snell G, Cork C, Nordmeyer R, Cornell E, Meigs G, Yegian D, Jaklevic J, Jin J, Stevens RC, Earnest T. Automated sample mounting and alignment system for biological crystallography at a synchrotron source. Structure. 2004;12:537–545. doi: 10.1016/j.str.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Cuff ME, Miller DJ, Korolev S, Xu X, Anderson WF, Edwards A, Joachimiak A, Savchenko A. Crystal structure of a predicted precorrin-8x methylmutase from Thermoplasma acidophilum. Proteins. 2005;58:751–754. doi: 10.1002/prot.20022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hansen C, Quake SR. Microfluidics in structural biology: smaller, faster… better. Curr Opin Struct Biol. 2003;13:538–544. doi: 10.1016/j.sbi.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 25.van der Woerd M, Ferree D, Pusey M. The promise of macromolecular crystallization in microfluidic chips. J Struct Biol. 2003;142:180–187. doi: 10.1016/s1047-8477(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 26.••.Hansen CL, Sommer MOA, Quake SR. Systematic investigation of protein phase behavior with a microfluidic formulator. Proc Natl Acad Sci USA. 2004;101:14431–14436. doi: 10.1073/pnas.0405847101. The authors present a new method for studying the phase diagram of protein crystallization on a microfluidic chip. Understanding phase behavior leads to the more accurate prediction of protein crystallization conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolland JP, Van Dam RM, Schorzman DA, Quake SR, DeSimone JM. Solvent-resistant photocurable ‘‘liquid teflon’’ for microfluidic device fabrication. J Am Chem Soc. 2004;126:2322–2323. doi: 10.1021/ja031657y. [DOI] [PubMed] [Google Scholar]
- 28.Santesson S, Nilsson S. Airborne chemistry: acoustic levitation in chemical analysis. Anal Bioanal Chem. 2004;378:1704–1709. doi: 10.1007/s00216-003-2403-2. [DOI] [PubMed] [Google Scholar]
- 29.Song H, Tice JD, Ismagilov RF. A microfluidic system for controlling reaction networks in time. Angew Chem Int Ed Engl. 2003;42:768–772. doi: 10.1002/anie.200390203. [DOI] [PubMed] [Google Scholar]
- 30.Thorsen T, Roberts RW, Arnold FH, Quake SR. Dynamic pattern formation in a vesicle-generating microfluidic device. Phys Rev Lett. 2001;86:4163–4166. doi: 10.1103/PhysRevLett.86.4163. [DOI] [PubMed] [Google Scholar]
- 31.Yi GR, Thorsen T, Manoharan VN, Hwang MJ, Jeon SJ, Pine DJ, Quake SR, Yang SM. Generation of uniform colloidal assemblies in soft microfluidic devices. Adv Mater. 2003;15:1300–1304. [Google Scholar]
- 32.Link DR, Anna SL, Weitz DA, Stone HA. Geometrically mediated breakup of drops in microfluidic devices. Phys Rev Lett. 2004;92:054503. doi: 10.1103/PhysRevLett.92.054503. [DOI] [PubMed] [Google Scholar]
- 33.Okushima S, Nisisako T, Torii T, Higuchi T. Controlled production of monodisperse double emulsions by two-step droplet breakup in microfluidic devices. Langmuir. 2004;20:9905–9908. doi: 10.1021/la0480336. [DOI] [PubMed] [Google Scholar]
- 34.Utada AS, Lorenceau E, Link DR, Kaplan PD, Stone HA, Weitz DA. Monodisperse double emulsions generated from a microcapillary device. Science. 2005;308:537–541. doi: 10.1126/science.1109164. [DOI] [PubMed] [Google Scholar]
- 35.Song H, Ismagilov RF. Millisecond kinetics on a microfluidic chip using nanoliters of reagents. J Am Chem Soc. 2003;125:14613–14619. doi: 10.1021/ja0354566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shestopalov I, Tice JD, Ismagilov RF. Multi-step synthesis of nanoparticles performed on millisecond time scale in a microfluidic droplet-based system. Lab Chip. 2004;4:316–321. doi: 10.1039/b403378g. [DOI] [PubMed] [Google Scholar]
- 37.Khan SA, Gunther A, Schmidt MA, Jensen KF. Microfluidic synthesis of colloidal silica. Langmuir. 2004;20:8604–8611. doi: 10.1021/la0499012. [DOI] [PubMed] [Google Scholar]
