Abstract
T cell differentiation in the thymus is driven by positive selection through the interaction of αβ T cell receptors (TCRs) with self-peptides bound to self-major histocompatibility complex molecules, yet the influence of the peptide sequence on this process remains unknown. To address this issue, we have compared CD4+ T cell differentiation between two sets of mouse lines in which MHC class II I-Ab molecules are occupied with either Eα chain-derived peptide (pEα) or its variant, p60K, with one amino acid substitution from leucine to lysine at P5 residue of TCR contacts. Here, we show that despite the comparable expression of I-Ab–peptide complex in the thymus, this substitution from leucine to lysine affects efficiency of positive selection, resulting in extremely small numbers of CD4+ T cells to be selected to mature on I-Ab–p60K complex. Furthermore, we show that, although I-Ab–pEα complex selects diverse T cells, T cell repertoire shaped by I-Ab–p60K complex is markedly constrained. Our findings thus suggest that positive selection is both specific and degenerate, depending on the amino acid residues at TCR contacts of the selecting self-peptides.
Although the diversity of αβ T cell receptors (TCRs) theoretically reaches 1015 by random rearrangement of five gene segments (Vα, Jα, Vβ, Dβ, and Jβ) and random nucleotide addition (1), mature T cells express highly selected TCRs in that they exhibit tolerance to self-antigenic peptides and restriction by self-MHC molecules. This mainly results from two reciprocal selection processes, positive and negative selection, acting during T cell development in the thymus. Positive selection is the process that induces the differentiation of CD4+CD8+ immature thymocytes into CD4−CD8+ or CD4+CD8− mature thymocytes only when their TCRs recognize self-MHC class I or class II molecules with sufficient, albeit weak, affinity and ensures that the specificity of mature T cell repertoire is directed against antigenic peptides bound to self-MHC molecules. On the other hand, negative selection plays a central role in tolerance induction by eliminating immature thymocytes bearing TCRs specific for self-peptides bound to self-MHC molecules.
Since the discovery that positive selection is influenced by the substitutions at amino acid residues of MHC molecules positioned toward the peptide-binding groove (2–5), the role of self-peptides in positive selection has been the subject of considerable debate. This was first directly addressed for CD8+ T cells using fetal thymic organ cultures derived from mutant mouse strains where a particular MHC class I–peptide complex is expressed by exogenously adding a given peptide to the culture (6–8). Subsequently, several groups developed in vivo experimental system focusing on the role of self-peptides in positive selection of CD4+ T cells by creating mouse strains in which all or a large fraction of the MHC class II molecules are occupied with a single peptide (9–13). With the use of in vitro and in vivo experimental systems, it was shown that limited numbers of self-peptides bound to a given MHC molecule fail to support positive selection of CD8+ or CD4+ T cells expressing a transgenic TCRαβ that are selected by the same MHC molecule with a normal array of self-peptides (14–19). In addition, CD4+ T cells selected to mature on such MHC class II molecules were shown to express highly restricted TCRα chains in the presence of a single rearranged TCRβ (20, 21). Thus, it is clear that specific recognition of self-peptides by TCRs could be involved during the process of positive selection. This is also supported by the result that the antibody recognizing MHC class II molecules in a peptide-specific manner prevents positive selection of thymocytes with a certain TCR specificity (22). However, it remains unclear to what extent specific TCR-peptide contacts are determinant in positive selection under physiological T cell differentiation. Because positive selection of sizable numbers of T cells is not driven in vitro by all of the peptides that comparably stabilize MHC molecule (6), the structure of the selecting self-peptides may affect efficiency of positive selection, probably by changing the extent of specific TCR-peptide contacts, versus less stringent TCR–MHC interaction, involved in this process. This possibility was first proposed by Schumacher and Ploegh (23) predicting that self-peptides with bulky or charged side chains at TCR contacts would select a far smaller set of T cells as compared with those bearing small side chains at these positions. Thus far, however, no experimental evidence supporting this model has been provided.
To address the question whether amino acid residues at TCR contacts of the selecting self-peptides affect positive thymocyte selection, we used the strategy to express a single MHC class II–peptide complex in vivo and compared CD4+ T cell differentiation directed by two distinct peptide ligands with one amino acid difference, leucine versus lysine, at P5 residue of TCR contacts. Here, we demonstrate that this amino acid substitution critically affects efficiency of positive selection and diversity of the selected T cell repertoire.
Materials and Methods
Generation of Transgenic Knockout Mice.
The gene encoding I-Aβb chain covalently bound to Eα chain-derived peptide (Eα52–68; pEα) has been described elsewhere (13). This construct (PDR-pEα) was modified by PCR to encode lysine at position 58 or 60 of pEα. The following oligonucleotides were used: primer A, GCTTCTTTCGAAGCTCAAAAGGCTTTAGCTAATATTGCTGTCGACAAGGCTGGT; primer B, GCTTCTTTCGAAGCTCAAGGTGCTAAGGCTAATATTGCTGTCGACAAGGCTGGT; primer C, GCTCGGTCACCGCGCGGTGCT. Briefly, the NSPV–BstEII fragment of PDR-pEα was replaced with the amplified DNA using primer C and primer A or primer B at these sites to develop the transgenic constructs for I-Aβb covalently bound to an analog peptide bearing lysine instead of glycine or leucine at position 58 or 60 of pEα (p58K or p60K), respectively. These constructs were injected into fertilized eggs from the mice lacking the expression of endogenous I-Aβb, invariant chain, and β2-microglobulin (TKO). Integration and transmission of the transgenes was assessed by Southern blot analysis using I-Aβ cDNA. In case of the F1 mice expressing both PDR-pEα and PDR-p60K, the genotype of the progeny was determined by amplifying tail DNA with the primers corresponding to the conserved sequence and probing with 32P kinase-labeled oligonucleotide specific for each construct. All of the mice were maintained under the specific pathogen-free condition, and mice hemizygous for the transgene were used, except for the case especially noted.
