Abstract
The introduction of molecular genetic methods has caused confusion about the nature of microbial species. Environmental DNA extraction has indicated the existence of a vast diversity of genotypes, but how this relates to functional and phenotypic diversity has not been sufficiently explored. It has been implied that genetic distance per se correlates with phenotypic differentiation and thus reflects subtle (but undiscovered) adaptive fine-tuning to the environment, and that microbes may show biogeographic patterns at the genetic level. Here, we argue that no theoretically based species concept exists; species represent only the basic unit in the taxonomic hierarchy. The significance of naming species is that it organizes biological information. The reason why microbial species collectively represent large genetic differences is owing to huge absolute population sizes, absence of allopatric speciation and low extinction rates. Microbial phenotypes are, therefore, ancient in terms of the geological time-scale and have been maintained through stabilizing selection. These problems are discussed with special reference to eukaryotic micro-organisms.
Keywords: eukaryotic micro-organisms, species concept, biogeography, genetic distances, ancient microbial phenotypes, species richness
1. Introduction
The taxonomy of eukaryotic micro-organisms has traditionally been based on morphological criteria. Some groups (e.g. ciliates) provide a wealth of morphological detail, other groups less so (e.g. naked amoebae), but this has in part been remedied by higher microscopic resolution, such as the use of transmission electron microscopy. At most taxonomic levels, classical taxonomy is supported by molecular data; for example, the monophyletic nature of classical species has generally been confirmed (Patterson 1999, this paper). In contrast to prokaryotes, the concept of ‘unculturable species’ is less relevant in that attempts to culture free-living eukaryotic microbes are usually successful. The reason is primarily that, in contrast to prokaryotes, single cells can be isolated manually, and requirements such as food and particular environmental factors can be directly observed. In contrast, many bacteria cannot grow or form colonies on solid media, and their lifestyle can only rarely be witnessed at the microscopic level.
Molecular genetics has shown that both eukaryotic and prokaryotic micro-organisms represent large intraspecific genetic differences, even for conservative genes such as those coding for rRNA. Extraction and amplification of environmental DNA suggests a large ‘cryptic’ diversity of eukaryotic microbes (e.g. López-Garcia et al. 2001; Moon-van der Stay et al. 2001; Dawson & Pace 2002; Edgcomb et al. 2002). These studies generally ignore the fact that only a fraction of described protists are represented in gene sequence databases, and much phenotype diversity may in fact be well known. Furthermore, it is often assumed that genetic distance in terms of rRNA genes necessarily correlates with phenotypic differentiation and represents subtle (but unobserved) adaptive differentiation. This is not necessarily so, and there has been no attempt to test this rigorously (Fenchel 2005). Claims that the observed diversity is ‘unculturable’ are simply postulates, insofar as only a gene sequence is available.
While most nominal microbial species have cosmopolitan distribution, it has been implied that different genotypes may show biogeography, and that allopatric speciation occurs (e.g. Nanney 2004). But again, no robust demonstration of this has been carried out.
Here, we discuss the species concept for eukaryotic microbes (protists). We argue that no theoretically based species concept exists. The significance of naming species is primarily that it organizes biological information, especially the functional and other phenotypic properties of organisms. Notwithstanding adaptive specializations within nominal species, large genetic distances are largely owing to the accumulation of selectively neutral mutations. This again is the result of huge absolute population sizes, absence of geographical isolation, and low rates of local and global extinction. For these reasons, microbial phenotypes are ancient with respect to the geological time-scale. It is an open question as to what extent asexual nominal species represent discrete units in terms of phenotypic properties, or whether a continuum of sets of properties occurs. This question cannot be resolved solely by gene sequencing, but requires studies of the physiological and morphological properties of the organisms.
Finally, we conclude that sequencing rRNA genes divorced from concomitant studies of phenotypes does not yield relevant biological information. What is important is, after all, the functional properties and diversity of nature.
2. Are species ‘real’?
A vast amount of literature targets the question of what a species is or is supposed to be, and taxonomic specialists often disagree on how to delimit species within some particular group (Mallet 2001). Underlying this discussion is a notion that species are ‘real’ in the sense that they represent an evolutionary unit. Zoologists have—in principle at least—generally accepted Mayr's (1970) biological species concept, although this is meaningful only for panmictic populations (gene pools) at a given place and time. In practice, clines and complex patterns of interfertility and sterility between sub-populations complicate matters, and interfertility between disjunct populations is rarely tested. The existence of sympatric or allopatric sibling species which are intersterile, but morphologically indistinguishable or almost so, is well documented, but in practice such species complexes have been unravelled only in a few cases. Botanists, on the other hand, have been reluctant to accept Mayr's biological species concept because related but phenotypically distinct ‘species’ often hybridize easily and because asexual or apomictic plant species are common. In contrast, asexual animals are sufficiently uncommon to be considered anomalous.
With Mallet (2001), we simply consider species to be the basic taxonomic unit within the taxonomic hierarchy (such as genera, families, etc.). They do not constitute evolutionary units. In the case of sexual outbreeders, the evolutionary unit is a panmictic population (a gene pool) living at a particular point in time and space. All prokaryotes and a large fraction of eukaryotic micro-organisms are sexless. Horizontal gene transfer (HGT) is a frequent phenomenon in some prokaryotes, but this aspect has received little attention in the case of eukaryotic micro-organisms. But assuming that HGT is absent or rare, then following a cell division two new clones are initiated that are in principle evolutionarily independent. The evolutionary unit is then a clone.