- 38.••.Zheng B, Ismagilov RF. A microfluidic approach for screening submicroliter volumes against multiple reagents by using preformed arrays of nanoliter plugs in a three-phase liquid/liquid/gas flow. Angew Chem Int Ed Engl. 2005;44:2520–2523. doi: 10.1002/anie.200462857. A detailed description of the use of microfluidic preformed arrays in sparse matrix screening of protein crystallization conditions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.••.Zheng B, Tice JD, Roach LS, Ismagilov RF. A droplet-based, composite PDMS/glass capillary microfluidic system for evaluating protein crystallization conditions by microbatch and vapor-diffusion methods with on-chip X-ray diffraction. Angew Chem Int Ed Engl. 2004;43:2508–2511. doi: 10.1002/anie.200453974. A description of optimization screens of protein crystallization conditions performed in nanoliter plugs by microbatch and vapor diffusion methods. The quality of crystals was analyzed by in situ X-ray diffraction in nanoliter plugs inside capillaries. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zheng B, Tice JD, Ismagilov RF. Formation of droplets of in microfluidic channels alternating composition and applications to indexing of concentrations in droplet-based assays. Anal Chem. 2004;76:4977–4982. doi: 10.1021/ac0495743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McPherson A. In situ X-ray crystallography. J Appl Cryst. 2000;33:397–400. [Google Scholar]
- 42.Pusey ML, Liu ZJ, Tempel W, Praissman J, Lin DW, Wang BC, Gavira JA, Ng JD. Life in the fast lane for protein crystallization and X-ray crystallography. Prog Biophys Mol Biol. 2005;88:359–386. doi: 10.1016/j.pbiomolbio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 43.Gavira JA, Toh D, Lopez-Jaramillo J, Garcia-Ruiz JM, Ng JD. Ab initio crystallographic structure determination of insulin from protein to electron density without crystal handling. Acta Crystallogr D Biol Crystallogr. 2002;58:1147–1154. doi: 10.1107/s0907444902006959. [DOI] [PubMed] [Google Scholar]
- 44.Ng JD, Gavira JA, Garcia-Ruiz JM. Protein crystallization by capillary counterdiffusion for applied crystallographic structure determination. J Struct Biol. 2003;142:218–231. doi: 10.1016/s1047-8477(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 45.Riekel C, Burghammer M, Schertler G. Protein crystallography microdiffraction. Curr Opin Struct Biol. 2005;15 doi: 10.1016/j.sbi.2005.08.013. in press. [DOI] [PubMed] [Google Scholar]
- 46.Li J, Edwards P, Burghammer M, Villa C, Schertler G. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 47.Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, Eisenberg D. Structure of the cross-spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chernov AA. Protein crystals and their growth. J Struct Biol. 2003;142:3–21. doi: 10.1016/s1047-8477(03)00034-0. [DOI] [PubMed] [Google Scholar]
- 49.•.Chen DL, Gerdts CJ, Ismagilov RF. Using microfluidics to observe the effect of mixing on nucleation of protein crystals. J Am Chem Soc. 2005;127:9672–9673. doi: 10.1021/ja052279v. The authors used plugs to understand the effect of mixing on the nucleation of protein crystals in microbatch crystallization trials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Song H, Bringer MR, Tice JD, Gerdts CJ, Ismagilov RF. Experimental test of scaling of mixing by chaotic advection in droplets moving through microfluidic channels. Appl Phys Lett. 2003;83:4664–4666. doi: 10.1063/1.1630378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ottino JM, Wiggins S. Applied physics - designing optimal micromixers. Science. 2004;305:485–486. doi: 10.1126/science.1099343. [DOI] [PubMed] [Google Scholar]
- 52.Chayen NE. Methods for separating nucleation and growth in protein crystallisation. Prog Biophys Mol Biol. 2005;88:329–337. doi: 10.1016/j.pbiomolbio.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 53.Saridakis E, Chayen NE. Systematic improvement of protein crystals by determining the supersolubility curves of phase diagrams. Biophys J. 2003;84:1218–1222. doi: 10.1016/S0006-3495(03)74936-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roach LS, Song H, Ismagilov RF. Controlling nonspecific protein adsorption in a plug-based microfluidic system by controlling interfacial chemistry using fluorous-phase surfactants. Anal Chem. 2005;77:785–796. doi: 10.1021/ac049061w. [DOI] [PMC free article] [PubMed] [Google Scholar]


