Abstract
We have investigated microbial mats of alkaline siliceous hot springs in Yellowstone National Park as natural model communities to learn how microbial populations group into species-like fundamental units. Here, we bring together empirical patterns of the distribution of molecular variation in predominant mat cyanobacterial populations, theory-based modelling of how to demarcate phylogenetic clusters that correspond to ecological species and the dynamic patterns of the physical and chemical microenvironments these populations inhabit and towards which they have evolved adaptations. We show that putative ecotypes predicted by the theory-based model correspond well with distribution patterns, suggesting populations with distinct ecologies, as expected of ecological species. Further, we show that increased molecular resolution enhances our ability to detect ecotypes in this way, though yet higher molecular resolution is probably needed to detect all ecotypes in this microbial community.
Keywords: species, ecotype, adaptation, cyanobacteria, hot spring
1. Introduction
Molecular analysis of many microbial communities has revealed evidence of closely related, yet ecologically distinct, genetic variants (Ward 2006). For instance, we have studied microbial populations inhabiting mat communities of alkaline siliceous hot springs (figure 1). In a 1998 review (Ward et al. 1998), we summarized work mainly conducted at Octopus Spring in Yellowstone National Park. 16S rRNA analysis revealed that natural populations of the two types of predominant mat phototrophs (unicellular cyanobacteria, Synechococcus spp.; filamentous green non-sulphur-like bacteria, Roseiflexus spp.) were distantly related to cyanobacteria (Synechococcus cf. lividus) and green non-sulphur bacteria (Chloroflexus aurantiacus) that can be readily cultivated from the mat. Each type of phototroph was represented by a set of 16S rRNA variants exhibiting less than 5% sequence divergence. The patterning of 16S rRNA sequences along the thermal gradient in the effluent channel suggested that these variants were representative of distinct temperature-adapted populations. Recently, Allewalt et al. (2006) succeeded in cultivating Synechococcus strains with 16S rRNA sequences matching predominant natural populations and in demonstrating temperature adaptations consistent with the in situ distribution of these genotypes. The 1998 review also summarizes our initial studies on the vertical distribution of 16S rRNA variants conducted at a 60°C site in Mushroom Spring, a nearby alkaline siliceous hot spring with very similar microbial populations (Ramsing et al. 2000). In that study, we observed that a subsurface Synechococcus population with distinctive pigmentation was coincident with one of the cyanobacterial 16S rRNA variants, suggesting the possibility that Synechococcus populations may also adapt to light and/or other parameters that vary in the vertical aspect of the mats.
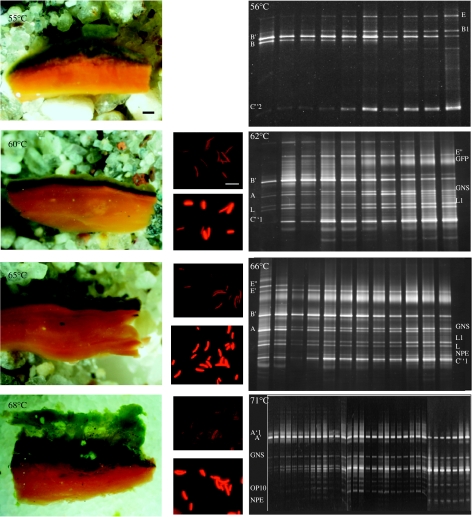
Figure 1.
Mushroom Spring showing cyanobacterial mat in foreground and spring in background. Inset shows DGGE analysis of PCR-amplified 16S rRNA segments for unsectioned samples collected at the four temperature sites. Band labels are discussed in text; letters and primes (e.g. A, B′ and NPE) indicate 16S rRNA genotypes and, except for genotype OP10, a number following these designations indicates the number of nucleotide differences from that genotype (e.g. A′1 and C″2 indicate sequences one and two nucleotides different from genotypes A′ and C″).
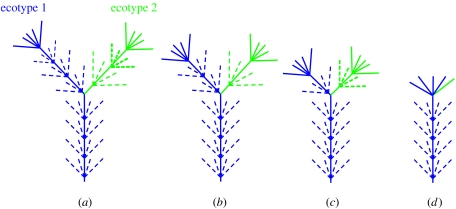
The existence of closely related, ecologically distinct variants suggested that these ‘ecotypes’ may have arisen via adaptive radiation in a manner similar to that of many plant and animal species (Ward et al. 2002). Further, if an ecological species concept was used, ecotypes could be considered species because they represent distinct evolutionary lineages and occupy unique niches (Ward 1998, 2006). The issue of whether ecotypes should be considered species may be debated, but irrespective of what we call these ecologically adapted populations, it seems clear that they are, as Mayr (1982) said of species, ‘the basic unit[s] of ecology’, the populations within communities that will uniquely occupy niches, respond as the environment changes and regulate community function. The empirical evolutionary and ecological patterns we have observed also match the predictions of evolutionary ecology theory (Ward & Cohan 2005). As shown in figure 2a, periodic selection theory predicts that an ecological population evolves through a series of periodic selection events. New populations with distinct ecologies arise in such a population through niche-invasion events. Each population then diverges owing to subsequent private periodic selection events and accumulation of random neutral mutations in each descendant lineage, leading to separate terminal phylogenetic clades; each of these clades groups together variants that occupy one of two distinct niches. This enables theory-based modelling approaches for ecotype identification through analysis of molecular sequence data to identify clades that correspond to ecotypes (Cohan 2002; Gevers et al. 2005; Cohan et al. in press, submitted). However, the ability to detect ecotypes is limited by the degree of their divergence from other closely related ecotypes (Palys et al. 1997; figure 2b–d). Ecotype populations that are younger (i.e. less and less diverged) will be harder to detect with sequence variation in molecules that evolve more slowly, such as 16S rRNA. In the microbial mats we study, for instance, Ferris et al. (2003) observed that a faster-evolving genetic locus, the intervening transcribed spacer (ITS) region separating the 16S and the 23S rRNA genes, had to be targeted in order to reveal the genetic distinction between Synechococcus populations with the same 16S rRNA sequence that were found at different depths. This raised the general question of how much genetic resolution is needed to detect all the ecotypes within a community.
Figure 2.