Within populations of sexual outbreeders, genetic variation is to some extent constrained within populations, and differentiation (speciation) requires some sort of genetic isolation. Geographical isolation between two sub-populations is one of several mechanisms by which this can take place. In asexuals, there are no such constraints, and it is not obvious that ‘species’ in the sense of discrete sets of phenotypes is the rule, rather than a continuum of phenotypic properties.
The problem was addressed by Hutchinson (1968). He studied the obligatory parthenogenetic bdelloid rotifers and found that they do come in discrete units (‘species’) with particular sets of phenotypic properties. The conclusion was that species represent adaptive peaks, being maintained by stabilizing selection. The existence of species—defined as discrete sets of phenotypic properties—is usually taken for granted by taxonomists studying protists (sexual or asexual) in accordance with common experience, although it may, perhaps, not be true always.
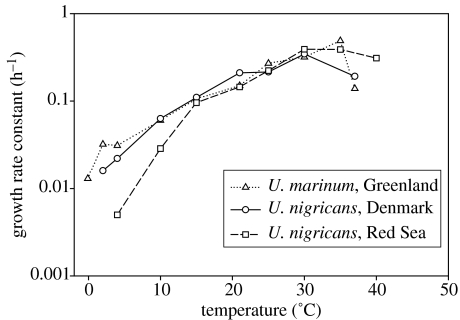
Under all circumstances, nominal microbial species, whether sexual or asexual, may show intraspecific variation with respect to particular phenotypic traits, e.g. with respect to the tolerance range of salinity and temperature (figure 1). But this also applies to animal and plant species. Notably, the magnitude of phenotypic variation within well-studied nominal species of eukaryotic micro-organisms is not different from that found within animal and plant species. There is, therefore, no close correlation between intraspecific genetic distance (e.g. of rRNA genes) and phenotypic differentiation. For example, the species complex within the ciliate genus Tetrahymena displays genetic distances with respect to rRNA genes exceeding that of all mammals, although these ciliates are phenotypically very similar. The Tetrahymena phenotype has existed since the beginning of the Mesozoic (Wright & Lynn 1997) or even earlier (Nanney 1982).
Figure 1.
Maximum growth rate constants of the ciliates Uronema marinum isolated from the east coast of Greenland and Uronema nigricans isolated from the Red Sea (Eilat) and from Denmark, as a function of temperature. The tropical isolate grows significantly slower at temperatures below 10°C, but it is more striking that all three strains show balanced growth within a temperature range that far exceeds that of the habitats from which they were isolated (T. Fenchel, unpublished data).
3. Why do we need species?
The taxonomic system serves two purposes. One of these is the reconstruction of evolution, i.e. the phylogenetic relations between different groups of organisms. In this respect, the methods of molecular genetics have revolutionized our understanding of prokaryote phylogeny. With respect to eukaryotic micro-organisms, they too have provided much better understanding of relationships at higher taxonomic levels. At the level of species, they have largely confirmed classical taxonomy in the sense that traditionally described species have mostly been shown to be monophyletic.
The other purpose of taxonomy is to organize biological information. The name of a taxonomic group at any level carries information about a number of correlated phenotypic properties. Assigning an organism to some taxonomic designation, such as for example ruminants, tells us it is a mammal, that it is devoid of teeth in the upper jaw, carries antlers or horns, and has paired hoofs and a rumen with a symbiotic consortium of microbes that degrade cellulose. The taxonomic system is a way to organize the information that is needed to understand the functional roles of organisms in nature, biodiversity and community structure. To expand our knowledge and understanding, it is necessary to study the phenotypic properties of organisms based on observations, descriptions and experiments. Describing biota solely on the basis of phylogenetic trees provides little relevant biological information, and at the same time threatens to dismiss a huge body of biological knowledge accumulated during the last two centuries (Wheeler 2004).
4. How many nominal species of protists are there?
Estimates of the global species inventory are uncertain for several reasons (e.g. May 1988). On one hand, many species may remain undiscovered, but on the other hand, many taxonomic groups are burdened by synonyms (for ciliates, see Finlay et al. 1996). Table 1 includes our literature-based estimate of the number of named free-living organisms belonging to what has traditionally been termed protozoa (a subset of the protists), i.e. all groups with phagotrophic members (some groups also include phototrophs), but excluding parasitic forms. Protist groups in which all members are obligatory phototrophs (e.g. diatoms) have been excluded, as well as some fungi-like unicellular forms (e.g. chytrids). The species numbers are arranged according to size classes.
Table 1.