Flow Cytometry and Cell Sorting.
All mAbs used for the staining, except for Y3P, were purchased from PharMingen. The expression of I-Ab molecules in the periphery was assessed by staining lymph node (LN) cells with FITC–anti-CD45R mAb and biotinylated Y3P followed by streptavidin-phycoerythrin (PE). CD4+ T cell differentiation was examined by staining thymocytes or LN cells with PE-anti-CD4 and FITC–anti-CD8 mAbs. To assess the expression of TCR Vβs on CD4+CD8− thymocytes, CD4+CD8− thymocytes were enriched as previously described (13) and stained with PE-anti-CD4, FITC–anti-TCR Vβ, and biotinylated anti-CD8 mAbs followed by streptavidin-Cy-Chrome. For cell sorting, CD4+CD8− thymocytes were enriched and stained with PE-anti-CD4 and FITC–anti-CD8 mAbs.
Immunohistochemical Analysis.
Frozen thymus sections were prepared using a cryostat and fixed in cold acetone. The sections were first incubated with Y3P, then reacted with peroxidase-conjugated anti-mouse IgG and visualized as described elsewhere (13).
T Cell Hybridoma Stimulation.
The following T cell hybridomas were used: BV2-3-1, specific for pigeon cytochrome c-derived peptide, 50V, restricted by I-Ab molecules; and 12-2, specific for I-Ab–p60K complex. The stimulation experiment was done by cultivating BV2-3-1 or 12-2 (1 × 105/well) with irradiated LN cells (1 × 106/well) in the presence or absence of 50V (AEGFSYTVANKNKGIT), or truncated variants of pEα-bearing leucine (60L, FEAQGALANIAVDK) or lysine (60K, FEAQGAKANIAVDK), respectively. Supernatant was recovered from the culture after 24 h, and IL-2 was quantified by the proliferative response of CTLL (5 × 103/well).
Mixed Lymphocyte Reaction.
LN CD4+ T cells were prepared by eliminating CD8+ T cells and B cells from LN cells or by purifying them using immunomagnetic beads (Dynal, Great Neck, NY). LN CD4+ T cells (2 × 105/well) were cultured with irradiated spleen cells (1 × 106/well) for 80 h, and 1 μCi of [3H]thymidine was added during the final 16 h of the culture.
Anchored PCR, and Cloning and Sequencing of PCR Products.
Details have been described elsewhere (21). Briefly, total cellular RNA extracted from the sorted CD4+CD8− thymocytes was primed with Cα-specific antisense oligonucleotide for cDNA synthesis. After treatment with RNase H and RNase I, cDNA was subject to homopolymeric tailing using terminal deoxynucleotidyltransferase and dCTP. This dC-tailed cDNA was used for the first PCR with Cα-specific primer and anchor primer. Then, the first PCR products were mixed and subject to the nested PCR in the presence of TaqStart antibody (CLONTECH). The mixture of the second PCR products was ligated to pGMT-T Easy Vector (Promega) and used for transformation of DH5α. Each colony was picked up and cultured in LB medium, and then the supernatant was subject to PCR using the nested primers. After removal of excess primers and dNTPs, the purified PCR products were labeled with dye termination cycle sequencing and analyzed on an ABI PRISM 377 DNA sequencer (Perkin–Elmer).
Immunoscope Analysis.
Poly(A)+ mRNA extracted from the sorted CD4+CD8− thymocytes was subject to cDNA synthesis. The number of CD3ɛ copies was determined using quantitative PCR, and cDNA samples with similar amounts of CD3ɛ copies were subjected to PCR using TCR Vβ14-specific and Cβ-specific primers as described elsewhere (24). After run-off extensions with six Jβ2-specific fluorescent antisense primers, the fluorescent products were loaded on polyacrylamide gels and subjected to electrophoresis in an ABI Prism 377 DNA sequencer. CDR3 length distribution and signal intensities were then analyzed with the immunoscope software (25).
Results
Expression of I-Ab–p60K Complex in Lymphoid Tissues.
Earlier, we reported three lines of transgenic mice that have been developed by introducing the gene encoding I-Aβb chain covalently bound to pEα into the mice lacking the expression of endogenous I-Aβb, invariant chain, and β2-microglobulin (B2L TKO, H3 TKO, and B2H TKO; refs. 13 and 21). A similar mouse line had been described by Ignatowicz et al. (9). Thus far, no evidence has been provided that I-Ab molecules expressed in such mice present other self- or foreign antigenic peptides than pEα to T cells (9, 13, 17). Thus, this strategy made it feasible to follow the fate of CD4+ T cells directed by a single MHC class II-peptide complex in vivo. Global amino acid replacement of pEα has revealed that no or only a few amino acid substitutions are tolerated at positions 57, 58, 60, and 62 in T cell activation (26), suggesting that four amino acid residues at these positions are directly involved in the recognition by TCRs. To assess whether positive thymocyte selection is influenced by the amino acid residues at TCR contacts of selecting self-peptides, we focused on the amino acid residues at position 58 and 60 and generated new transgenic TKO mouse lines that express I-Ab molecules covalently bound to p58K or p60K-bearing lysine instead of glycine or leucine at these positions as a single species (58K TKO, 60K-2 TKO, and 60K-5 TKO).
