Abstract
Forkhead box (Fox) proteins play critical roles in the regulation of differentiation, proliferation, immunity and aging of cells. Most studies on Fox proteins are limited to cultured cells and rodent. The aim of the current study is to detect by immunohistrochemistry whether FoxO1, FoxO3a and FoxO4 proteins are localized in the stomach and intestine of the pig. The results showed that FoxO4 exists in the mucosa in all parts of the stomach and intestine; FoxO3a exists mainly in the lamina propria and muscularis of some parts. However, FoxO1 is not detectable in all parts of the stomach and intestine. Collectively, the results of the present study indicate that there exists a distinct expression pattern of Fox proteins, and that FoxO4 is a primary forkhead transcriptional factor localized in the gastrointestinal tracts of the pig.
Keywords: Forkhead, FoxO, Gastrointestinal tract, Immunohistochemistry, Swine
INTRODUCTION
Pigs are omnivorous animals and constitute a good animal model for studying human digestive processes. The gastrointestinal tracts of pigs are similar to that of the human in size and weight and are the ideal organs to transplant to human. Recently, pigs are widely used in the fields of study of oncology, immunology and for some disease models, for example, pig’s virosis gastroenteritis as an infantile virural diarrhea model, swine mycoplasma arthritis as a human arthritis model and double albuminemia, which has only been observed in humans and pigs (Wang et al., 2005; Zhang et al., 2001). Forkhead box (Fox) proteins are one kind of transcriptional factors and tumor suppressors which play a critical role in many physiological processes, including cell apoptosis, development and proliferation which may all impact with the generation of the tumor (Accili and Arden, 2004). Studies of Fox proteins constitute a very exciting field since they have been nomenclature formally in 2000, but most of them are limited to cell culture, invertebrate and rodent studies including caenorhabditis elegans, fruit fly, mouse and rat (Liu and Lehmann, 2006; Skurk et al., 2005; Weinkove et al., 2006). Fox proteins induce cell death mainly through two downstream pathways of FasL and Bim, and connect with cell cycle through the downstream pathways of p27kip1, cyclin G2, cyclin B, p130, and are related to DNA repairs through the downstream pathway of GADD45 (Barthélémy et al., 2004; Martinez-Gac et al., 2004; Kobayashi et al., 2005; Yamamura et al., 2006). This is why Fox proteins are called the “survival and death switch” (Burgering and Kops, 2002). In the past decade, scientists engaged in the expression and mechanism of PI3K/PKB, upstream of FoxO in the gastrointestinal cancers (Michl and Downward, 2005). So, researches about FoxO, downstream of PI3K/PKB, on gastrointestinal cancers therapy are immediately important. However, data on Fox proteins in the gastrointestinal tracts of pigs are lacking.
Three members of FoxO family, i.e., FoxO1 (FKHR), FoxO3a (FKHRL1) and FoxO4 (AFX) are mostly studied in vertebrates (Arden and Biggs, 2002). Recently, a novel FoxO member, FoxO6 was found in the mouse brain (Jacobs et al., 2003; Hoekman et al., 2006). However, there are only antisera against FoxO1, FoxO3a and FoxO4 ready available to date. Therefore, the aim of the present study is to examine whether these three FoxO members are specifically localized in the gastrointestinal tracts of the pig using immunohistochemistry.
MATERIALS AND METHODS
Reagents
Antibodies for FoxO1 (Cat. No. 9462, Lot. 2), FoxO3a (Cat. No. 06-951, Lot. 27567) and FoxO4 (Cat. No. 9472, Lot. 1) were obtained from Cell Signaling Technology, Inc. (Beverly, MA, USA). All other chemicals were reagent grade and were obtained from Fisher Scientific (Pittsburgh, PA, USA).
Animals
Four commercial pigs of Landrace-Duroc F1 cross breed commercial pigs including two castrated males and two other were females from Dingshan Hopgen Farm (Nanjing, China) were used in the current experiment. The experimental pigs were about 4 months old with 87~90 kg of body weights. The gastrointestinal samples were obtained within 30 min after sacrifice, fixed in 4% paraformaldehyde, and processed for immunohistochemical analyses of FoxO protein levels. The experimental protocol was approved in accordance with the Guide for the Care and Use of Laboratory Animals prepared by the Institutional Animal Care and Use Committee, Nanjing Agricultural University.