Number of species and size range of major protozoan groups.
| size range (mm) | geometric mean for each size interval (mm) | Prymnesiida (haptomonads) | cryptomonads | heterokont flagellates | anaerobic flagellates | choanoflagellates | other heterotrophic flagellates | euglenids | heliozoa | amoebae with uncertain affinities | naked amoebae (Lobosea and Heterolobosea) | dinoflagellates | testate amoebae | ciliates marine and non-marine | Radiolarian—Acantharia, Polycystina, Phaeodaria | myxomycete sporangia | foraminifera | sum protozoa |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0.001–0.00178 | 0.001 | |||||||||||||||||
| 0.00178–0.00316 | 0.002 | 7 | 11 | 1 | 2 | 2 | 23 | |||||||||||
| 0.00316–0.00562 | 0.004 | 59 | 24 | 35 | 11 | 17 | 19 | 4 | 1 | 1 | 8 | 10 | 3 | 192 | ||||
| 0.00562–0.01 | 0.008 | 105 | 69 | 105 | 26 | 26 | 46 | 42 | 8 | 4 | 17 | 116 | 5 | 569 | ||||
| 0.01–0.0178 | 0.013 | 99 | 65 | 236 | 23 | 41 | 40 | 343 | 34 | 17 | 67 | 398 | 30 | 8 | 1401 | |||
| 0.0178–0.0316 | 0.024 | 39 | 18 | 165 | 15 | 12 | 26 | 977 | 75 | 21 | 110 | 794 | 80 | 203 | 2535 | |||
| 0.0316–0.0562 | 0.042 | 16 | 105 | 2 | 7 | 4 | 837 | 34 | 14 | 70 | 736 | 210 | 480 | 7 | 2522 | |||
| 0.0562–0.1 | 0.075 | 8 | 47 | 1 | 5 | 2 | 202 | 9 | 12 | 48 | 340 | 149 | 699 | 41 | 3 | 12 | 1578 | |
| 0.1–0.178 | 0.133 | 35 | 152 | 5 | 13 | 31 | 78 | 22 | 569 | 93 | 30 | 197 | 1225 | |||||
| 0.178–0.316 | 0.237 | 11 | 40 | 2 | 8 | 24 | 30 | 459 | 187 | 122 | 593 | 1476 | ||||||
| 0.316–0.562 | 0.421 | 2 | 10 | 1 | 4 | 14 | 207 | 112 | 309 | 812 | 1471 | |||||||
| 0.562–1 | 0.75 | 1 | 5 | 2 | 179 | 75 | 247 | 893 | 1402 | |||||||||
| 1–1.78 | 1.33 | 1 | 1 | 4 | 4 | 122 | 48 | 204 | 649 | 1033 | ||||||||
| 1.78–3.16 | 2.37 | 2 | 1 | 73 | 45 | 56 | 533 | 710 | ||||||||||
| 3.16–5.62 | 4.21 | 2 | 7 | 10 | 195 | 314 | ||||||||||||
| 5.62–10 | 7.5 | 4 | 10 | 81 | 95 | |||||||||||||
| 10–17.8 | 13.3 | 1 | 4 | 7 | 23 | 35 | ||||||||||||
| 17.8–31.6 | 23.7 | 2 | 12 | 14 | ||||||||||||||
| 31.6–56.2 | 42.1 | 1 | 1 | |||||||||||||||
| 309 | 200 | 750 | 80 | 110 | 139 | 2610 | 170 | 111 | 396 | 2502 | 499 | 2999 | 623 | 998 | 4000 | 16 596 |
In total, this includes about 16 600 protozoan species with ciliates and foraminifera as the most species rich groups. Earlier published estimates have been somewhat higher in that they included fossils (foraminifera) and used inflated estimates of radiolarian species diversity (Anderson 1983).
Excluding a few giant protozoa (the xenophyophorans and some foraminifera and myxomycetes), there are about 16 000 species based on phenotypic (mainly morphological) criteria. These have a size range from slightly greater than 2 μm to 2 mm, i.e. a size range of ×1000. The number of metazoan species ranging between 2 mm and 2 m would, in contrast, exceed 1 million. Since the great majority of protist species are aquatic, it might be more reasonable to compare them with the number of named aquatic metazoan species, which is about 180 000. Even so, protozoan species numbers with a size range of ×1000 constitute only about 8% of named aquatic metazoa covering a similar range of body lengths.
There is no way of knowing whether this ratio between protozoan and metazoan species numbers is a good estimate. But the difference is so great that it is difficult not to accept it as a fact that there are more large species than small species. In fact, most species (terrestrial as well as aquatic) measure around 1 cm (May 1988; Fenchel & Finlay 2004). The reason for so few very large species is that they have small absolute population sizes, making them prone to higher extinction rates over evolutionary time.
The relative importance of different mechanisms determining why ecological communities include a particular number of species and how they partition available resources is still debated. But there is no reason to believe that resource or habitat partitioning should be fundamentally different for organisms that differ greatly in size. We believe that the relatively high global number of larger species is owing to allopatric speciation and endemic species, brought about by historical contingencies over evolutionary time. In contrast, we have argued (Finlay & Fenchel 2004; Fenchel & Finlay 2004) that owing to huge absolute population sizes and thus high dispersal potential and low extinction rates, most protist species have cosmopolitan distribution.
5. Why do microbes show large intraspecific genetic variation?
Simple methods to study enzyme polymorphism became available in the 1960s, and it was found that genetic variation within most animal species was much higher than previously imagined, although species with small absolute population sizes (e.g. some large mammals) proved to be genetically monomorphic. It was initially also assumed that such genetic variation reflected adaptive fine-tuning to the environment (Lewontin 1974) and many attempts were made to demonstrate this. Kimura (1983), on the other hand, argued that most of this variation reflects adaptively neutral or near neutral mutations (‘near neutral’ meaning that stochastic drift exceeds directional selection) that may spread in populations over time through genetic drift. Initially, this interpretation was resisted (e.g. Lewontin 1974), largely because Kimura's ideas were—incorrectly—suspected of being at variance with Darwinian evolution. The theory of neutral molecular evolution does acknowledge that evolution is driven by adaptive mutations and directional selection, but it also recognizes that favourable mutations are rare in stable environments compared with neutral and near neutral mutations. Today, Kimura's theory of neutral molecular evolution is generally accepted.