Schematics showing the evolution of an ecotype (blue) through a series of periodic selection events (dashed fans exhibiting extinct variants) and the divergence of a new distinct ecotype (green) through a niche-invasion mutation and subsequent private periodic selection events, to form two distinct clades of extant variants (solid fans at ends of lineages). a–d depict progressively ‘younger’ ecotypes with progressively smaller degrees of divergence, with d depicting a nascent ecotype in a population whose diversity has yet to be purged by periodic selection.
Here, we map the patterning of molecular variants in the Mushroom Spring microbial mat in two dimensions. The first follows the flow of effluent water above the mat away from the source pool and includes gradients of decreasing temperature and any chemical change that may occur with distance from the source. The second follows the vertical dimension in the microbial mats, which includes steep and fluctuating gradients of light and chemical variables, as measured by microsensor analysis. We used both 16S rRNA and ITS region sequences to provide two levels of molecular resolution. In addition, we conducted a theory-based analysis of ecotypes as modelled from the sequence data. Our results confirm that ecotypes predicted by the theory-based model exhibit unique distributions in the mat. They also point to the need for yet higher resolution analysis to resolve less divergent ecotypes.
2. Material and methods
(a) Study sites
Our study was focused primarily on four temperature-defined sites along the effluent channel of a microbial mat found in Mushroom Spring, an alkaline siliceous hot spring in the Lower Geyser Basin, Yellowstone National Park, Wyoming, USA (Ward et al. 1998; Ramsing et al. 2000). These sites exhibited temperature ranges of 68–71, 65–66, 60–62 and 55–56°C over several collections made on different dates. However, we provide the actual temperatures measured in connection with different sets of data reported below. In order to provide additional information on Synechococcus A/B type ITS variation, we also used samples collected at nearby Octopus Spring.
(b) Sample collection and analysis
Samples for denaturing gradient gel electrophoresis (DGGE) analysis of PCR-amplified bacterial 16S rRNA genes (Escherichia coli positions 1070–1392) were collected on 22 August 1999 using a no. 4 cork borer. The top layer of the mat was sliced from core samples using a clean razor blade as previously described (Ramsing et al. 2000). DGGE patterns were generated for microbial populations using the entire vertical interval of mat core slice samples, which included the green layers and 0.5–1.0 mm of the orange underlayer. The core slice samples varied from 1 to 3.5 mm thickness depending on the temperature-defined site from which the samples were collected. DGGE patterns were also generated for 100 μm thick vertical intervals subsampled parallel to the mat surface using a cryotome as previously described (Ramsing et al. 2000). Purified DGGE bands and clones were sequenced and sequences analysed as previously described (Ferris et al. 2003).
The samples for microscopy and clone library construction were collected from similar temperature-defined sites (68, 65 and 60°C) on 17 July 2001. Subsamples from near the mat surface and from a distinct dark-green subsurface layer were obtained using a dissecting microscope and a sterile no. 27 gauge syringe needle. These clone libraries were given the following designations: 6##, 68°C top layer; 7##, 68°C bottom layer; T##, 65°C top layer; B##, 65°C bottom layer; 10##, 60°C top layer; and 11##, 60°C bottom layer, where ## indicates additional clone name designators after the prefix shown. Details of microscopic methods and methods for DNA extraction, PCR amplification of the 16S rRNA and ITS regions (primers 781f (A and B) and 23cyR, which amplified approximately 750 bp of the 3′ end of the 16S rRNA gene along with the adjoining ITS region), cloning, sequencing and sequence analysis have been previously described (Ferris et al. 2003). Additional whole mat samples were collected from sites averaging 60 and 65°C in Mushroom Spring on 2 October 2003 and Octopus Spring (where flow surges occurring at approximately 5 min intervals caused temperatures to vary from 53.5 to 63.5°C and 58 to 67°C) on 5 November 2004 and used to prepare 16S rRNA/ITS clone libraries as described earlier, with the exception that no separation of layers was done other than to restrict analysis to the top 2–3 mm. These clone libraries were given the following designations: M60, Mushroom Spring 60°C; M65, Mushroom Spring 65°C; OH, Octopus Spring high temperature; and OL, Octopus Spring low temperature.
(c) Nucleotide sequence accession numbers
The sequences have been submitted to GenBank with accession numbers DQ979027–DQ979319, and AF550092–AF550127.
(d) Community phylogeny analysis
The 16S rRNA/ITS clone variants from this study were analysed using the ‘community phylogeny’ method described in detail by Cohan (2006) and Cohan et al. (in press, submitted). This method assumes the periodic selection model of microbial evolution. Briefly, a Monte Carlo simulation seeks to find the best trio of values for parameters that control the evolution of a phylogeny, as expressed by the relationship between the number of sequence clusters in a phylogeny and the sequence relatedness used to define clusters. The three parameters are the net rate of ecotype formation (Ω), the rate of periodic selection per ecotype per unit time (σ) and the number of ecotypes (n). Individual ecotypes were demarcated by determining the number of ecotypes that yields the greatest likelihood of a fit to the observed clade sequence diversity, then finding the largest clades that were consistent with a single ecotype (i.e. the confidence interval for n included 1). In this process, we checked to see if a given group could be expanded to include even larger clades.
(e) Microsensor analysis
Measurements of O2 distribution and oxygenic photosynthesis in the microbial mat were done in situ on 22 August 1999 with O2 microsensors (Revsbech 1989). The microsensors were connected to a picoampere meter (PA2000; Unisense A/S, Aarhus, Denmark), and the measuring signals were recorded on a strip chart recorder (Servogor 110); both instruments were battery driven. The microsensors were linearly calibrated from sensor readings in the anoxic part of the mat and in the overlying spring water, the O2 content of which was determined by Winkler titration measurements with 50 ml Winkler bottles. Measurements were done under a cloudless sky with downwelling scalar irradiance ranging from 1800 to 2300 μmol photons m−2 s−1, as determined with a quantum scalar irradiance meter (Biospherical Instruments QSL-101) placed in the spring next to the sampling site. The microsensors were positioned in vertical steps of 100–200 μm by a manually operated micromanipulator (MM33; Märtzhaüser GmbH, Wetzlar, Germany), taking care that no self-shadowing affected the measurements. Light–dark shifts to measure photosynthetic rates were done by temporarily shading the measuring area with a non-transparent box (Revsbech & Ward 1984).