Immunohistochemistry
After fixation, the stomach and intestine samples were embedded in paraffin and 8 μm sections were cut and mounted on slides. The sections were then processed for immunohistochemical analysis similar to the method used in our previous reports (Shi and LaPolt, 2003; Li et al., 2005a). Briefly, sections were incubated overnight at room temperature with a polyclonal rabbit immunoaffinity-purified antiserum directed against the full length FoxO1, FoxO3a or FoxO4 proteins (0.5 mg/ml IgG). The specific protein immunoreactivity was visualized by the ABC Kit Elite and 0.05% 3,3′-diaminobenzidine tetrachloride (DAB; Sigma Chemical Co., St. Louis, MO, USA) in 10 mmol/L PBS-buffered saline containing 0.01% H2O2 for 5 min. Specificity of the antibody was examined using normal rabbit serum instead of primary antibody. In order to identify structural components and cell types within the stomach and intestine, the section was counter-stained with hematoxylin and mounted with coverslips. Relative levels of immunostaining between animals and cell types were evaluated by three independent observers. Three individuals blinded to the experimental design were asked to examine the pictures and assign the intensity of staining using a scale of 0~10 with 0 indicating lack of brown immunoreactivity for FoxO proteins and 10 reflecting intense dark brown staining. All observers evaluated all slides and observations outside of the 5th to 95th percentile of the remaining observations for that treatment group were considered outlying data and were excluded from analysis. At last, we calculated the mean of them. If the immunostained cells were 1~3 the result was +, 4~6 was ++, more than 7 was +++.
RESULTS
In order to detect the localization of FoxO family in the gastrointestinal tracts of pigs, we took samples from the fundic gland, non-glandular zones, pyloric gland of stomachs, duodenum, jejunum, ileum of small intestines, colon, cecum, and rectum of large intestines and performed immunohistochemsitry of FoxO1, FoxO3a and FoxO4 proteins.
Localization of FoxO4, FoxO1 and FoxO3a in the Stomach
1. Fundic part
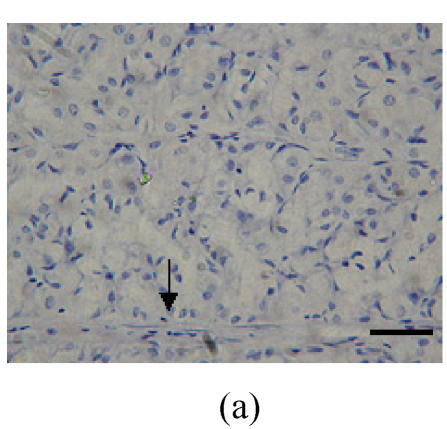
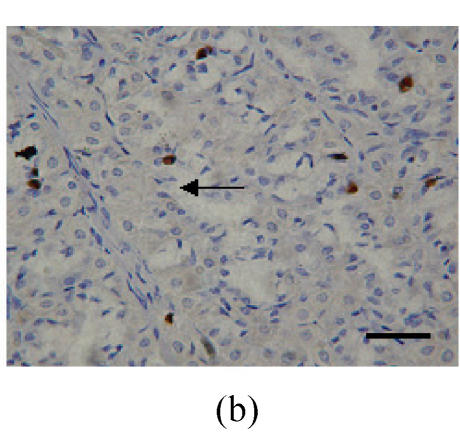
Localizations of FoxO4 in fundic part were distinct, and primarily concentrated in the nuclei of cells in the mucosa (Figs.1a and 1b; Table 1). However, there was no detectable localization of FoxO1 and FoxO3a in the fundic part (Table 1).
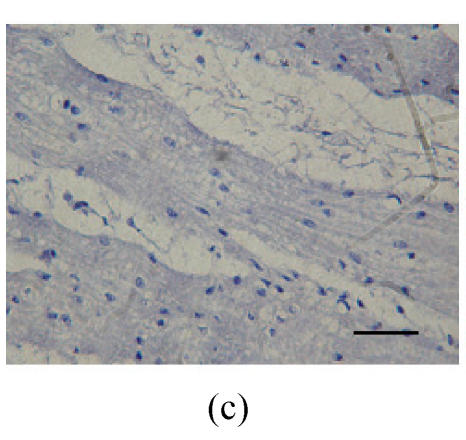
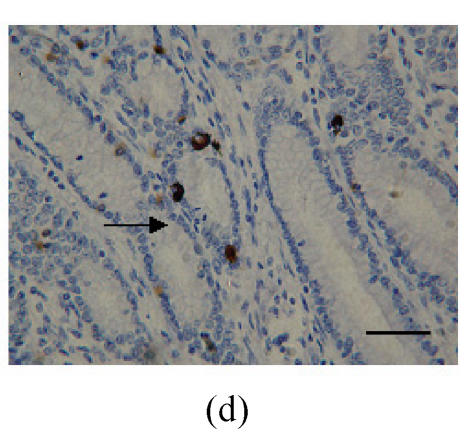
Fig. 1.