Because the expression level of MHC–peptide complexes in the thymus is a critical parameter that determines T cell differentiation (8, 13–15, 27), we first examined the expression of I-Ab molecules bound to p58K, p60K, or pEα using the mAb, Y3P, specific for I-Ab molecules irrespective of the binding peptides (28). In 58K TKO, the transgene-encoding I-Ab molecules were expressed in the thymic medulla (Fig. 1A). However, this line lacked I-Ab expression in the thymic cortex where positive selection takes place (29–32). In contrast, both 60K-5 TKO and 60K-2 TKO expressed I-Ab molecules in the thymic cortex and medulla, although the expression level in 60K-2 TKO was remarkably lower than that in 60K-5 TKO (Fig. 1B). Simultaneous staining experiments revealed that expression level and expression pattern of I-Ab molecules in 60K-5 TKO and 60K-2 TKO was similar to that in B2L TKO or H3 TKO, respectively (Fig. 1B).
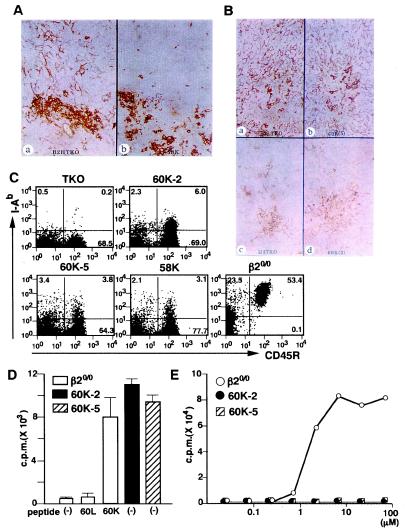
Figure 1.
Expression of I-Ab–p60K complex in the thymus and periphery in 60K-5 TKO and 60K-2 TKO. (A and B) Frozen thymus sections prepared from each mouse line were simultaneously stained with Y3P. The results represent two independent experiments using different mice for each line. A: B2H TKO (a), 58K TKO (b); B, B2L TKO (a); 60K-5 TKO (b); H3 TKO (c); 60K-2 TKO (d). (C) LN cells from TKO, transgenic TKO, and β20/0 mice were stained with Y3P and anti-CD45R mAb and analyzed by flow cytometry. (D) A T cell hybridoma, 12-2, specific for I-Ab–p60K complex, was cultured with irradiated LN cells from β20/0, 60K-2 TKO, and 60K-5 TKO in the presence (10 μM) or absence of the truncated variants of pEα bearing leucine (60L) or lysine (60K) at position 60. (E) A T cell hybridoma, BV2-3-1, specific for pigeon cytochrome c-derived analog (50V) restricted by I-Ab molecules was cultured with irradiated LN cells from β20/0, 60K-2 TKO, and 60K-5 TKO in the presence of the indicated concentration of 50V.
When LN cells from 60K-2 TKO, 60K-5 TKO, and 58K TKO were analyzed using Y3P and the anti-CD45R mAb, weak but definite expression of I-Ab molecules was observed with both B cell and non-B cell populations (Fig. 1C). The LN cells from 60K-5 TKO and 60K-2 TKO stimulated a T cell hybridoma, 12-2, specific for I-Ab–p60K complex in the absence of the exogenous p60K (Fig. 1D). However, LN cells from both lines were unable to present other I-Ab-binding peptide, 50V, to T cells at any concentration tested (Fig. 1E). These results indicate that, in 60K-5 TKO and 60K-2 TKO, almost all I-Ab molecules are occupied with p60K and expressed in appropriate form to be recognized by a given TCR.
Positive Selection Directed by I-Ab–p60K Complex.
Having established two sets of mouse lines in which I-Ab molecules bound to a single peptide with one amino acid difference at a TCR contact are expressed at comparable levels in the thymus, we compared CD4+ T cell differentiation directed by these ligands. Significant numbers of CD4+CD8− thymocytes were selected to mature in B2L TKO and H3 TKO, whereas in both 60K-5 TKO and 60K-2 TKO, the percentages of mature CD4+ T cells in the thymus and periphery reached only the level seen in TKO or 58K TKO mice, which essentially lack positive selection of CD4+ T cells (Fig. 2 A and B). The expression of invariant chain restores, to some extent, the diversity of self-peptides bound to I-Ab molecules in these mice (9, 13). When CD4+ T cell differentiation was analyzed in transgenic and nontransgenic double knockout (DKO) mouse lines that express invariant chain but still lack the expression of endogenous I-Aβb chain and β2-microglobulin, considerable numbers of CD4+ T cells were found in the thymus and periphery in 60K-2 DKO and 60K-5 DKO, but not in DKO and 58K-DKO (Fig. 2C).
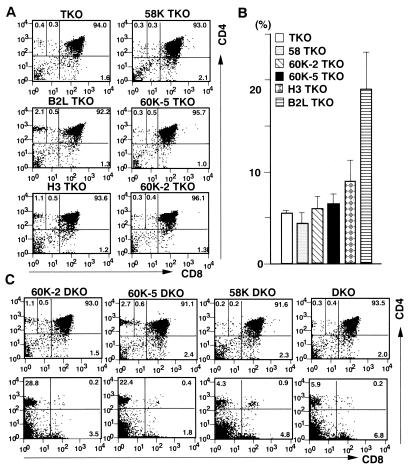
Figure 2.