Measurements of spectral light penetration in the mat were done with fibre-optic scalar irradiance microprobes (Kühl et al. 1997; Kühl 2005) on freshly collected samples in the laboratory. Undisturbed mat samples were taken from the field site in small glass-coring devices and transported to the laboratory within 1 h for further analysis on the same day. The samples were incubated in spring water and illuminated vertically from above with a fibre-optic halogen lamp (KL-1500; Schott GmbH, Mainz, Germany) that had the heat filter removed to ensure sufficient intensity in the near infrared (NIR) spectral range greater than 700 nm. Visible downwelling irradiance (400–700 nm) was approximately 1000 μmol photons m−2 s−1. The scalar irradiance microprobe was inserted in steps of 100–200 μm vertical distance from above into the mat at a zenith angle of 145° using a manually operated micromanipulator. Reference spectra of incident downwelling irradiance were recorded with the microprobe tip positioned over a black-light trap at a distance from the light source identical to that of the mat surface.
3. Results
(a) Distribution of Synechococcus 16S rRNA genotypes along the effluent channel
DGGE band patterns were generated using the entire vertical interval of mat core slice samples from the four temperature sites (figure 1 inset). A distinct distribution of Synechococcus genotypes was found, with genotypes B and B′ co-occurring at 56°C, B′ and A co-occurring at 62°C (band intensity for B′ greater than that for A) and 66°C (band intensity for A greater than that for B′), and only the A′ genotype occurring at 71°C. This pattern was previously observed for the distribution of Synechococcus genotypes in the Octopus Spring effluent channel (Ferris & Ward 1997).
(b) Vertical distribution of Synechococcus cells and genotypes
Samples from each temperature-defined site exhibited similar colour patterns with depth (figure 3, column 1). The uppermost layer was yellow-green, with an underlying dark-green coloured band, followed by orange laminations beneath. The thickness and consistency of these layers differed among sites, with a diffuse upper yellow-green layer at 55°C, thinner (approx. 1 mm) green layers at 60 and 65°C and much thicker green layers at 68°C.
Figure 3.
Detailed views of vertical profiles within samples of each temperature site. (Column 1, left-most) cross-section views of mat layers (bar is 1 mm); (Column 2) photomicrographs showing autofluorescence of native Synechococcus populations sampled from the upper yellow-green and deep dark-green layers (bar is 10 μm); (Column 3) DGGE profiles of samples collected at successive 100 μm intervals progressing from top (left-most lane) downwards (to right). Band labels are discussed in text and described in the legend of figure 1.
Synechococcus cells collected from the mat surface showed less intense per-cell autofluorescence than did those sampled from the dark-green subsurface band (figure 3, column 2), suggesting that Synechococcus populations in these two layers had distinct pigment content and were either acclimated or adapted to low light intensity. These alternatives were tested using molecular analysis, reasoning that one differently acclimated population would belong to a single genetic group, while populations with distinct adaptations should also be genetically distinct (like the two ecotypes in figure 2a–c). We initially used DGGE analysis of PCR-amplified 16S rRNA genes within 100 μm thick horizontal slices through the vertical aspect (figure 3, column 3). We are completely aware that PCR may be biased and that DGGE band intensity may not truly reflect population abundance. However, we consider these analyses to be at least semi-quantitative on the basis of similar results from multiple forms of analysis (see Ward et al. 1998 and below). At 56°C, genotype B was detected in the top 600 μm, while genotype B′ was detected throughout the vertical profile to a depth of 1000 μm. It is interesting that a B-like genotype (band B1, figure 3, column 3), which has never been observed before in our studies of these mats, was detected. The DGGE band intensities for this genotype appeared to increase with depth in the mat. At 62°C, the B′ population showed a similar distribution throughout the vertical profile, with the band intensity declining with depth; genotype A band intensity increased with depth, maximizing 600–900 μm below the mat surface, as had been previously observed (Ramsing et al. 2000). These patterns may be an indication of differences in light adaptation of the populations contributing B, B′ and A genotypes. At 66°C, genotypes B′ and A were present throughout the vertical interval with no apparent pattern with depth. Unlike the unsectioned sample (figure 1 inset), the B′ bands were more intense than the A bands, which may have been owing to differences in primer bias on subsamples compared with whole samples. At 71°C, the A′-like genotype also showed no apparent depth stratification.
Since 16S rRNA analysis did not distinguish between the alternatives of acclimation and adaptation at 66 and 68°C sites, and because mat topography might have mixed layers from different depths upon cryotome sectioning, we separately cloned and sequenced a portion of the 16S rRNA gene and the adjacent faster-evolving ITS sequence region from the uppermost yellow-green and deeper dark-green layers from several sites. The 60° site was included as a case where 16S rRNA sequence variation may suffice to detect different ecotypes in the vertical aspect of the mat (i.e. subsurface A population). The 56°C site was not studied, as the yellow-green and deep-green layers were more diffuse and difficult to sample separately. Except as noted earlier, the frequencies of ITS clones that could be assigned to different 16S rRNA-defined genotypes (table 1) reflected the patterns of band intensities noted in DGGE analysis (figure 1 inset and figure 3, column 3), as follows:
mainly B′ and A genotypes at 60°C; B′ genotypes more abundant in surface layer and A genotypes predominantly from the dark-green layer,
B′ and A genotypes at 65°C; A genotypes more frequent and equally distributed with depth; B′ genotypes only from the surface sample and
only A′ genotypes at 68°C; equally distributed with depth.
Table 1.
Distribution of 16S rRNA/ITS clones from libraries constructed from different temperatures and depth intervals.