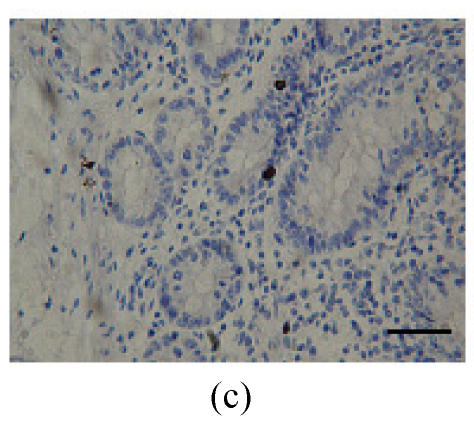
Immunohistochemical localization of FoxO proteins in the stomach of pigs (Bar=50 m). (a) Fundic gland control (×400). ↓: Muscularis mucosa; (b) Localization of FoxO4 in the fundic gland (×400). Localizations of FoxO4 in fundic part are very distinct, and primarily concentrated in the cell nuclei of the gastric mucosa. ←: Fundic gland; (c) Localization of FoxO4 in the non-glandular zones (×400). Compared with the non-glandular zones of control, FoxO4 staining was not observed; (d) Localization of FoxO4 in the pyloric gland (×400). FoxO4 in the pyloric gland was very similar to that in the fundic part, with the localizations concentrated in the cell nuclei of the mucosa. →: Pyloric gland
Table 1.
Expressions of FoxO4, FoxO1 and FoxO3a in the mucosa of the gastrointestinal tracts in the pig
| Control | FoxO4 | FoxO1 | FoxO3a | ||
| Fundic gland | − | +++ | − | − | |
| Pyloric gland | − | +++ | − | − | |
| Non-glandular zones | − | − | − | − | |
| Duodenum | Intestinal gland | − | +++ | − | − |
| Lamina propria | − | − | − | − | |
| Jejunum | Intestinal gland | − | ++ | − | − |
| Lamina propria | − | + | − | + | |
| Ileum | Intestinal gland | − | +++ | − | − |
| Lamina propria | − | − | − | − | |
| Colon | Intestinal gland | − | + | − | − |
| Lamina propria | − | − | − | + | |
| Cecum | Intestinal gland | − | + | − | − |
| Lamina propria | − | − | − | + | |
| Rectum | Intestinal gland | − | ++ | − | − |
| Lamina propria | − | − | − | + | |
2. Non-glandular zones
Compared with the non-glandular zones as control, FoxO4 (Fig.1c; Table 1) and FoxO1 were not observed, and FoxO3a showed scattered localization in the muscularis layer.
3. Pyloric gland
FoxO4 in the pyloric gland was very similar to immunocytochemical localization in the fundus, and the localizations were concentrated in the nuclei of cells in the mucosa (Fig.1d; Table 1). However, there was no FoxO1 and FoxO3a localization observed in this part.
Localization of FoxO4, FoxO1 and FoxO3a in the small intestine
1. Duodenum
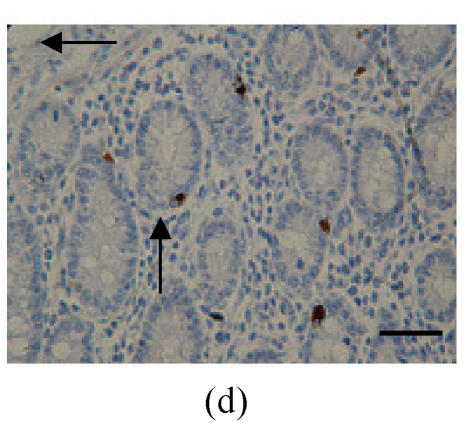
FoxO4 staining was very obvious in the intestinal gland of the Duodenum. The localization patterns were all basically in the base of the mucosa, close to the muscularis mucosa. These immunostained positive cells were almost all absorptive cells and goblet cells, but positively staining cells were observed in lamina propria (Figs.2a and 2b; Table 1). Same as for the above parts, there was no localization of FoxO1 and FoxO3a observed.