CD4+ T cell differentiation in transgenic knockout mouse lines with or without invariant chain expression. (A and B) Thymocytes and LN cells were prepared from the indicated transgenic TKO mouse lines at 6–7 weeks old and stained with anti-CD4 mAb and anti-CD8 mAb. CD4 versus CD8 plots of thymocytes (A) and the proportion of LN CD4+ T cells to total LN cells (B) are shown. For each line, at least three mice were analyzed. Each value for the percentage of CD4+CD8− thymocytes was as follows. TKO: 0.3, 0.4, 0.3, 0.5; 58K TKO: 0.4, 0.3, 0.5; B2L TKO: 1.5, 2.3, 2.1, 2.0; 60K-5 TKO: 0.7, 0.3, 0.6, 0.3; H3 TKO: 1.1, 1.0, 0.9; 60K-2 TKO: 0.3, 0.4, 0.5, 0.4, 0.6. (C) Thymocytes and LN cells were prepared from the indicated transgenic DKO mouse lines at 6–7 weeks old and stained with anti-CD4 mAb and anti-CD8 mAb. CD4 versus CD8 plots of thymocytes or LN cells are shown in upper and lower panels, respectively. The mean ± SD for the percentage of CD4+CD8− thymocytes and LN CD4+ T cells in each line are as follows: 60K-2 DKO (n = 3), 1.1 ± 0.0% and 23.6 ± 4.4%; 60K-5 DKO (n =3), 2.3 ± 0.4% and 26.5 ± 5.3%; 58K DKO (n = 2), 0.2% and 4.3%; DKO (n = 3), 0.4 ± 0.2% and 5.0 ± 1.3%.
The functional expression of the transgenes in 60K-5 DKO and 60K-2 DKO raises the possibility that mature CD4+ T cells found in 60K-5 TKO and 60K-2 TKO may be qualitatively different from those in TKO or 58K TKO. To address this issue, we cultured LN CD4+ T cells from transgenic TKO mice with spleen cells from β2-microglobulin-deficient mice, β20/0 expressing wild-type I-Ab molecules with a normal array of self-peptides. Neither LN CD4+ T cells from TKO nor those from 58K TKO responded to β20/0 spleen cells. In contrast, LN CD4+ T cells from both 60K-5 TKO and 60K-2 TKO, similar to the case with B2L TKO or H3 TKO, elicited robust proliferative response to spleen cells from β20/0 mice, but not to syngenic or TKO spleen cells (Fig. 3A). These results suggest that CD4+ T cells are selected to mature on I-Ab–p60K complex even in 60K-5 TKO and 60K-2 TKO through positive and negative selection, although the frequency of mature CD4+ T cells surviving both selection processes is extremely low.
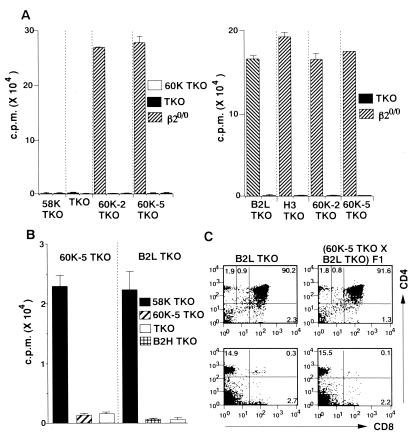
Figure 3.
Functional and phenotypic analysis for positive selection directed by I-Ab–p60K complex. (A) B cell- and CD8+ T cell-depleted LN cells prepared from 58K TKO, TKO, 60K-2 TKO, and 60K-5 TKO were cultured with irradiated spleen cells from 60K-5 TKO, TKO, or β20/0, and [3H]thymidine incorporation was measured. (B) Purified LN CD4+ T cells prepared from 60K-5 TKO and B2L TKO were cultured with irradiated spleen cells from 58K TKO, 60K-5 TKO, TKO, or B2H TKO, and [3H]thymidine incorporation was measured. (C) (60K-5 TKO × B2L TKO) F1 mice were obtained by crossing 60 K-5 TKO with B2L TKO homozygous for the transgene. Thymocytes and LN cells were prepared from the littermates with the indicated genotype at 7–8 weeks old and stained with anti-CD4 mAb and anti-CD8 mAb. CD4 versus CD8 plots of thymocytes and LN cells are shown in upper and lower panels, respectively. The mean ± SD for the percentage of CD4+CD8− thymocytes and LN CD4+ T cells in each line are as follows: (60K-5 TKO × B2L TKO) F1 (n = 3), 1.9 ± 0.3% and 15.4 ± 1.5%; B2L TKO (n = 3), 2.0 ± 0.4% and 15.2 ± 0.6%.
This brings into question which process, positive or negative selection, is responsible for the poor CD4+ T cell differentiation in 60K-5 TKO and 60K-2 TKO. When LN CD4+ T cells from 60K-5 TKO and B2L TKO expressing the single I-Ab–peptide ligand at a higher and comparable level in the thymus were cultured with 58K TKO spleen cells, definite proliferative response was observed with both CD4+ T cells (Fig. 3B), indicating that even CD4+ T cells recognizing the structurally related peptide ligand are selected to mature in the thymus in these mice. Therefore, it seems unlikely that negative selection directed by I-Ab–p60K complex eliminates large numbers of CD4+ T cells expressing TCRs with various specificity and various affinity for this ligand. This was confirmed by the result that, in (60K-5 TKO × B2L TKO) F1 mice expressing both I-Ab–p60K complex and I-Ab–pEα complex, the comparable number of mature CD4+ T cells to that in B2L TKO was found in the thymus and periphery (Fig. 3C).