| sample | number of 16S rRNA/ITS clones of 16S rRNA genotype | ||
|---|---|---|---|
| B′ | A | A′ | |
| 60°C surface | 13 | 4 | 2 |
| 60°C deep-green | 8 | 20 | 0 |
| 65°C surface | 9 | 9 | 0 |
| 65°C deep-green | 0 | 10 | 0 |
| 68°C surface | 0 | 0 | 19 |
| 68°C deep-green | 0 | 0 | 18 |
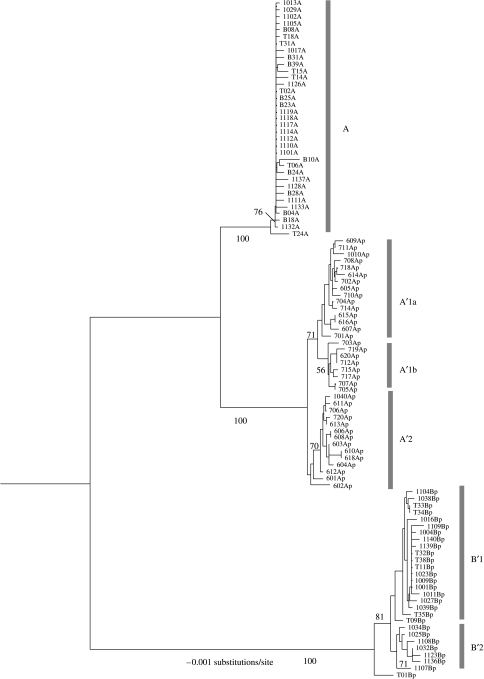
Phylogenetic analysis of ITS variants (figure 4; table 2) revealed three obvious clades that correspond to 16S rRNA genotypes B′, A and A′. The A clade exhibited little variation though one variant did branch outside a clade containing the rest, supported by a bootstrap value of 76%. The A′ clade contains variants almost exclusively recovered from the 68°C sample. There are two A′ subclades supported by bootstrap values greater than or equal to 70% (A′1 and A′2); subclade A′1 might be divided into two subclades (A′1a and A′1b), but the bootstrap support for this is lower (56%). Subclade A′1a showed a nearly equivalent number of variants obtained from surface and subsurface samples, while subclades A′1b and A′2 contained more variants from either the bottom or the top mat layers, respectively, as reported by Ferris et al. (2003). The B′ clade exhibited two possible subclades, one with more variants from the top layer (B′1) and the other with approximately equal numbers of variants from top and bottom layers (B′2). These evolutionary and ecological patterns suggest that at the 68°C site, there are at least two genetically distinct A′ Synechococcus populations, possibly adapted to different vertically positioned niches with different light regimes. At the 60 and 65°C sites, there may be a distinct B′ ecotype associated with top mat layers.
Figure 4.
Neighbour-joining phylogenetic tree based on ITS sequence variation. Clades and subclades corresponding A, A′ and B′ Synechococcus lineages defined by 16S rRNA variation are indicated. These clades were mostly demarcated based on bootstrap values greater than or equal to 70%. The tree was rooted with the ITS sequence of Synechococcus strain C9. Scale bar indicates substitutions per site.
Table 2.
Temperature and vertical distribution of variants in clades with good bootstrap support.
| clade | 68°C | 65°C | 60°C | |||
|---|---|---|---|---|---|---|
| top | bottom | top | bottom | top | bottom | |
| A | 7 | 10 | 3 | 15 | ||
| A′1a | 6 | 8 | 1 | |||
| A′1b | 1 | 7 | ||||
| A′2 | 11 | 2 | 1 | |||
| B′1 | 6 | 9 | 5 | |||
| B′2 | 3 | 4 | ||||
(c) Community phylogeny analysis of Synechococcus ecotypes
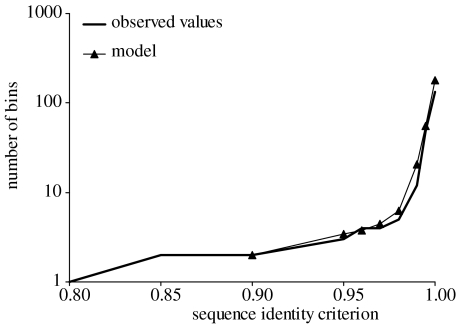
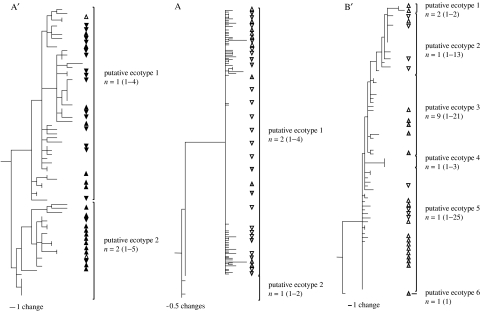
The results of community phylogeny analysis are presented in figures 5 and 6, and table 3. The fit of the model to observed values of the number of sequence clusters versus the sequence identity criterion was excellent (figure 5). Ten putative ecotypes were identified in the Synechococcus A/B clade phylogeny, with each of the major clades, A′, A and B′, divided into more than one putative ecotype (figure 6). The A′, A and B′ clades exhibit biased distribution towards 68, 65 and 60°C, respectively (χ42=150.78, p<0.0001), which is consistent with the notion that these major clades correspond to ecologically specialized groups of populations with different temperature adaptations. Two putative ecotypes were identified within the A′ clade, and these were associated with different depths in the 68°C mat, with the A′1 clade members in higher abundance in the lower depth and the A′2 clade members in higher abundance at the mat surface (χ12=9.11, p<0.0025), as reported by Cohan et al. (submitted). The analysis did not separate the A′1a and A′1b subclades described earlier (figure 4), though in this and other cases more than a single putative ecotype is within the 95% confidence range, suggesting that more data might resolve such clades. Community phylogeny predicted two putative ecotypes in the A clade, one of which (A2) is defined by variants that were observed only at 65°C (χ12=9.44, p<0.0025). Putative ecotype A2 was found only in Mushroom Spring in October 2003, suggesting possible temporal variation in ecotypes. Community phylogeny demarcated six ecotypes within the B′ clade. The B′ putative ecotypes are heterogeneous in their association with temperature and with surface and subsurface layers, with putative ecotype B′5 being associated most strongly with the surface layer (χ42=9.71, p<0.05).
Figure 5.
Community phylogeny analysis of ITS variants from 2001 to 2004 Mushroom Spring and Octopus Spring samples showing comparison of observed and modelled distributions of the number of sequence clusters (bins) versus sequence identity criterion.
Figure 6.
Parsimony phylogenetic trees showing clades demarcated as ecotypes by community phylogeny analysis of ITS variants of the Synechococcus A, A′ and B′ clades. n is the most probable number of ecotypes and values in parentheses indicate the lower and upper 95% confidence interval bounds of ecotype number, respectively. Clones originating from the upper and lower parts of the photic zone are indicated by upward- and downward-pointing triangles, respectively. Shading indicates temperature: white, 60°C; grey, 65°C; and black, 68°C.
Table 3.