Fig. 2.
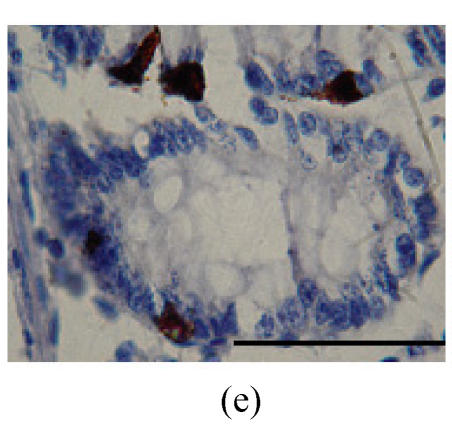
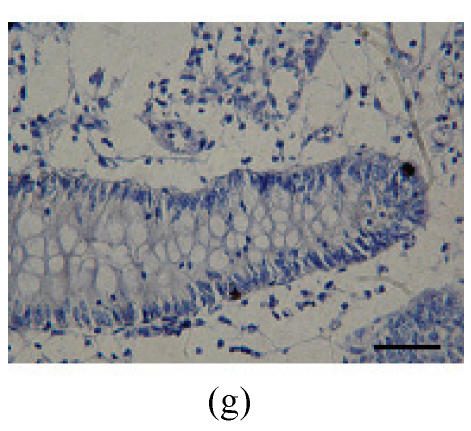
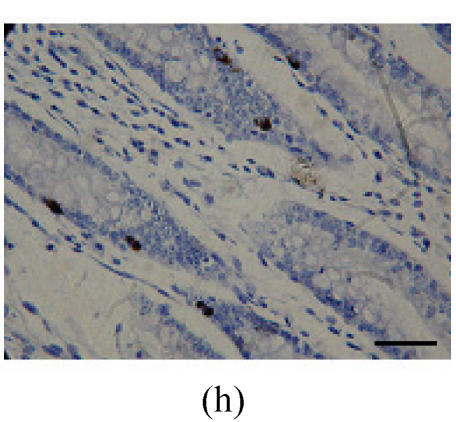
Immunohistochemical localization of FoxO proteins in the intestines of pigs (Bar=50 m). (a) Duodenum control (×400). →: Muscularis mucosa, ↓: Submucosa; (b) Localization of FoxO4 in the duodenum (×400). FoxO4 was obvious in the intestinal gland of duodenum. The localization was basically all at the base of the mucosa, close to the muscularis mucosa. These immunostained cells were almost all in absorptive cells, and some goblet cells of one part, and none in the lamina propria; (c) Localization of FoxO4 in the jejunum (×400). Some FoxO4 staining was observed in the mucosa intestinal gland; with these localizations all in absorptive cells, little in goblet cells, and little in the lamina propria; (d) Localization of FoxO4 in the ileum (×400). FoxO4 observed is exist in the intestinal gland nuclei may be absorptive cells and goblet cells. All of the localization is in the gland or near the gland, but not in the lamina propria. ←: Muscularis mucosa, ↑: Intestinal gland; (e) Localization of FoxO4 in the colon (×1000). FoxO4 was found mainly in the cell nuclei of the intestinal glands, and around the intestinal gland, with less localization in the lamina propria, and no expression in the submucosa and muscularis. Both absorptive cells and goblet cells possibly show FoxO4; (f) Localization of FoxO3a in the colon (×1000). FoxO3a staining was shown here in the cell nuclei of the lamina propria, but not in intestinal gland cells; and also in the muscularis of blood vessels viewed microscopically at 100 magnification; (g) Localization of FoxO4 in the cecum (×400). FoxO4 found here in nuclei of the intestinal gland was possibly absorptive cells and/or goblet cells; (h) Localization of FoxO4 in the rectum (×400). The localization of FoxO4 in nuclei of the intestinal gland of the mucosa is very obvious; the cells may be absorptive cells, goblet cells, and/or endocrine cells
2. Jejunum
FoxO4 proteins were localized clearly in the intestinal gland of the mucosa, with these localizations possibly all in absorptive cells, little in goblet cells, and little in the lamina propria (Fig.2c; Table 1). Under high magnification, FoxO3a was non-existent in the intestinal gland but was present in the lamina propria.