T Cell Repertoire Shaped by I-Ab–p60K Complex.
To characterize T cell repertoire shaped by I-Ab–p60K complex, we first compared TCR Vβ usage in CD4+CD8− thymocytes using a panel of mAbs. The frequency of CD4+CD8− thymocytes expressing TCR Vβ4 or Vβ14 was significantly higher in both 60K-2 TKO and 60K-5 TKO than that in TKO (Table 1). Supporting the results on mixed lymphocyte reaction (Fig. 3A), these results indicate that CD4+CD8− thymocytes in 60K-2 TKO and 60K-5 TKO are qualitatively different from those in TKO. However, when TCR Vβ usage was compared between 60K-2 TKO or 60K-5 TKO and H3 TKO or B2L TKO, no remarkable difference could be deduced, although the frequency of CD4+CD8− thymocytes expressing TCR Vβ14 slightly decreased in both 60K-2 TKO and 60K-5 TKO (Table 1).
Table 1.
Comparison of TCR Vβ usage in CD4+CD8− thymocytes
| Vβ | TKO (n = 4) | 60K-2 TKO (n = 4) | 60K-5 TKO (n = 4) | H3 TKO (n = 3) | B2L TKO (n = 4) | β20/0 (n = 3) |
|---|---|---|---|---|---|---|
| 2 | 3.1 ± 0.9 | 2.4 ± 0.3 | 2.8 ± 0.5 | 3.7 ± 0.5 | 4.4 ± 0.3 | 5.8 ± 0.3 |
| 4 | 4.4 ± 0.9 | 13.0 ± 1.0 | 12.0 ± 0.7 | 12.9 ± 1.0 | 12.7 ± 0.5 | 8.3 ± 0.3 |
| 5 | 11.0 ± 1.5 | 7.1 ± 0.9 | 7.4 ± 1.1 | 6.3 ± 0.4 | 6.1 ± 0.6 | 2.8 ± 0.2 |
| 6 | 5.1 ± 0.8 | 7.1 ± 0.8 | 6.7 ± 0.7 | 6.2 ± 0.4 | 6.3 ± 0.4 | 8.7 ± 0.5 |
| 8 | 15.1 ± 2.0 | 16.6 ± 0.6 | 16.8 ± 0.3 | 17.7 ± 0.5 | 18.0 ± 0.4 | 18.2 ± 0.5 |
| 10 | 5.6 ± 1.4 | 3.6 ± 0.2 | 5.0 ± 1.7 | 6.9 ± 1.9 | 7.1 ± 1.9 | 7.5 ± 0.6 |
| 11 | 5.7 ± 0.2 | 4.6 ± 0.6 | 4.8 ± 0.2 | 4.4 ± 0.3 | 5.2 ± 0.3 | 4.5 ± 0.1 |
| 14 | 3.7 ± 0.8 | 6.9 ± 1.1 | 8.5 ± 1.3 | 11.1 ± 1.4 | 12.7 ± 0.7 | 10.2 ± 0.2 |
For each experiment, one mouse was used for H3 TKO, B2L TKO, and β20/0, and two or three mice were used for 60K-2 TKO, 60K-5 TKO, and TKO. The data represent the mean ± SD of the percentages.
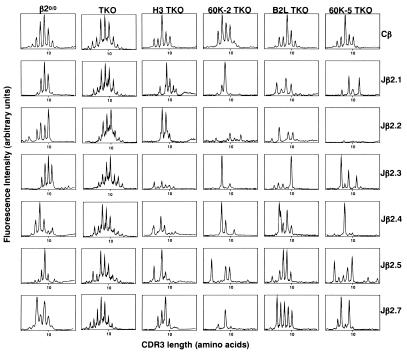
We then collected CD4+CD8− thymocytes from several mice corresponding to each genotype and analyzed TCRα repertoire using anchored PCR followed by sequencing of the cloned PCR products. This approach has the advantage in selecting the clones originating from different templates by difference in the anchored position, even though they encode the same TCRα chain. Although no obvious bias for Vα usage was observed with any mouse lines, the frequency of clones encoding Vα10 in 60K-5 TKO and 60K-2 TKO decreased or increased, respectively, as compared with that in their counterparts, B2L TKO or H3 TKO (Fig. 4A). When Vα10-bearing clones, which originate from different templates and do not overlap with 143 different sequences obtained from TKO, were analyzed for their complementarity determining region three (CDR3) loops, the extremely perturbed CDR3 length distribution was found with the clones obtained from 60K-2 TKO or 60K-5 TKO, over 40% of which had a CDR3 loop with 12 amino acids (60K-2 TKO, 13 of 28 clones; 60K-5 TKO, 4 of 10 clones) (Fig. 4B). All these clones, except two whose Vα10 subfamily could not be identified due to the shortness of the PCR products, encoded AV10S3, Jα25, and a CDR3 loop with similar amino acid composition (Fig. 4C). In contrast, such structural features were not observed with Vα10-bearing clones selected by I-Ab–pEα complex or wild-type I-Ab molecules as well as those obtained from TKO (Fig. 4B; data not shown). When we analyzed Vβ14 rearrangements in combination with Jβ2 subfamilies using immunoscope technique, it was also found that skewing of the CDR3 length distribution was somewhat prominent in 60K-2 TKO and 60K-5 TKO as compared with their counterparts (Fig. 5). These results indicate that CD4+ T cells selected to mature on I-Ab–p60K complex express restricted TCRs in terms of length and/or amino acid composition of their CDR3 loops, thereby suggesting that T cell repertoire shaped by I-Ab–p60K complex is less diverse than that by I-Ab–pEα complex.
