Abstract
The claim that eukaryotic micro-organisms have global geographic ranges, constituting a significant departure from the situation with macro-organisms, has been supported by studies of morphological species from protistan kingdoms. Here, we examine this claim by reviewing examples from another kingdom of eukaryotic microbes, the Fungi. We show that inferred geographic range of a fungal species depends upon the method of species recognition. While some fungal species defined by morphology show global geographic ranges, when fungal species are defined by phylogenetic species recognition they are typically shown to harbour several to many endemic species. We advance two non-exclusive reasons to explain the perceived difference between the size of geographic ranges of microscopic and macroscopic eukaryotic species when morphological methods of species recognition are used. These reasons are that microbial organisms generally have fewer morphological characters, and that the rate of morphological change will be slower for organisms with less elaborate development and fewer cells. Both of these reasons result in fewer discriminatory morphological differences between recently diverged lineages. The rate of genetic change, moreover, is similar for both large and small organisms, which helps to explain why phylogenetic species of large and small organisms show a more similar distribution of geographic ranges. As a consequence of the different rates in fungi of genetic and morphological changes, genetic isolation precedes a recognizable morphological change. The final step in speciation, reproductive isolation, also follows genetic isolation and may precede morphological change.
Keywords: microbial species recognition, microbial geographic range, phylogenetic species recognition, endemism
1. Introduction
In a series of publications, Bland Finlay & Thomas Fenchel have demonstrated that a large number of morphologically recognized species of free-living microbial eukaryotes smaller than 1–10 mm have global distributions that show no correlation between geographic and genetic distances (e.g. Finlay 2002; Fenchel & Finlay 2004). They contrast the global distributions of these microbial morphospecies with the more narrow endemic ranges of large eukaryotic plants and animals, and postulate how the global distribution of small organisms affects microbial community structure (Finlay & Fenchel 2004).
Finlay & Fenchel's claim of global ranges for eukaryotic microbes echoes the thoughts of Baas-Becking (Baas-Becking 1934) who famously proclaimed, ‘Everything is everywhere, the environment selects’. The logic of this claim is that due to the omnipresence of microbes, where the same environment is created in different locations, the same microbial community will develop with the same microbial species. Baas-Becking developed his hypothesis about cosmopolitan species from studies of bacteria in cultivation, and Finlay & Fenchel have extended it from studies of small aquatic animals and protists at two aquatic sites, one freshwater site in the UK and another marine site in Denmark. They have generalized their claim to include all small eukaryotes, both aquatic and terrestrial. Recently, their hypothesis has been challenged with molecular genetic evidence from a variety of microbes (LaChance 2004), including prokaryotes (Whitaker et al. 2003), diatoms (Telford et al. 2006) and protists (Katz et al. 2005; Foissner 2006). Here, we challenge this ‘everything is everywhere’ hypothesis by reviewing the evidence from a kingdom of small eukaryotes, the Fungi. The data from the fungi show that reliance on morphological species recognition (MSR) can confound assessment of the range of small eukaryotic species. We consider three reasons to account for the observation that fewer microbial species are recognized by morphology compared to measures of genetic or reproductive isolation than is the case for macrobes. First, microbes, although morphologically diverse, may have fewer morphological characters than macrobes. Second, microbial morphology is more difficult to assess than macrobial morphology. Third, the rate of change of morphological characters may be greater in macrobes than in microbes. We formulate a testable hypothesis concerning the rate of taxonomic character evolution that helps to account for the disparity in the geographical ranges of morphological species between small and large organisms.
Finlay & Fenchel distinguish between microbes that are free living and those that are not free living due to their symbiotic relationships with other life, whether parasite or mutualist. They exclude those microbes that associate with larger organisms from their hypothesis of global distribution because symbiotic microbes are likely to follow the distribution of the larger partner. Given the many interactions among organisms, one might fairly ask, is there any organism that could be considered free of entanglements with other organisms and thereby free living? These biological interactions constitute the part of the environment which is most susceptible to rapid change, and their change would create differences in otherwise chemically and physically identical environments and narrow the range of seemingly free-living microbes. Only autotrophic organisms can be considered energetically autonomous, but even microscopic algae have their zooplankton predators (McCauley & Murdoch 1990) and parasites, among them viruses (Dunigan et al. 2006) and fungi (e.g. Polyphagus species; Sparrow 1960, p. 449), whose presence would influence the distribution of each species.