3. Ileum
FoxO4 existed in the nuclei of absorptive cells and goblet cells in the intestinal gland. All of the localized protein was in the gland or near the gland, but not in the lamina propria (Fig.2d; Table 1). FoxO1 and FoxO3a were undetectable in this part of the small intestine; however, FoxO3a was found in the muscularis of blood vessels in this area.
Localization of FoxO4, FoxO1 and FoxO3a in the large intestine
1. Colon
FoxO4 was found mainly in nuclei of cells in the intestinal gland, and around the intestinal gland, with less localization in the lamina propria, and non-existent in the submucosa and muscularis (Fig.2e; Table 1). Strong staining for FoxO3a was found in nuclei of cells in the lamina propria, but not in the intestinal gland cells. Under high magnification, some FoxO3a were observed in the muscularis of blood vessels (Fig.2f; Table 1).
2. Cecum
FoxO4 was found in nuclei of cells in the intestinal gland (Fig.2g; Table 1), while FoxO3a was observed in nuclei of cells in the lamina propria and muscularis of blood vessel. Localization of FoxO1 was not observed.
3. Rectum
Localization of FoxO4 in nuclei of cells in the intestinal gland of mucosa was very obvious; and it might exist in absorptive cells, goblet cells, and endocrine cells (Fig.2h; Table 1). FoxO3a staining was observed in the lamina propria, but there was no staining for FoxO1.
DISCUSSION
To our knowledge this is the first report of localization of FoxO4, FoxO1 and FoxO3a in the stomach and intestine of pigs. The results of our current study clearly showed that FoxO4 exists in the nuclei of cells in the mucosa of the fundic gland, pyloric gland, non-glandular zones, duodenum, jejunum, ileum, colon, cecum, rectum; and that FoxO3a exists mainly in the nuclei of lamina propria cells in the mucosa of jejunum, colon, cecum and rectum, and probably in the muscularis. FoxO1 was not found to be localized immunohistochemically in the stomach and intestine of pigs. This distinct localization pattern of FoxO proteins implies that functions of FoxO proteins, especially FoxO4, are involved in the processes of the digestive system of pigs.
Earlier researches showed that expression of FoxO4 blocked cell-cycle progression at phase G1, and is dependent on the cell-cycle inhibitor p27kip1 (Yang et al., 2005). Protein kinase B (PKB) phosphorylates and down-regulates the FoxO4 then enhances the transcription of p27kip1 (Fukuoka et al., 2003). It is known that p27kip1 inhibits the activity of cyclinE/CDK2, which allows the cell-cycle progression through G1 (Graff et al., 2005). Some researchers found that the mutant of FoxO4 (FoxO4A3), which maintains the activity to transactivate p27kip1, may be used as an anticancer agent for HER2-overexpressing cancers (Yang et al., 2005). In the mammal, the molecular signaling FoxOs are located at the intersection of three important signaling pathways: Smad, PI3K and FoxG1 (Seoane et al., 2004). Smad binds to FoxO, and actively inhibits the growth-related gene, p21cip1 (Li et al., 2005b; 2006); however, the binding of FoxG1 to the compound FoxO-Smad inhibits the expression of p21cip1 (Seoane et al., 2004). This interaction requires further study to examine the expression patterns of FoxO4 related to metabolism and proliferation.
Our previous study showed that FoxO1 protein exists in the rat ovary (Shi and LaPolt, 2003; Li et al., 2005a). However, the present study did not find any significant localization of FoxO1 in the gastrointestinal tract of pigs. This implies that this member of FoxO family has tissue-specific expression patterns.
From the present study, we found different localization patterns of FoxO3a in the different parts of the stomach and intestine in the pig. This may imply some relation with the cell-cycle. Some researches have shown that independent actions of Akt, Ikappa B kinase (IKK-β) can cause proteolysis of FoxO3a via the Ub-dependent proteasome pathway (Hu et al., 2004). TGF-β-active Smad protein then binds to FoxO and activates the growth inhibitory gene p21cip1 (Hu et al., 2004). In addition, IKK-β was shown to interact with and phosphorylate FoxO3a and that the IKKs might serve as a potential drug target in anticancer therapy (Finnberg and El-Deiry, 2004). Active forms of FoxO3a increase cyclin G2 mRNA levels and then block the cell cycle progression (Martinez-Gac et al., 2004). Some other studies proved that active FoxO3a could induce expression of IL-2-regulated genes, such as the cdk inhibitor p27kip1 and the proapoptotic Bcl-2 family member Bim (Stahl et al., 2002). On the other hand, active FoxO3a leads to accumulation of Bcl6 and down-regulation of cyclin D2 at protein and mRNA levels (Fernandez de Mattos et al., 2004). More interestingly, in contrast to FoxO4, FoxO3a does not exist in the intestinal gland, which may show that it has no relation to the gland of the intestinal tract.