With the advent of polymerase chain reaction (PCR) and methods for sequencing DNA, it became apparent that nominal species of microbes display a much higher degree of genetic differentiation than do macroscopic organisms, and especially this applies to conservative genes such as those coding for ribosomal RNA. In a sense, the present debate on the significance of intraspecific genetic variation in microbes is a revival of the discussion in the 1960s and 1970s.
When a group of phenotypically similar organisms show large genetic differentiation, the null hypothesis must be that the phenotype has been conserved over a long geological period. With few exceptions, e.g. foraminifera (Pawlowski et al. 2003), protists generally do not leave recognizable fossils, so it is impossible to calibrate the molecular clock. Nevertheless, based on various other estimates, it has been suggested that ciliate species can be traced back to the beginning of the Mesozoic or Late Palaeozoic (Nanney 1982; Wright & Lynn 1997). The finding of Triassic amber with fossil protists from freshwater also suggests the presence of some morphospecies that are still common today (Schönborn et al. 1999).
It is quite possible that, in some cases, lineages in phylogenetic trees of nominal species will turn out to be correlated with particular physiological adaptations which are not reflected in any morphologic traits. This will be discussed in more detail later. But in the absence of further evidence, it must be concluded that the substantial genetic distances within nominal species reflect ancient and conserved phenotypes. The underlying reason for a slow species turnover in geological time is probably related to their huge absolute population sizes. The consequence is great potential for dispersal and global distribution, and at the same time a vanishingly low probability of local and global extinctions.
6. Biogeography for protists
The majority of nominal protist species can be found worldwide, wherever their particular habitat requirements are met, and this has been recognized for more than a century. However, some morphologically distinct species are confined to particular climatic zones, such as the tropics or polar sea ice, and in several cases this has been shown to correlate with a limited temperature range for growth and survival (Dragesco 1968; Lee & Fenchel 1972). Such organisms have a pantropical or bipolar distribution, and there is no reason to assume allopatric speciation, but only specialized habitat requirements. Whether all nominal species occur everywhere on Earth where their required habitat is realized is difficult to prove or disprove, and some examples of real biogeography of limnic or terrestrial protists may turn out to be correct (Tyler 1996; Foissner 1999; Wilkinson 2001).
There are some corollaries for the cosmopolitan distribution of microbes. In aquatic habitats, the local diversity of unicellular eukaryotes is very high, but on a global scale the number of small species is low relative to larger organisms. For example, a 2 ha shallow marine area and a 1 ha pond each included about 1000 eukaryotic species, and about two-thirds were protists. In addition, there was a close correlation between the decreasing body size for different taxonomic groups and the fraction of the global species pool present at these sites (Fenchel & Finlay 2004).
Recognition of large genetic distances within nominal species has raised the question whether different genotypes have biogeography, in the sense that they represent allopatric differentiation and ‘cryptic species’ that are morphologically indistinguishable, but different with respect to physiological properties. It is likely that adaptive genetic differences occur within nominal species in different types of habitats or climates (cf. figure 1), as in macroscopic organisms with wide distribution ranges. So far, however, studies have concentrated on differences in the sequences of rRNA genes that must represent neutral mutations, and here the rate of nucleotide replacements is low. This has the advantage that it constrains the age of differentiation. The problem then is (i) whether differentiation of rRNA genes reflects any geographical pattern and (ii) whether such differentiation correlates with phenotypic differentiation.
Demonstration of a geographical pattern of genotypes is not simple, and most studies are based on an inadequate number of samples to draw any useful conclusions. Studies on rRNA gene sequences of the asexual bodonid flagellates (e.g. Bodo designis and Bodo saltans) that had been collected worldwide (Von der Heyden et al. 2004; Koch & Ekelund 2005) primarily demonstrated that no two genotypically identical isolates were found, implying that the number of existing genotypes must be vast. No geographical pattern was apparent in the sense that genetic distance correlated with geographical distance. There was some indication that marine isolates of B. designis constituted a clade that was separate from freshwater isolates. A more comprehensive study by Šlapeta et al. (2006) on the plankton flagellate Micromonas pusilla also showed a large number of genotypes, but all seem to have a global distribution.
Lowe et al. (2005) studied isolates of the heterotrophic dinoflagellate Oxyrrhis marina collected in different oceans and climates. No geographical pattern was evident, and while the isolates differed somewhat with respect to their salinity tolerance range, there was no correlation with rRNA genotype.
In sexual outbreeding species, we would expect to find discrete groups of genotypes. The ciliate genus Tetrahymena is presently constituted by 39 such groups. Some of these are morphologically distinguishable and show some other minor phenotypic differences. Most are outbreeders, but some asexual strains also exist. Nanney and co-workers (Nanney 1982, 2004; Nanney et al. 1998) have studied this complex in detail and they claim that some of the ‘species’ have restricted geographical distribution. However, biased and insufficient samplings render this conclusion premature (for discussion see Fenchel & Finlay (2004)). Darling et al. (2000) and Montresor et al. (2003) found that several species of subpolar planktonic foraminifera and dinoflagellates with bipolar distribution have identical rRNA gene sequences in the Arctic and Antarctic, although minor genetic differentiation was found in one nominal species (Darling et al. 2004). These findings do not, of course, exclude the possibility that the antipodal populations are presently isolated and have differentiated with respect to other genes, but they constrain the time of isolation to the Pleistocene.