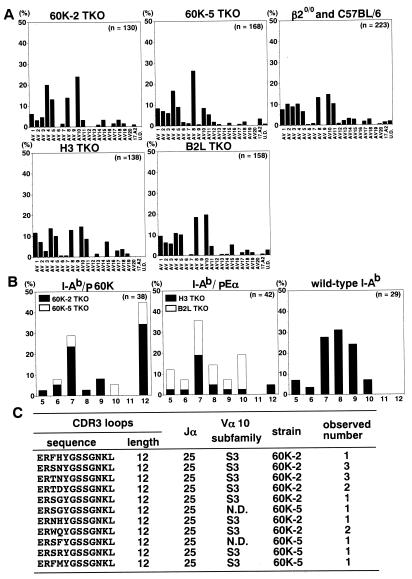
Figure 4.
TCRα repertoire shaped by I-Ab–p60K complex. (A) CD4+CD8− thymocytes (3 × 105) were collected from several mice per each line, and TCRα transcripts were analyzed using anchored PCR followed by sequencing of the cloned PCR products. The results are shown as the percentages of the Vα usage of clones originating from different templates. The clones encoding undefined TCR Vα are indicated as UD. (B and C) Vα10-encoding clones, which originate from different templates and do not overlap 143 different Vα10-encoding sequences collected from TKO CD4+CD8− thymocytes, were analyzed for their CDR3 loops. The CDR3 length distribution of these clones selected by I-Ab–p60K complex, I-Ab–pEα complex, or wild-type I-Ab molecules is shown in B. Vα10 subfamily, Jα subfamily, and predicted CDR3 amino acid sequences are shown in C, regarding the clones with CDR3 of 12 amino acids obtained from 60K-2 TKO or 60K-5 TKO. The clone, of which Vα10 subfamily was not determined because of its shortness, is indicated as ND.
Figure 5.
Immunoscope analysis for Vβ14 rearrangements in combination with Jβ2 subfamilies. The fluorescent run-off product profiles on Vβ14-Jβ2 rearrangements are compared among transgenic TKO mouse lines, TKO, and β20/0, using cDNA obtained from the CD4+CD8− thymocytes. The intensity of fluorescence is represented in arbitrary units as a function of CDR3 size in amino acids. The profiles are representative of three independent experiments for each mouse line.
Discussion
In the present study, we have compared CD4+ T cell differentiation between two sets of mouse lines, 60K-2 TKO or 60K-5 TKO and H3 TKO or B2L TKO, in which almost all I-Ab molecules are occupied with either p60K or pEα and expressed in the thymus at comparable levels. Although considerably large numbers of CD4+CD8− thymocytes were selected to mature in H3 TKO and B2L TKO, the number of mature CD4+ T cells in the thymus and periphery was extremely small in both 60K-2 TKO and 60K-5 TKO. It is clear that this apparently “impaired” CD4+ T cell differentiation results from neither inappropriate expression of the I-Ab molecules in the thymus nor a position effect of the transgenes causing some T cell defects. In addition, the result on (60K-5 TKO × B2L TKO) F1 mice argues against the possibility that overwhelming negative selection directed by I-Ab–p60K complex causes such poor CD4+ T cell differentiation. Our results thus indicate that the amino acid substitution from leucine to lysine at position 60 of the pEα critically affects CD4+ T cell differentiation through the process of positive selection.
Several experiments, including this study, have failed to detect any replacement of the covalently bound peptide by other self- or foreign antigenic peptides under the condition lacking invariant chain expression (9, 13, 17). Nevertheless, in light of the result by Barton and Rudensky suggesting that positive thymocyte selection is driven by low-abundance peptides bound to MHC molecules even when they are expressed at undetectable levels (33), it is formally possible that pEα or p60K covalently attached to I-Ab molecules, depending on the affinity of each peptide to I-Ab, allows replacement by other endogenous peptides at different efficiencies, resulting in the differential CD4+ T cell maturation. However, this possibility is unlikely because this amino acid difference at position 60 of TCR contacts does not affect binding affinity of the peptide to I-Ab molecules (26). Therefore, we conclude that the amino acid substitution from leucine to lysine affects positive selection by changing the structure of the self-peptide involved in this process.
Although several groups have reported that antigen-specific T cells selected by a given MHC–peptide complex express somewhat different TCRs from those in normal mice (34–36), no definite evidence for the imprint of intrathymic self-peptides on mature T cell repertoire has been obtained without introducing TCR transgenes (6, 9–13, 25). However, such imprint was easily visible in CD4+ T cell repertoire selected by I-Ab–p60K complex. This seems likely to reflect specific TCR–peptide interaction required to survive positive selection directed by this ligand. Recently, crystal structure of an MHC class II–peptide complex associated with its TCR has been resolved (37), in which it is shown that P5 residue located at the center of the core peptide sequence plays a critical role in the recognition by TCR through the interaction with CDR3α. Since amino acid residue at position 60 of the pEα corresponds to P5 residue (26), our finding that the amino acid substitution from leucine to lysine at this position affects the CDR3α of the selected TCRs suggests that the mode of TCR recognition is essentially the same in T cell activation and positive selection. However, despite the fact that TCR recognition is relatively flexible in the periphery (38), this amino acid substitution has a profound effect on positive selection. Although the molecular basis for this remains unclear, it might be attributed to low TCR expression on immature thymocytes, lack of cell division during positive selection, and differential expression of adhesion molecules between antigen-presenting cells and thymic cortical epithelial cells.