In summary, FoxO4 localization is observed in the nuclei of cells in the mucosa, and FoxO3a localized in the nuclei of cells in the lamina propria. The different localization patterns indicate different mechanisms involved, which may regulate cellular apoptosis (Hosaka et al., 2004). From our study, the localization of FoxO in the gastrointestinal tracts may indicate that FoxO have some relations with functions of gastrointestinal tracts. However, the mechanisms of FoxO4 and FoxO3a involved in the gastrointestinal tract require further study.
Acknowledgments
We express our gratitude to Dr. Reinhold J. Hutz (Department of Biological Sciences, University of Wisconsin-Milwaukee, USA) for reading the original manuscript and for valuable suggestions.
Footnotes
Project supported by the National Basic Research Program (973) of China (No. 2004CB117500) and the National Natural Science Foundation of China (Nos. 30571335 and 330471253)
References
- 1.Accili D, Arden KC. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell. 2004;117(4):421–426. doi: 10.1016/S0092-8674(04)00452-0. [DOI] [PubMed] [Google Scholar]
- 2.Arden KC, Biggs WH. Regulation of the FoxO family of transcription factors by phosphatidylinositol-3 kinase-activated signaling. Arch Biochem Biophys. 2002;403(2):292–298. doi: 10.1016/S0003-9861(02)00207-2. [DOI] [PubMed] [Google Scholar]
- 3.Barthélémy C, Henderson CE, Pettmann B. FoxO3a induces motoneuron death through the Fas pathway in cooperation with JNK. BMC Neurosci. 2004;5(1):48. doi: 10.1186/1471-2202-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burgering BM, Kops GJ. Cell cycle and death control: long live forkheads. Trends Biochem Sci. 2002;27(7):352–360. doi: 10.1016/S0968-0004(02)02113-8. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez de Mattos S, Essafi A, Soeiro I, Pietersen AM, Birkenkamp KU, Edwards CS, Martino A, Nelson BH, Francis JM, Jones MC, et al. FoxO3a and BCR-ABL regulate cyclin D2 transcription through a STAT5/BCL6-dependent mechanism. Mol Cell Biol. 2004;24(22):10058–10071. doi: 10.1128/MCB.24.22.10058-10071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Finnberg N, El-Deiry WS. Activating FOXO3a, NF-kappaB and p53 by targeting IKKs: an effective multi-faceted targeting of the tumor-cell phenotype? Cancer Biol Ther. 2004;3(7):614–616. doi: 10.4161/cbt.3.7.1057. [DOI] [PubMed] [Google Scholar]
- 7.Fukuoka M, Daitoku H, Hatta M, Matsuzaki H, Umemura S, Fukamizu A. Negative regulation of forkhead transcription factor AFX (FoxO4) by CBP-induced acetylation. Int J Mol Med. 2003;12(4):503–508. [PubMed] [Google Scholar]
- 8.Graff P, Amellem O, Seim J, Stokke T, Pettersen EO. The role of p27 in controlling the oxygen-dependent checkpoint of mammalian cells in late G1. Anticancer Res. 2005;25(3B):2259–2267. [PubMed] [Google Scholar]
- 9.Hoekman MF, Jacobs FM, Smidt MP, Burbach JP. Spatial and temporal expression of FoxO transcription factors in the developing and adult murine brain. Gene Expr Patterns. 2006;6(2):134–140. doi: 10.1016/j.modgep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Hosaka T, Biggs WH3rd, Tieu D. Disruption of forkhead transcription factor (FOXO) family members in mice reveals their functional diversification. Proc. Natl. Acad. Sci. USA. 2004;101(9):2975–2980. doi: 10.1073/pnas.0400093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117(2):225–237. doi: 10.1016/S0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs FM, van der Heide LP, Wijchers PJ, Burbach JP, Hoekman MF, Smidt MP. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem. 2003;278(38):35959–35967. doi: 10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi Y, Furukawa-Hibi Y, Chen C, Horio Y, Isobe K, Ikeda K, Motoyema N. SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int J Mol Med. 2005;16(2):237–243. [PubMed] [Google Scholar]