7. An example: Cyclidium glaucoma
Cyclidium glaucoma OFM 1786 is a 25–30 μm long ciliated protozoon. It is morphologically well defined (figure 2) among about 50 described congeners (Kahl 1931), but future revisions may show that some are synonyms. A detailed morphological study is provided by Bardele (1983). The species is a filter-feeder that collects suspended bacteria by drawing water through a parallel array of long cilia that functions as a sieve. It is ubiquitous in fresh water, in the sea and in the hyperhaline lakes. Ten millilitres of water from any natural aquatic habitat will frequently yield the species when the sample is enriched with suspended bacteria. We (Finlay et al. in press) have collected isolates worldwide. These are morphologically identical in all details, although cell size and the number of kineties change somewhat for given clones when grown at different salinities (kineties are longitudinal rows of cilia with associated fibrillar systems). Conjugation has been observed in at least some isolates (G. Esteban 2004, personal communication), but we have maintained clone cultures for many years, so it appears that the species can be maintained asexually for several thousand generations.
Figure 2.
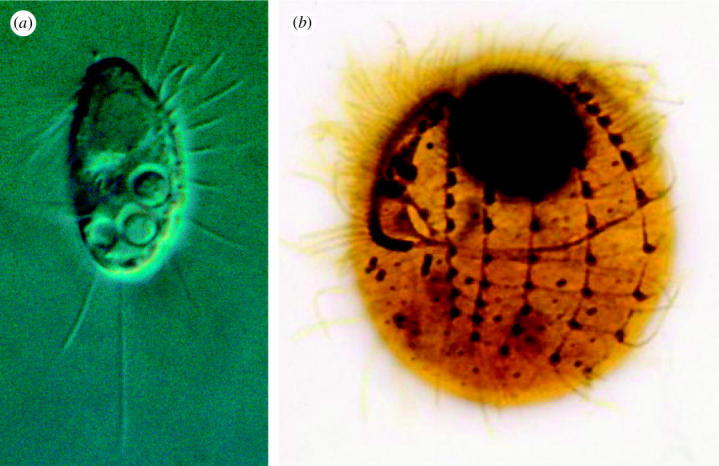
Cyclidium glaucoma. (a) Living cell. (b) Silver stained specimen showing the longitudinal kineties and the complex oral ciliature. The dark sphere at the top is the macronucleus. The living cell measures about 25 μm.
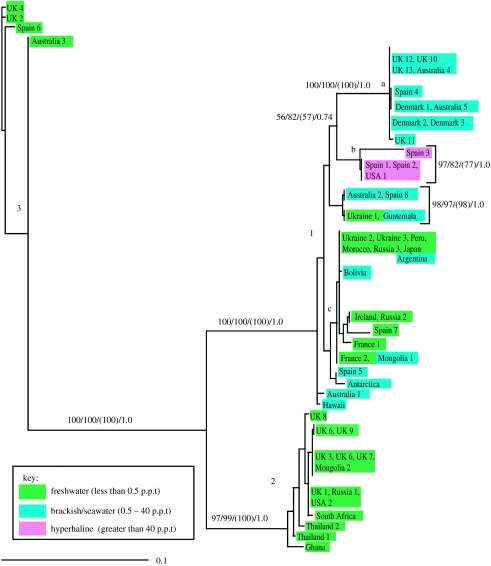
Figure 3 shows an unrooted phylogenetic tree of C. glaucoma based rRNA-gene sequences of 56 isolates, representing 37 distinct genotypes. The species appears to include three major lineages (1–3) and lineage 1 seems to include 3 subgroups (a–c). There is no obvious geographical pattern: identical genotypes have been collected in, for example, Australia and Northern Europe, and in Argentina, Japan, Morocco and Russia. The number of extant genotypes is unknown and probably very large. Owing to the many distinct genotypes, a more limited sampling effort could lead to a false impression of endemism. There is some correlation between salinity preference and genotype, although this is not very clear cut. Thus, all members of group 1a so far found have been isolated in seawater or brackish water and all three isolates from saline lakes or salt pans belong to group 1b. On the other hand, group 1c includes marine as well as freshwater isolates. Groups 2 and 3 are represented only by freshwater isolates. However, our data are biased in favour of freshwater samples, so this picture may not hold.
Figure 3.
Unrooted maximum-likelihood tree of nuclear ribosomal sequences of Cyclidium glaucoma (partial SSU rDNA and partial LSU rDNA). Scale bar measures substitutions per site. Support values from left to right: maximum-likelihood bootstrap/neighbour-joining bootstrap/maximum parsimony with reduced taxon sampling/posterior probabilities (data from Finlay et al. (in press)).