Temperature and vertical distribution of variants in putative ecotypes. (Upper and lower samples are from Mushroom Spring, 17 July 2001. Total is the sum of those variants and additional variants detected in unsectioned top 2 mm samples from Mushroom Spring, 2 October 2003, and Octopus Spring, 5 November 2004.)
| putative ecotype | 68°C | 65°C | 60°C | all temperatures | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| upper photic zone | lower photic zone | entire photic zone | upper photic zone | lower photic zone | entire photic zone | upper photic zone | lower photic zone | entire photic zone | total upper photic zone | total lower photic zone | |
| A′1 | 7 | 15 | 22 | 0 | 0 | 20 | 1 | 0 | 5 | 8 | 15 |
| A′2 | 11 | 2 | 13 | 0 | 0 | 7 | 1 | 0 | 3 | 12 | 2 |
| A1 | 0 | 0 | 0 | 7 | 10 | 91 | 3 | 15 | 65 | 10 | 25 |
| A2 | 0 | 0 | 0 | 0 | 0 | 14 | 0 | 0 | 0 | 0 | 0 |
| B′1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 2 | 0 |
| B′2 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 13 | 1 | 4 |
| B′3 | 0 | 0 | 0 | 4 | 0 | 4 | 1 | 0 | 17 | 5 | 0 |
| B′4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | ||
| B′5 | 0 | 0 | 0 | 3 | 0 | 3 | 8 | 4 | 22 | 11 | 4 |
| B′6 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | |
(d) Vertical profiles of physical/chemical parameters and oxygenic photosynthesis
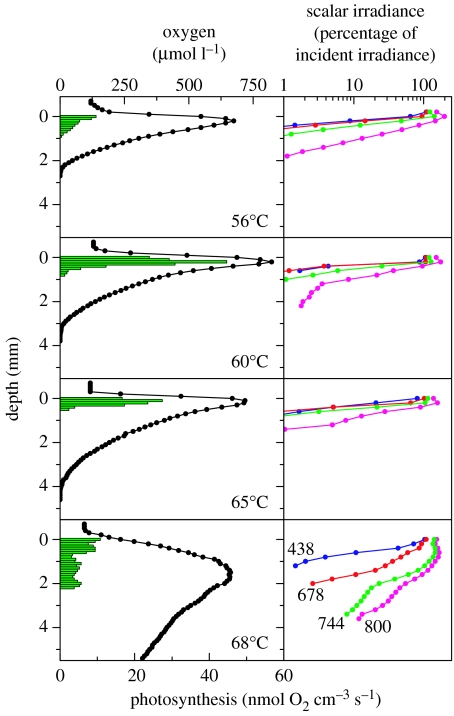
Steep gradients of oxygen concentration were found in the mats under high solar irradiance (figure 7), showing a supersaturation of oxygen in the mats owing to intense cyanobacterial photosynthesis at all temperature sites. While the 68°C mat was completely oxygenated, oxygen penetration decreased from about 4 mm at 65°C to 2.5 mm at 56°C, pointing to a higher potential for bacterial oxygen consumption with decreasing temperature. The photic zone for oxygenic photosynthesis was limited by the strong absorption of visible light in the upper 1–2 mm of the mat, while NIR light penetrated more efficiently to deeper layers. The 68°C mat had a more loose and translucent structure, which caused less light attenuation and the thickest photic zone. With decreasing temperature, the mat structure became more compact and the cyanobacterial layers were thinner and more densely pigmented. Especially at 65 and 60°C, this causes a very high specific activity of gross photosynthesis.
Figure 7.
Microprofiles of oxygen, oxygenic photosynthesis and light at photosynthetically useful wavelengths (numbers on curves indicate nanometres) measured with microsensors through the vertical aspect of mats at each temperature site.
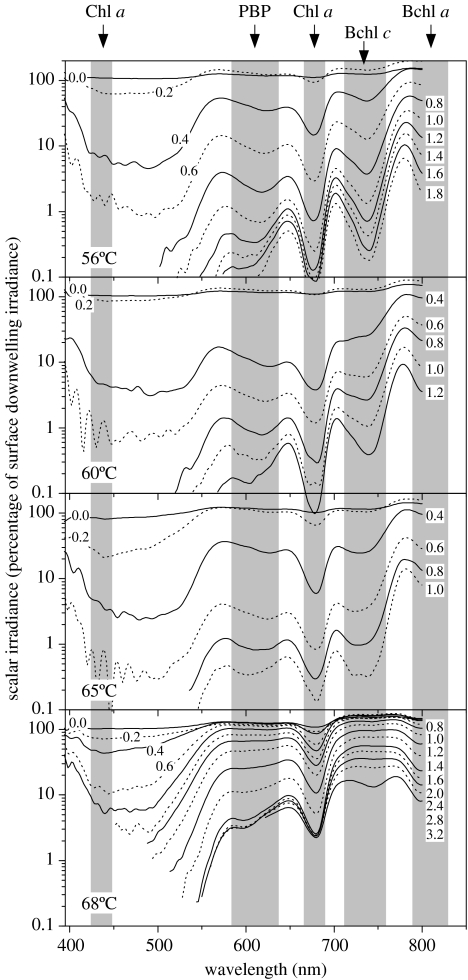
The spectral light field in the mats showed dramatic shifts with depth (figure 8). In the surface 0.2–0.6 mm thick layers of the mats, intense scattering caused light-trapping effects resulting in local maxima of scalar irradiance reaching greater than 150% of the incident irradiance. This effect was most pronounced at wavelengths outside the absorption bands of the major photopigments in the mat, especially in the NIR part of the spectrum. The translucent mat at 68°C showed a much lower attenuation of light than in the mats at lower temperatures. However, in all mats, blue light was attenuated most efficiently, while far-red and NIR light penetrated deeper. In the visible part of the spectrum, distinct troughs and shoulders in the spectra corresponded to in vivo absorption maxima of chlorophyll (Chl) a and the phycobiliproteins. A tendency was seen towards a shift from predominantly phycocyanin absorption at 620 nm in the upper parts of the cyanobacterial layers towards more absorption around 580–610 nm in deeper parts. In the 68°C mat, phycocyanin absorption was much weaker than in the other mats. In the NIR part of the spectrum, clear spectral signatures of bacteriochlorophyll (Bchl) c and Bchl a were seen in all samples. Owing to the limited spectral range of the spectrometer used, bacteriochlorophylls with absorption maxima at wavelengths greater than 800 nm could not be detected. Interestingly, shifts in different absorption maxima within the spectral range of Bchl c were observed both with increasing depth in the individual mat samples and in between mats from different temperature sites. This may point to a diversity of Bchl c-containing phototrophs in the mats, which will be investigated in another study.
Figure 8.