- 14.Li X, Jiang Y, Wang Z, Liu G, Hutz RJ, Liu W, Shi F. Regulation of FoxO1 transcription factor by nitric oxide and cyclic GMP in cultured rat granulosa cells. Zool Sci. 2005;22(12):1339–1346. doi: 10.2108/zsj.22.1339. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Xie Z, Shi F. Involvement of forkhead box proteins in apoptosis and oncogenesis. Prog Biochem Biophs. 2005;32(7):600–606. [Google Scholar]
- 16.Li X, Xie Z, Shi F. FOXO in the regulation of differentiation, proliferation, immunity and aging of cells. Chin J Clin Rehabil. 2006;10(9):158–162. [Google Scholar]
- 17.Liu Y, Lehmann M. FOXO-independent suppression of programmed cell death by the PI3K/Akt signaling pathway in Drosophila. Dev Genes Evol. 2006;216(9):531–535. doi: 10.1007/s00427-006-0063-x. [DOI] [PubMed] [Google Scholar]
- 18.Martinez-Gac L, Marques M, Garcia Z, Campanero MR, Carrera AC. Control of cyclin G2 mRNA expression by forkhead transcription factors: novel mechanism for cell cycle control by phosphoinositide 3-kinase and forkhead. Mol Cell Biol. 2004;24(5):2181–2189. doi: 10.1128/MCB.24.5.2181-2189.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michl P, Downward J. PI3K/AKT signaling in gastrointestinal cancers. Z Gastroenterol. 2005;43(10):1133–1139. doi: 10.1055/s-2005-858638. [DOI] [PubMed] [Google Scholar]
- 20.Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117(2):211–223. doi: 10.1016/S0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 21.Shi F, LaPolt PS. Relationship between FoxO1 protein levels and follicular development, atresia, and luteinization in the rat ovary. J Endocrinol. 2003;179(2):195–203. doi: 10.1677/joe.0.1790195. [DOI] [PubMed] [Google Scholar]
- 22.Skurk C, Izumiya Y, Maatz H, Razeghi P, Shiojima I, Sandri M, Sato K, Zeng L, Schiekofer S, Pimentel D, et al. The FoxO3a transcription factor regulates cardiac myocyte size downstream of AKT signaling. J Biol Chem. 2005;280(21):20814–20823. doi: 10.1074/jbc.M500528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stahl M, Dijkers PF, Kops GJ, Lens SM, Coffer PJ, Burgering BM, Medema RH. The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J Immunol (Baltimore, Md: 1950) 2002;168(10):5024–5031. doi: 10.4049/jimmunol.168.10.5024. [DOI] [PubMed] [Google Scholar]
- 24.Wang T, Huo YJ, Shi F, Xu RJ, Hutz RJ. Effects of intrauterine growth retardation on development of the gastrointestinal tract in neonatal pigs. Biol Neonate. 2005;88(1):66–72. doi: 10.1159/000084645. [DOI] [PubMed] [Google Scholar]
- 25.Weinkove D, Halstead JR, Gems D, Divecha N. Long-term starvation and ageing induce AGE-1/PI 3-kinase-dependent translocation of DAF-16/FoxO to the cytoplasm. BMC Biol. 2006;4(1):1. doi: 10.1186/1741-7007-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamura Y, Lee WL, Inoue K, Ida H, Ito Y. RUNX3 cooperates with FOXO3a to induce apoptosis in gastric cancer cells. J Biol Chem. 2006;281(8):5267–5276. doi: 10.1074/jbc.M512151200. [DOI] [PubMed] [Google Scholar]
- 27.Yang H, Zhao R, Yang HY, Lee MH. Constitutively active FOXO4 inhibits Akt activity, regulates p27 Kip1 stability, and suppresses HER2-mediated tumorigenicity. Oncogene. 2005;24(11):1924–1935. doi: 10.1038/sj.onc.1208352. [DOI] [PubMed] [Google Scholar]
- 28.Zhang DW, Li XY, Chen LT. Animal models and pathological changes in gastroesophageal reflux. Chin J Pediatr Surg. 2001;22(2):112–114. [Google Scholar]