Whether a single MHC–peptide complex can drive the positive selection of large numbers of T cells or select limited numbers of T cells is currently controversial. At this stage, we cannot completely exclude the possibility that other self-peptides than pEα expressed in association with I-Ab molecules at undetectable levels contribute to positive selection in B2L TKO and H3 TKO. However, in light of the present results that CD4+ T cell differentiation is remarkably different between two sets of transgenic TKO mouse lines expressing the I-Ab molecule covalently bound to p60K or pEα at comparable levels in the thymus, it seems unlikely that non-Eα peptides select the majority of CD4+ T cells in these lines. Therefore, although it is clear that T cell repertoire shaped by I-Ab–pEα complex does not include all of the TCR specificity seen in normal mice (17, 21), self-peptides like pEα with uncharged and relatively small amino acid residues at TCR contacts, especially at P5, may serve as “superselectors” when they are expressed at an appropriate level in the thymus (13). In contrast, I-Ab–p60K complex selects only small numbers of CD4+ T cells and shapes less diverse T cell repertoire, suggesting that the substitution from leucine to lysine at P5 residue results in requirement for more stringent TCR–peptide interactions to survive positive selection directed by this ligand. Therefore, the process of positive selection may be both specific and degenerate, depending on individual self-peptides involved in this process.
In summary, we have shown that the substitution from leucine to lysine at P5 residue of a selecting self-peptide critically affects positive selection of CD4+ T cells. Our findings suggest that diversity of T cell repertoire depends not only on the diversity but also on the structure of peptide ligands mediating positive selection, which would establish a critical role of self-peptides in shaping T cell repertoire.
Acknowledgments
We thank Drs. D. Mathis and C. Benoist for mice deficient in wild-type I-Aβb or invariant chain, Dr. K. Ogasawara for a T cell hybridoma, BV2-3-1, and Dr. M. Kimoto for a B cell hybridoma, Y3P. This work was supported by the Japan Science and Technology Corporation and a grant from the Ministry of Education, Science, Sports, and Culture of Japan.
Abbreviations
- TCR
T cell receptor
- pEα
Eα chain-derived peptide 52–68
- p58K and p60K
pEα variant peptides bearing lysine instead of glycine or leucine at position 58 or 60
- TKO
mice lacking endogenous I-Aβb, invariant chain, and β2-microglobulin
- DKO
mice lacking endogenous I-Aβb and β2-microglobulin
- β20/0
mice lacking β2-microglobulin
- CDR
complementarity determining region
- LN
lymph node
- PE
phycoerythrin
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250470797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250470797
References
- 1.Davis M M, Bjorkman P J. Nature (London) 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Nikolic-Zugic J, Bevan M J. Nature (London) 1990;344:65–67. doi: 10.1038/344065a0. [DOI] [PubMed] [Google Scholar]
- 3.Berg L J, Frank G D, Davis M M. Cell. 1990;60:1043–1053. doi: 10.1016/0092-8674(90)90352-f. [DOI] [PubMed] [Google Scholar]
- 4.Jacobs H, von Boehmer H, Melief C J, Bernes A. Eur J Immunol. 1990;20:2333–2337. doi: 10.1002/eji.1830201024. [DOI] [PubMed] [Google Scholar]
- 5.Sha W C, Nelson C A, Newberry R D, Pullen J K, Pease L R, Russel J H, Loh D Y. Proc Natl Acad Sci USA. 1990;87:6186–6190. doi: 10.1073/pnas.87.16.6186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashton-Rickardt P G, Van Kaer L, Schumacher T N M, Ploegh H L, Tonegawa S. Cell. 1993;73:1041–1049. doi: 10.1016/0092-8674(93)90281-t. [DOI] [PubMed] [Google Scholar]
- 7.Hogquist K A, Gavin M A, Bevan M J. J Exp Med. 1993;177:1469–1473. doi: 10.1084/jem.177.5.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sebzda E, Wallace V A, Mayer J, Yeung R S M, Mak T W, Ohashi P S. Science. 1994;263:1615–1618. doi: 10.1126/science.8128249. [DOI] [PubMed] [Google Scholar]
- 9.Ignatowicz L, Kappler J, Marrack P. Cell. 1996;84:521–529. doi: 10.1016/s0092-8674(00)81028-4. [DOI] [PubMed] [Google Scholar]
- 10.Miyazaki T, Wolf P, Tourne S, Waltzinger C, Dierich A, Barois N, Ploegh H, Benoist C, Mathis D. Cell. 1996;84:531–541. doi: 10.1016/s0092-8674(00)81029-6. [DOI] [PubMed] [Google Scholar]
- 11.Martin W D, Hicks G G, Mendiratta S K, Leva H I, Ruley H E, Van Kaer L. Cell. 1996;84:543–550. doi: 10.1016/s0092-8674(00)81030-2. [DOI] [PubMed] [Google Scholar]