Microsensor measurements of light intensity versus wavelength through the vertical aspect of mats (numbers on curves denote depth in millimetres below the mat surface) at each temperature site. Troughs in the transmitted light spectra indicate absorption maxima of characteristic photopigments in the mats: Chl a, chlorophyll a; PBP, phycobiliproteins; Bchl c, bacteriochlorophyll c; Bchl a, bacteriochlorophyll a.
4. Discussion
Our analysis of the Mushroom Spring mat system showed highly similar patterns of Synechococcus 16S rRNA variants and their distribution along flow and vertical gradients compared to those we observed in the Octopus Spring mat system. These patterns suggested that individual 16S rRNA variants represented distinct temperature-adapted ecotype populations. Our DGGE analysis of the vertical distribution of 16S rRNA variants revealed evidence suggesting possible adaptation to light as well. Ramsing et al. (2000) first reported the subsurface position of the Synechococcus A 16S rRNA variant at approximately 60°C. In the present study, we repeated that observation and also found patterns in a 56°C sample suggesting that different B-like populations may have distributions limited to near-surface or subsurface microenvironments. The lack of variation of 16S rRNA genotypes A and A′ in higher temperature samples with depth contrasted with the obvious differences in pigmentation between surface and subsurface populations and led to the hypothesis that these populations might be less divergent (i.e. younger) ecotypes that could not be resolved by a slowly evolving marker like 16S rRNA.
The use of a more rapidly evolving marker, ITS, for analysis of community composition increased our ability to resolve ecotype populations within these hot spring mat communities because diverse variants within each 16S rRNA-defined population (A, A′ and B′) could be seen to cluster separately. More importantly, our ITS datasets allowed a test of the theory-based community phylogeny analysis, and several predicted putative ecotypes could indeed be observed to have unique distributions, characteristic of organisms that occupy unique niches. This was observed for the three major clades defined by 16S rRNA sequences, which showed a biased distribution to different temperatures, as well as for some putative ecotypes within each of these clades. Closely related ecotypes within the A′ clade (A′1 and A′2) and within the B′ clade (B′5 versus others) were shown to differ in their associations with the vertical aspect of the photic zone. Closely related ecotypes also differed in temperature distribution (A1 versus A2). Some of the ecotype subclades we observed cannot yet be correlated with distributions relative to flow-related or vertical gradients.
The question arises as to whether we have detected all the Synechococcus ecotypes in the mats, especially since there is evidence of possible additional subclustering in our ITS phylogeny. Thus, there is clearly a need to attempt to identify ecotypes using higher resolution molecular markers. In our present work, we are taking a genomic approach to study the population genetics of mat Synechococcus populations using protein-encoding genes. We have obtained (i) the complete genome sequences of Synechococcus isolates with A and B′ genotypes (GenBank accession numbers CP000239 and CP000240, respectively), which exhibit distinct adaptations to temperature, and (ii) metagenomic databases for the Octopus Spring and Mushroom Spring mats (www.tigr.org). By comparing metagenomic sequences to these reference genome sequences, we can observe the populations of variants at each protein-encoding locus. Further, for each locus, we can determine the degree of divergence of these variants from the homologous genes in the reference genomes. This will allow us to select genes that offer the maximum resolution for ecotype detection. Such genes can be targeted in community phylogeny analysis of putative ecotypes, as exemplified by our analysis of ITS sequence data presented here, and by cultivation-independent multilocus sequence typing approaches we are developing for even higher resolution. The species-like attributes of putative ecotypes detected in this way can be evaluated through analysis of distribution of their unique allelic variants. For example, the putative ecotypes A′1 and A′2 correspond to populations distributed within the deeper dark-green layer or near the mat surface in the 68°C Mushroom Spring mat sample.
Ecotypes adapted to temporally varying environmental parameters might not exhibit unique spatial distribution, but would be expected to exhibit unique spatio-temporal patterns of gene expression. For instance, diurnally and nocturnally active populations might live in the same spatial location, but express genes differently as light and/or other covarying parameters (e.g. O2, CO2 and pH) change through the diel cycle. It should be possible to study this, as the divergence of ecotypes should be paralleled by the divergence of allelic variants for all homologous genes in the genomes of the members of a population. For instance, as Synechococcus ecotypes evolved, there should have been a divergence of genes involved in photosynthesis leading to unique sets of allelic variants in photosynthesis genes in each ecotype population. By studying the spatio-temporal patterns of expression of such genes, it should be possible to determine where and when photosynthesis genes from each population are expressed. In this regard, it is noteworthy that we have recently succeeded in conducting in situ gene expression studies in Octopus Spring and Mushroom Spring mat samples (Steunou et al. 2006). Reverse transcriptase- and quantitative-PCR have been used to quantitatively evaluate how the expression of Synechococcus A and B′ photosynthesis, fermentation and nitrogen fixation genes varies during an afternoon to evening light transition. We are confident that such studies will ultimately demonstrate how the ecotype populations regulate guild functions within microbial communities.
It is easy to see how environmental variation can provide sufficient opportunity for niche partitioning in this mat system. Microsensor measurements reveal that the microenvironment of the deep-green subsurface layer is highly different from that at the surface of the mat in terms of oxygen (figure 7) but also other chemical parameters like pH (Kühl, unpublished results). At all temperatures, there is a steep light gradient, such that light intensity in the deep-green layer is a small fraction of that at the surface (approx. 5% of the incident light remains). Furthermore, there are changes in light quality with depth. As noted earlier, there appears to be a shift within the spectral range of phycobiliprotein absorption between the surface and the deeper mat layers. At 68°C, where light penetrated more deeply, there are two subsurface photosynthesis maxima, which might be associated with Synechococcus ecotypes with adaptations to less intense and spectrally altered light (e.g. putative ecotype A′1). Similarly, at 65 and 60°C, putative ecotype B′5 may represent a surface-associated population. One wonders if higher resolution analysis will eventually subdivide the B genotype into more than one ecotype with distinct temperature adaptations.