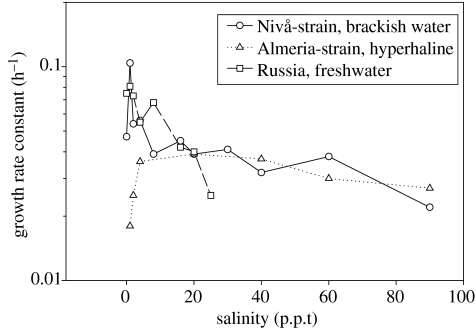
Examples of the growth response of different isolates to different salinities are shown in figure 4. The freshwater strain grew best in fresh water, and did not grow above 25 parts per thousand (p.p.t). Three other tested fresh water strains (not shown in the graph) stopped growing at salinities ranging between less than 15 and 30 p.p.t. A strain isolated in brackish water could grow at all salinities from freshwater to 90 ppt and, surprisingly, performed better at a somewhat lower salinity than the water from which it was isolated (12–20 p.p.t). An isolate from a salt pan in Spain (with a salinity of 80 p.p.t) grew at all salinities, but its performance was significantly poorer at salinities below 5 p.p.t.
Figure 4.
Growth responses of different isolates of Cyclidium glaucoma to different salinities.
It is probably a common characteristic for the isolates to be unusually euryhaline. However, it is also likely that some degree of specialization to life in either fresh- or seawater has probably evolved independently on more than one occasion. Osmoregulation depends on a number of mechanisms, including active water secretion, active ion transport through the cell membrane and regulation of the pool of intracellular dissolved amino acids. The genetic basis for these functions is not known, but it stands to reason that changes in the capacity of these mechanisms may evolve faster than substitutions in rRNA genes on which the phylogenetic tree is based. Although not tested, it is possible that strains from different latitudes show a differential temperature range. But again, since tropical isolates occur within all major groups, any correlation with the position in the phylogenetic tree appears unlikely. Other specializations among these genotypes can also be imagined, such as differential utilization of different bacterial food organisms, but this has not been tested. The characteristic swimming behaviour of the species is similar for all strains.
It is not possible to calibrate the molecular clock for ciliates with any precision, but the maximum genetic distance of the C. glaucoma phylogenetic tree is 0.27 substitutions per site. This suggests that the C. glaucoma phenotype has remained unaltered over a very long period of geological time.
It cannot be excluded that sequencing rRNA genes of congeners would reveal that some are embedded in the C. glaucoma phylogenetic tree. Sequencing of rRNA genes of an isolate of Cyclidium citrullus Cohn 1865 indicates that it may have evolved from lineage 1 in the C. glaucoma tree (Finlay et al. in press). Cyclidium citrullus can be identified on the basis of morphological details, and it also has a different swimming behaviour (T. Fenchel 2003, unpublished observations).
8. Conclusion
Our C. glaucoma data show that phenotypic criteria are appropriate for defining species. The ciliate was originally described 220 years ago, it is morphologically well defined relatively to other Cyclidium species and different isolates of the species cannot be distinguished microscopically in spite of large genetic distances. Its ecology and physiological properties such as food requirements, feeding mechanisms and growth rates have been described, and so far there is no reason to question the hypothesis that it is monophyletic. Our data show that the species includes genetically based differences with respect to salinity tolerance, but tolerance ranges are broadly overlapping. There is a correlation with rRNA genotype, but figure 3 also shows several examples of genotypes that occur in both freshwater and brackish water.
The name of a species embodies a wealth of biological information, whereas rRNA sequences do not, per se, provide any such information. We suggest that the use of a classical taxonomic approach (describing species on the basis of phenotypic properties) is not only applicable to eukaryotic micro-organisms, but also the only useful approach. Natural selection and the dynamics of ecological systems tend to create discrete groups of organisms with discrete sets of phenotypic traits that we refer to as species. Why this is so, and what determines local and global numbers of such species are central and relevant questions in ecology and evolutionary biology.
We have emphasized the utility of the ‘phenotype’ as a means of gaining better understanding of microbial diversity in the natural world. The reason for this is that it is usually the phenotype (and especially the morphotype) that enables a species to be characterized and identified. The phenotype also reveals the roles played by a species in the natural environment—what it feeds on, what feeds on it, which species interact with it and what are its preferred habitat requirements. With knowledge of the phenotype and the naming of species, we can organize biological information—especially the functional and other phenotypic properties of organisms—in a meaningful fashion. This practice has been in existence for at least 200 years, but the arrival of molecular markers such as rDNA and mitochondrial cytochrome oxidase subunit I have the potential to sow confusion with the potential discovery of an almost infinite variety of genotypes drawn from the vast pool of largely selectively neutral mutations that have accumulated over historical time.
In the case of rDNA, it is now apparent that there is no close correlation between intraspecific genetic (rDNA) distance and phenotypic differentiation. In the case of DNA ‘barcoding’ using the 650-base sequence of a single gene—mitochondrial cytochrome oxidase subunit I as a marker for identifying biological specimens—it has been claimed that the tool will ‘revolutionize’ species identification and enable enumeration of global species richness. But DNA barcoding generates nothing more than data. It does not provide anything that could be called ‘knowledge’ (Ebach & Holdrege 2005a,b). What, for example, does it tell us about the physiology, or ecological function of an organism? On this, the barcode is silent. How will barcoding replace traditional taxonomy? What will it add to the global knowledge of organisms collected over hundreds of years? Can we seriously relegate species and their histories to barcodes? Barcoding generates a sequence of bases—but that is all it generates. Real knowledge is acquired only after describing and understanding the phenotype.