- 12.Fung-Leung W-P, Surh C D, Liljedahl M, Pang J, Leturcq D, Peterson P A, Webb S R, Karlsson L. Science. 1996;271:1278–1281. doi: 10.1126/science.271.5253.1278. [DOI] [PubMed] [Google Scholar]
- 13.Fukui Y, Ishimoto T, Utsuyama M, Gyotoku T, Koga T, Nakao K, Hirokawa K, Katsuki M, Sasazuki T. Immunity. 1997;6:401–410. doi: 10.1016/s1074-7613(00)80283-6. [DOI] [PubMed] [Google Scholar]
- 14.Hogquist K A, Jameson S C, Heath W R, Howard J L, Bevan M J, Carbone F R. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 15.Ashton-Rickardt P G, Bandeira A, Delaney J R, Van Kaer L, Pircher H-P, Zinkernagel R M, Tonegawa S. Cell. 1994;76:651–663. doi: 10.1016/0092-8674(94)90505-3. [DOI] [PubMed] [Google Scholar]
- 16.Tourne S, Miyazaki T, Oxenius A, Klein L, Fehr T, Kyewski B, Benoist C, Mathis D. Immunity. 1997;7:187–195. doi: 10.1016/s1074-7613(00)80522-1. [DOI] [PubMed] [Google Scholar]
- 17.Grubin C E, Kovats S, deRoos P, Rudensky A Y. Immunity. 1997;7:197–208. doi: 10.1016/s1074-7613(00)80523-3. [DOI] [PubMed] [Google Scholar]
- 18.Surh C D, Lee D-S, Fung-Leung W-p, Karlsson L, Sprent J. Immunity. 1997;7:209–219. doi: 10.1016/s1074-7613(00)80524-5. [DOI] [PubMed] [Google Scholar]
- 19.Hu Q, Walker C R B, Girao C, Opferman J T, Sun J, Shabanowitz J, Hunt D F, Ashton-Rickardt P G. Immunity. 1997;7:221–231. doi: 10.1016/s1074-7613(00)80525-7. [DOI] [PubMed] [Google Scholar]
- 20.Sant'Angelo D B, Waterbury P G, Cohen B E, Martin W D, Van Kaer L, Hayday A C, Janeway C A., Jr Immunity. 1997;7:517–524. doi: 10.1016/s1074-7613(00)80373-8. [DOI] [PubMed] [Google Scholar]
- 21.Fukui Y, Hashimoto O, Inayoshi A, Gyotoku T, Sano T, Koga T, Gushima T, Sasazuki T. J Exp Med. 1998;188:897–907. doi: 10.1084/jem.188.5.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldwin K K, Reay P A, Wu L, Farr A, Davis M M. J Exp Med. 1999;189:13–23. doi: 10.1084/jem.189.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schumacher T N M, Ploegh H L. Immunity. 1994;1:721–723. doi: 10.1016/s1074-7613(94)80013-8. [DOI] [PubMed] [Google Scholar]
- 24.Pannetier C, Cochet M, Darche S, Casrouge A, Zoller M, Kourilsky P. Proc Natl Acad Sci USA. 1993;90:4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gapin L, Fukui Y, Kanellopoulos J, Sano T, Cosrouge A, Malier V, Beaudoing E, Gautheret D, Claverie J M, Sasazuki T, et al. J Exp Med. 1998;187:1871–1883. doi: 10.1084/jem.187.11.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gyotoku T, Fukui Y, Sasazuki T. Eur J Immunol. 1998;28:4050–4061. doi: 10.1002/(SICI)1521-4141(199812)28:12<4050::AID-IMMU4050>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 27.Cook J R, Wormstall E-M, Hornell T, Russel J, Connolly J M, Hansen T H. Immunity. 1997;7:233–241. doi: 10.1016/s1074-7613(00)80526-9. [DOI] [PubMed] [Google Scholar]
- 28.Janeway C A, Conrad P, Lerner E, Babich J, Wettstein P, Murphy D. J Immunol. 1984;132:662–669. [PubMed] [Google Scholar]
- 29.Berg L J, Pullen A M, Fazekas de St. Growth B, Groth B, Mathis D, Benoist C, Davis M M. Cell. 1989;58:1035–1046. doi: 10.1016/0092-8674(89)90502-3. [DOI] [PubMed] [Google Scholar]
- 30.Benoist C, Mathis D. Cell. 1989;58:1027–1033. doi: 10.1016/0092-8674(89)90501-1. [DOI] [PubMed] [Google Scholar]
- 31.Bill J, Palmer E. Nature (London) 1989;341:649–650. doi: 10.1038/341649a0. [DOI] [PubMed] [Google Scholar]
- 32.Laufer T M, Dekoning J, Markowitz J S, Lo D, Glimcher L H. Nature (London) 1996;383:81–85. doi: 10.1038/383081a0. [DOI] [PubMed] [Google Scholar]
- 33.Barton G M, Rudensky A Y. Science. 1999;283:67–70. doi: 10.1126/science.283.5398.67. [DOI] [PubMed] [Google Scholar]
- 34.Nakano N, Rooke R, Benoist C, Mathis D. Science. 1997;275:678–683. doi: 10.1126/science.275.5300.678. [DOI] [PubMed] [Google Scholar]
- 35.Ignatowicz L, Rees W, Pacholczyk R, Ignatowicz H, Kushnir E, Kappler J, Marrack P. Immunity. 1997;7:179–186. doi: 10.1016/s1074-7613(00)80521-x. [DOI] [PubMed] [Google Scholar]
- 36.Liu C-P, Parker D, Kappler J, Marrack P. J Exp Med. 1997;186:1441–1450. doi: 10.1084/jem.186.9.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinherz E L, Tan K, Tang L, Kern P, Liu J-h, Xiong Y, Hussey R E, Smolyar A, Hare B, Zhong R, et al. Science. 1999;286:1913–1921. doi: 10.1126/science.286.5446.1913. [DOI] [PubMed] [Google Scholar]
- 38.Kersh G J, Allen P M. Nature (London) 1996;380:495–498. doi: 10.1038/380495a0. [DOI] [PubMed] [Google Scholar]