Our analyses also showed the patterning of many non-cyanobacterial community members relative to flow and vertical gradients (figures 1 and 3, column 3). In every case, the most closely related GenBank sequences were greater than 97% identical. As had been noted in previous work (Ramsing et al. 2000), sequences corresponding to the anoxygenic phototroph Roseiflexus spp. (C″1 and C″2) were detected at different temperatures and their DGGE band intensities often increased with depth in the mat. Chloroflexus-like DGGE bands (NPE, New Pit Enrichment which was found in a culture enriched under photoheterotrophic conditions from ‘New Pit’ Spring; see Ward et al. 1997) were observed in the 66 and 71°C samples, consistent with molecular evidence from oligonucleotide probes (Ruff-Roberts et al. 1994) and targeted cloning and sequencing studies (Nübel et al. 2002). Sequences closely related to those of green sulphur bacteria (E′ and E″) were detected and, as previously reported, sometimes showed a subsurface distribution (Ramsing et al. 2000). Sequences most closely related to that of Acidobacterium GFP1 (GFP Green Finger Pool, in figures 1 and 3) were detected in 62 and 66°C temperature sites. GFP1 was discovered in a 16S rRNA cloning and sequencing study of a small pool unofficially named ‘Green Finger Pool’ (64°C, pH 9) in the Sentinal Meadows group, Yellowstone National Park (Barns et al. 1999; S. Barns 2006, personal communication). We have recently determined that the organism contributing this 16S rRNA sequence is another anoxygenic phototrophic mat inhabitant (Bryant et al. submitted). These observations are interesting, since relatively large quantities of NIR light penetrate through the mat surface environment. Roseiflexus spp. are known to contain predominantly Bchl a, mostly absorbing at wavelengths we could not detect in this study, and relatively little Bchl c. Yet, absorption at wavelengths characteristic of Bchl c was strong in the mat, consistent with the presence of green sulphur and/or green non-sulphur bacteria and the newly discovered acidobacterial phototroph. Shifts in the spectral maxima in the Bchl c region of the spectrum at different depths and temperatures may foretell the stratification and/or subtle adaptations of such anoxygenic phototrophs. Observed shifts in the Bchl c absorption maxima from longer (approx. 745 nm) towards shorter (approx. 735 nm) wavelengths and vice versa with depth in the mats may thus point to changing predominance of green non-sulphur bacteria, green sulphur bacteria and phototrophic acidobacteria in different parts of the mat community. Evidently, there is a need for a closer examination of the diversity and the distribution of anoxygenic phototrophs in the Octopus and the Mushroom Spring mat communities.
Non-photosynthetic micro-organisms with sequences identical or closely related to the OS-L sequence (L and L1 in figures 1 and 3), detected in previous cloning studies of Octopus Spring mat samples (Ward et al. 1992), as well as aerobic chemoorganotrophic enrichment cultures (Santegoeds et al. 1996), were observed in 62 and 66°C samples. We retrieved sequences closely related to uncultivated bacteria previously observed in hot spring environments, including Thermodesulfovibrio sp. (AF321208, AF255603 and AB231858; see Skirnisdottir et al. 2000), uncultivated members of the Chloroflexi (GNS green non-sulphur, in figure 3; AY193183 and AY293486) and candidate division OP-10 (Obsidian Pool, AF027089; see Hugenholtz et al. 1998).
In conclusion, the patterns of distribution of molecular variants we observed strongly correlate with the predictions of evolutionary ecology theory, suggesting that this theory-based approach is successful in identifying the ecotype populations that group variants according to their evolved ecological character. Conversely, the patterning of ecotypes predicted by theory-based analyses like community phylogeny can be used to test whether putative ecotypes have the properties expected of true ecotypes (e.g. unique spatio-temporal distributions or gene expression). Another important observation from our results that was echoed by many speakers at this meeting is that we must be concerned about the ability of molecular markers to resolve the individuals that form ecotype clusters. Just as ITS variation could be used to magnify the variation represented by single 16S rRNA variants, we must be concerned that yet higher resolution genetic markers might be needed because ITS may not vary sufficiently to detect all the ecotypes in a microbial community. Staley (2006) suggested in his contribution to this meeting that we should define species as ‘irreducible phylogenetic clusters’, meaning that higher resolution analysis would not further subdivide a clade into more than one. His suggestion is much like the stable ecotype model presented in figure 2, except that he does not commit himself as to why the terminal clusters formed (i.e. owing to ecological adaptation, or, for instance, physical isolation). Staley's idea cautions us that we must continue to increase resolution to a point where the hypothesis of irreducibility can be tested to be sure that a cluster represents one and only one species. Even then, detection of species as clusters must be regarded with caution. One challenge is that nascent species that have not yet diverged much from the parent population that spawned them (e.g. figure 2d) may be imbedded within a single phylogenetic cluster. Another is that previous geographic isolation may lead to more than one cluster per ecotype within a community, if recent human transport has recently brought formerly isolated populations into the same community (Ward & Cohan 2005). Studies of the distribution and expression of allelic variants unique to such clusters may help us resolve some of these situations.
Acknowledgments
We appreciate long-term support from the National Science Foundation Ecology (BSR-9708136) and Frontiers in Integrative Biology Research (EF-0328698) Programs, the NASA Exobiology Program (NAG5-8824), the Montana State University Thermal Biology Institute (NASA NAG5-8807) and the National Park Service (especially personnel at Yellowstone National Park). Support from the Danish Natural Science Research Council (M.K.) is acknowledged. Anni Glud and Melanie Melendrez are thanked for excellent technical assistance in the field and laboratory. F.M.C. acknowledges research funding from Wesleyan University.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Species and speciation in micro-organisms’.
Present address: Department of Pediatrics, Louisiana State University Health Science Centre & Research Institute for Children, New Orleans, LA 70118, USA.
Present address: Institute of Biogeochemistry and Marine Chemistry, University of Hamburg, Bundesstrasse 55, 20146 Hamburg, Germany.
References
- Allewalt J.A, Bateson M.M, Revsbech N.P, Slack K, Ward D.M. Temperature and light adaptations of Synechococcus isolates from the microbial mat community in Octopus Spring, Yellowstone National Park. Appl. Environ. Microbiol. 2006;72:544–550. doi: 10.1128/AEM.72.1.544-550.2006. doi:10.1128/AEM.72.1.544-550.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barns S.M, Takala S.L, Kuske C.R. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl. Environ. Microbiol. 1999;65:1731–1737. doi: 10.1128/aem.65.4.1731-1737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, D. A., et al Submitted. A phototrophic acidobacterium discovered by metagenomics.
- Cohan F.M. What are bacterial species? Ann. Rev. Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. doi:10.1146/annurev.micro.56.012302.160634 [DOI] [PubMed] [Google Scholar]
- Cohan F.M. Toward a conceptual and operational union of bacterial systematics, ecology, and evolution. Phil. Trans. R. Soc. B. 2006;361:1985–1996. doi: 10.1098/rstb.2006.1918. doi:10.1098/rstb.2006.1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohan, F. M., Koeppel, A. & Krizanc, D. In press. Sequence-based discovery of ecological diversity within Legionella In Legionella (ed. N. P. Cianciotto, et al). Washington, DC: Am. Soc. Microbiol. Press.