Barcoding can be used as a tool for sorting new collections based on barcode sequences, and it may work well for species that are already much studied (Check 2005). But in groups that are less well studied, misidentifications are rather common. Barcoding technology is developing fast, but it may struggle to distinguish ‘species’ that are very similar. One possible advantage of barcoding is that it may, in time, help to resolve species clusters. The barcodes of 480 Costa Rican butterflies previously grouped as one species were shown to condense into sequence clusters suggesting 10 distinct species (Check 2005). This has implications for the likely dimensions of global species richness, and it brings us back full circle to the fundamental problem of the dichotomy separating molecular and morphological definitions of ‘species’. Some barcodes come from known species, and others are new to science, but the crucial point is that the buck stops with the expert taxonomists—the only people who have the knowledge to resolve the link between barcode clusters and species.
There is also a presumption that DNA sequence information will make a huge difference to our knowledge and understanding of the natural world. In ‘barcode world’, taxonomists of the future might conduct large biotic surveys without the need to learn or use keys based on morphology. The real work of course will still fall to the traditional taxonomists—the people who provide useful ‘knowledge’ rather than snippets of (possibly unique) nucleotide sequence. In Wheeler's (2004) words, ‘it is time to reinvest in traditional sources of biodiversity exploration including morphology’ (p. 574).
Acknowledgments
This work was supported by the Danish Natural Science Research Council and the Natural Environment Research Council (UK). We are grateful to Susan Brown, Kerstin Hoef-Emden and Genoveva F. Esteban for their assistance in preparing this paper.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Species and speciation in micro-organisms’.
References
- Anderson O.R. Springer; New York, NY: 1983. Radiolaria. [Google Scholar]
- Bardele C.D. Mapping of highly ordered membrane domains in the plasma membrane of the ciliate Cycidium glaucoma. J. Cell. Sci. 1983;6:1–30. doi: 10.1242/jcs.61.1.1. [DOI] [PubMed] [Google Scholar]
- Check E. Cowrie study strikes a blow for traditional taxonomy. Nature. 2005;438:722–723. doi: 10.1038/438722b. doi:10.1038/438722b [DOI] [PubMed] [Google Scholar]
- Darling K.F, Wade C.M, Stewary I.A, Kroon D, Dingle R, Brown A.J.L. Molecular evidence for genetic mixing of Arctic and Antarctic subpolar populations of planktonic foraminifers. Nature. 2000;405:43–47. doi: 10.1038/35011002. doi:10.1038/35011002 [DOI] [PubMed] [Google Scholar]
- Darling K.F, Kucera M, Pudsey C.J, Wade C.M. Molecular evidence links cryptic diversification in polar protists to Quaternary climate dynamics. Proc. Natl Acad. Sci. USA. 2004;101:7657–7662. doi: 10.1073/pnas.0402401101. doi:10.1073/pnas.0402401101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson S.C, Pace N.R. Novel kingdom-level eukaryotic diversity in anoxic environments. Proc. Natl Acad. Sci. USA. 2002;99:7657–7662. doi: 10.1073/pnas.062169599. doi:10.1073/pnas.062169599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragesco J. A propos de Neobursaridium gigas Balech 1941: Sténothermnie, inclusions, ultrastructure des trichocystes. Protistologica. 1968;4:157–167. [Google Scholar]
- Ebach M.C, Holdrege C. More taxonomy, not DNA barcoding. BioScience. 2005a;55:823–824. doi:10.1641/0006-3568(2005)055[0823:MTNDB]2.0.CO;2 [Google Scholar]
- Ebach M.C, Holdrege C. DNA barcoding is no substitute for taxonomy. Nature. 2005b;434:697. doi: 10.1038/434697b. doi:10.1038/434697b [DOI] [PubMed] [Google Scholar]
- Edgcomb V.P, Kysela D.T, Teske A, Gomez A, de V, Sogin M.L. Benthic eukaryotic diversity in the Guaymas Basin hydrothermal vent environment. Proc. Natl Acad. Sci. USA. 2002;99:7658–7662. doi: 10.1073/pnas.062186399. doi:10.1073/pnas.062186399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T. Cosmopolitan microbes and their “cryptic” species. Aquat. Microb. Ecol. 2005;41:49–54. [Google Scholar]
- Fenchel T, Finlay B.J. The ubiquity of small species: patterns of local and global diversity. BioScience. 2004;54:777–784. doi:10.1641/0006-3568(2004)054[0777:TUOSSP]2.0.CO;2 [Google Scholar]
- Finlay B.J, Fenchel T.F. Cosmopolitan metapopulations of free-living microbial eukaryotes. Protist. 2004;155:237–244. doi: 10.1078/143446104774199619. doi:10.1078/143446104774199619 [DOI] [PubMed] [Google Scholar]
- Finlay B.J, Corliss J.O, Esteban G, Fenchel T. Biodiversity at the microbial level: the number of free-living ciliates in the biosphere. Q. Rev. Biol. 1996;71:221–237. doi:10.1086/419370 [Google Scholar]
- Finlay, B. J., Esteban, G. F., Brown, S., Fenchel, T. & Hoef-Embden, K. In press. Multiple cosmopolitan ecotypes within a microbial eukaryotic morphospecies. Protist [DOI] [PubMed]
- Foissner W. Protist diversity: estimates of the near imponderable. Protist. 1999;150:363–368. doi: 10.1016/S1434-4610(99)70037-4. [DOI] [PubMed] [Google Scholar]
- Hutchinson G.E. When are species necessary? In: Lewontin R.C, editor. Population, biology and evolution. Syracuse University Press; Syracuse, NY: 1968. pp. 177–186. [Google Scholar]
- Kahl A. Verlag von Gustav Fischer; Jena, Germany: 1931. Urtiere oder Protozoa I: Wimpertiere oder Ciliata (Infusoria). 2. Holotricha. [Google Scholar]
- Kimura M. Cambridge University Press; Cambridge, UK: 1983. The neutral theory of molecular evolution. [Google Scholar]
- Koch T.A, Ekelund F. Strains of the heterotrophic flagellate Bodo designis from different environments vary considerably with respect to salinity preference and SSU rRNA gene composition. Protist. 2005;156:97–112. doi: 10.1016/j.protis.2004.12.001. doi:10.1016/j.protis.2004.12.001 [DOI] [PubMed] [Google Scholar]
- Lee C.C, Fenchel T. Studies on ciliates associated with sea ice from Antarctica. II. Temperature responses and tolerances in ciliates from Antaractic, temperate and tropical zones. Arch. Protistenk. 1972;114:237–244. [Google Scholar]
- Lewontin R.C. Columbia University Press; New York, NY: 1974. The genetic basis of evolutionary change. [Google Scholar]
- López-Garcia P, Rodriquez-Valera F, PedrósAlió C, Moreira D. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature. 2001;409:603–607. doi: 10.1038/35054537. [DOI] [PubMed] [Google Scholar]
- Lowe C.D, Day A, Kemp S.J, Montagnes D.J.S. There are high levels of functional and genetic diversity in Oxyrrhis marina. J. Eukaryot. Microbiol. 2005;52:250–257. doi: 10.1111/j.1550-7408.2005.00034.x. [DOI] [PubMed] [Google Scholar]
- Mallet J. Species, concepts of. In: Levin S, editor. Encyclopedia of biodiversity. Academic Press; San Diego, CA: 2001. pp. 427–440. [Google Scholar]
- May R.M. How many species are there on Earth? Science. 1988;241:1441–1449. doi: 10.1126/science.241.4872.1441. [DOI] [PubMed] [Google Scholar]
- Mayr E. Harvard University Press; Cambridge, MA: 1970. Populatons, species and evolution. [Google Scholar]
- Montresor M, Lovejoy C, Orsini L, Procaccini G, Roy S. Bipolar distribution of the cyst-forming dinoflagellate Polarella glacialis. Polar Biol. 2003;26:186–194. [Google Scholar]
- Moon-van der Stay S.Y, De Wachter R, Vaulot D. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature. 2001;409:607–610. doi: 10.1038/35054541. doi:10.1038/35054541 [DOI] [PubMed] [Google Scholar]
- Nanney D.L. Genes and phenes in Tetrahymena. BioScience. 1982;32:783–788. doi:10.2307/1308971 [Google Scholar]
- Nanney, D. L. 2004 Tetrahymena biogeography. www.life.uiuc.edu/nanney/biogeography.html
- Nanney D.L, Park C, Preparata R.M, Simon E. Comparison of sequence differences in a variable 23S rRNA domain among sets of cryptic species of ciliated protozoa. J. Eukaryot. Microbiol. 1998;51:402–416. doi: 10.1111/j.1550-7408.1998.tb05075.x. [DOI] [PubMed] [Google Scholar]
- Pawlowski J, Holzmann M, Berney C, Fahrni J, Gooday A.J, Cedhagen T, Habura A, Bowser S.S. The evolution of early Foraminifera. Proc. Natl Acad. Sci. USA. 2003;100:11494–11498. doi: 10.1073/pnas.2035132100. doi:10.1073/pnas.2035132100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson D.J. The diversity of eukaryotes. Am. Nat. 1999;154:S96–S124. doi: 10.1086/303287. doi:10.1086/303287 [DOI] [PubMed] [Google Scholar]
- Schönborn W, Dörfelt H, Foissner W, Krienitz L. A fossilized microcenosis in Triassic amber. J. Eukaryot. Microbiol. 1999;46:571–584. [Google Scholar]
- Šlapeta J, López-García P, Moreira D. Global dispersal and ancient cryptic speciation in the smallest marine eukaryotes. Mol. Biol. Evol. 2006;23:23–29. doi: 10.1093/molbev/msj001. doi:10.1093/molbev/msj001 [DOI] [PubMed] [Google Scholar]
- Tyler P.A. Endemism in freshwater algae. Hydrobiologia. 1996;226:127–135. [Google Scholar]
- Von der Heyden S, Chao E.E, Vickerman K, Cavalier-Smith T. Ribosomal RNA phylogeny of bodonid and diplonemid flagellates and the evolution of euglenozoa. J. Eukaryot. Microbiol. 2004;51:402–416. doi: 10.1111/j.1550-7408.2004.tb00387.x. doi:10.1111/j.1550-7408.2004.tb00387.x [DOI] [PubMed] [Google Scholar]
- Wheeler Q.D. Taxonomic triage and the poverty of phylogeny. Phil. Trans. R. Soc. B. 2004;359:571–583. doi: 10.1098/rstb.2003.1452. doi:10.1098/rstb.2003.1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D.M. What is the upper size limit for cosmopolitan distribution in free-living microorganisms? J. Biogeogr. 2001;28:285–291. [Google Scholar]
- Wright A.-D.G, Lynn D.H. Maximum ages of ciliate lineages estimated using a small subunit rRNA molecular clock: crown eukaryotes date back to the Paleoproterozoic. Arch. Protistenk. 1997;148:329–341. [Google Scholar]