- Cohan, F. M. et al Submitted. From sequences to ecotypes—a bacterial systematics based on population dynamics.
- Ferris M.J, Ward D.M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris M.J, Kühl M, Wieland A, Ward D.M. Different light-adapted ecotypes in a 68°C Synechococcus mat community revealed by analysis of 16S–23S intervening transcribed spacer variation. Appl. Environ. Microbiol. 2003;69:2893–2898. doi: 10.1128/AEM.69.5.2893-2898.2003. doi:10.1128/AEM.69.5.2893-2898.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, et al. Reevaluating prokaryotic species. Nat. Rev. Microbiol. 2005;3:1–7. doi: 10.1038/nrmicro1236. doi:10.1038/nrmicro1236 [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Itulle C.P, Hershberger K.L, Pace N.R. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl M. Optical microsensors for analysis of microbial communities. Methods Enzymol. 2005;397:166–199. doi: 10.1016/S0076-6879(05)97010-9. doi:10.1016/50076-6879(05)97010-9 [DOI] [PubMed] [Google Scholar]
- Kühl M, Lassen C, Revsbech N.P. A simple light meter for measurements of PAR (400–700 nm) with fiber-optic microprobes: application for P vs. I measurements in microbenthic communities. Aquat. Microb. Ecol. 1997;13:197–207. [Google Scholar]
- Mayr E. Harvard University Press; Cambridge, MA: 1982. The growth of biological thought: diversity, evolution and inheritance. [Google Scholar]
- Nübel U, Bateson M.M, Vandieken V, Kühl M, Ward D.M. Microscopic examination of distribution and phenotypic properties of phylogenetically diverse Chloroflexaceae-related bacteria in hot spring microbial mats. Appl. Environ. Microbiol. 2002;68:4593–4603. doi: 10.1128/AEM.68.9.4593-4603.2002. doi:10.1128/AEM.68.9.4593-4603.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palys T, Nakamura L.K, Cohan F.M. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- Ramsing N.B, Ferris M.J, Ward D.M. Highly ordered vertical structure of Synechococcus populations within the one-millimeter thick photic zone of a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 2000;66:1038–1049. doi: 10.1128/aem.66.3.1038-1049.2000. doi:10.1128/AEM.66.3.1038-1049.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revsbech N.P. An oxygen microelectrode with a guard cathode. Limnol. Oceanogr. 1989;34:474–478. [Google Scholar]
- Revsbech N.P, Ward D.M. Microelectrode studies of interstitial water chemistry and photosynthetic activity in a hot spring microbial mat. Appl. Environ. Microbiol. 1984;48:270–275. doi: 10.1128/aem.48.2.270-275.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff-Roberts A.L, Kuenen J.G, Ward D.M. Distribution of cultivated and uncultivated cyanobacteria and Chloroflexus-like bacteria in hot spring microbial mats. Appl. Environ. Microbiol. 1994;60:697–704. doi: 10.1128/aem.60.2.697-704.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santegoeds C.M, Nold S.C, Ward D.M. Denaturing gradient gel electrophoresis used to monitor the enrichment culture of aerobic chemoorganotrophic bacteria from a hot spring cyanobacterial mat. Appl. Environ. Microbiol. 1996;62:3922–3928. doi: 10.1128/aem.62.11.3922-3928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirnisdottir S, Hreggvidsonn G.O, Hjörleifsdottir S, Marteinsson V.T, Petursdottir S.K, Holst O, Kristjansson J.K. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl. Environ. Microbiol. 2000;66:2835–2841. doi: 10.1128/aem.66.7.2835-2841.2000. doi:10.1128/AEM.66.7.2835-2841.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley J.T. The bacterial species dilemma and the genomic–phylogenetic species concept. Phil. Trans. R. Soc. 2006;361:1899–1909. doi: 10.1098/rstb.2006.1914. doi:10.1098/rstb.2006.1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steunou A.-S, Bhaya D, Bateson M.M, Melendrez M, Ward D.M, Brecht E, Peters J.W, Kühl M, Grossman A. In situ analysis of nitrogen fixation and metabolic switching in unicellular thermophilic cyanobacteria inhabiting hot spring microbial mats. Proc. Natl Acad. Sci. USA. 2006;103:2398–2403. doi: 10.1073/pnas.0507513103. doi:10.1073/pnas.0507513103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D.M. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1998;1:271–277. doi: 10.1016/s1369-5274(98)80029-5. doi:10.1016/S1369-5274(98)80029-5 [DOI] [PubMed] [Google Scholar]
- Ward D.M. A macrobiological perspective on microbial species. Microbe. 2006;1:269–278. [Google Scholar]
- Ward D.M, Cohan F.M. Microbial diversity in hot spring cyanobacterial mats: pattern and prediction. In: Inskeep W.P, McDermott T.R, editors. Geothermal biology and geochemistry in Yellowstone National Park. Montana State University Thermal Biology Institute; Bozeman, MT: 2005. pp. 185–202. [Google Scholar]
- Ward D.M, Bateson M.M, Weller R, Ruff-Roberts A.L. Ribosomal analysis of microorganisms as they occur in nature. Adv. Microb. Ecol. 1992;12:219–286. [Google Scholar]
- Ward D.M, Santegoeds C.M, Nold S.C, Ramsing N.B, Ferris M.J, Bateson M.M. Biodiversity within hot spring microbial mat communities: molecular monitoring of enrichment cultures. Antonie van Leeuwenhoek. 1997;71:143–150. doi: 10.1023/a:1000131426164. doi:10.1023/A:1000131426164 [DOI] [PubMed] [Google Scholar]
- Ward D.M, Bateson M.M, Ferris M.J, Nold S.C. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol. Mol. Biol. Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D.M, Papke R.T, Nübel U, McKitrick M.C. Natural history of microorganisms inhabiting hot spring microbial mat communities: clues to the origin of microbial diversity and implications for micro- and macro-biology. In: Staley J.T, Reysenbach A.-L, editors. Biodiversity of microbial life: foundation of Earth's biosphere. Wiley; New York, NY: 2002. pp. 25–48. [Google Scholar]