Finlay & Fenchel use morphological characters to recognize species of the small eukaryotes that they study (Fenchel & Finlay 2006). We may ask, can microbial species be recognized solely by morphology or phenotype? Finlay & Fenchel are aware that MSR in microbes has been challenged by other methods, e.g. species recognition by reproductive isolation (biological species recognition, BSR) or by genetic isolation (phylogenetic species recognition, PSR) and make two arguments defending MSR over BSR or PSR (Finlay 2002). First, they cite an example of PSR finding several genetically isolated species in a morphospecies of foraminifera (de Vargas et al. 1999), and note that a posteriori, the three phylogenetic species were found to have morphologically diagnosable characters. We would argue the contrary that PSR allows biologists to sort individual organisms into species and then to find which of many variable phenotypic characters correlate with the phylogenetic species. The useful phenotypic characters are those that change on the time-scale of the speciation events which define the species. A fungal example is provided by the morphological species, Aspergillus flavus. Only after this fungus was shown to embrace at least four phylogenetic species (Geiser et al. 1998) was it possible to find diagnostic phenotypic characters for each phylogenetic species (Geiser et al. 2000). Second, Finlay & Fenchel also note that members of different sibling phylogenetic species from different geographic regions may still be able to mate (Coleman et al. 1994; Stoeck et al. 1998), implying that they are truly one biological species. Here, we would argue that geographic separation is an important means of reproductive isolation and that if such populations can be recognized as genetically diverged, they cannot be used to support the hypothesis that ‘everything is everywhere’. The fungus Schizophyllum commune provides a good example of this point and will be discussed in the fungal examples featured later. Third, Finlay & Fenchel claim that the morphological richness of the protists that they examine is sufficient for species-level taxonomy. We argue that MSR must be challenged by methods of species recognition that measure genetic or reproductive isolation. Microbial eukaryotes may have many morphological features, but they neither have as many morphological characters as do larger eukaryotes nor are the characters as easily observed. Equally important is the rate of change of morphological characters, which must be appropriate for species recognition. If the rate of change of morphological characters is slow when compared with genetic or reproductive isolation, fewer species will be recognized by morphology and their ranges will appear to be larger.
2. The Fungi, a kingdom of microbial eukaryotes
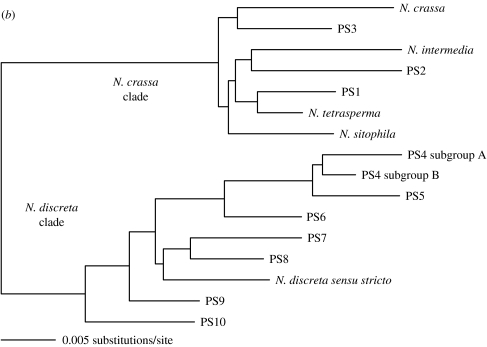
The Fungi, a large and important kingdom, has not yet been considered in this debate over global distribution of microbial eukaryotes. The fungi, however, provide organisms to specifically test the hypothesis of Finlay & Fenchel. First, there are large numbers of saprobic (free living) fungi that inhabit a wide range of environments. Second, almost all fungi have propagules under 1 mm (in fact, almost all spores are two orders of magnitude smaller than this limit), and many exhibit structures adapted to promote wide dissemination of those propagules. Third, although mycologists have traditionally used MSR and BSR to infer the taxonomy of fungi, in no other group of microbial eukaryotes has more effort been devoted to species recognition by phylogenetic methods using nucleic acid variation than in fungi (Taylor et al. 2006). Fungi, therefore, provide an ideal group of organisms in which to compare MSR, BSR and PSR. Here, we challenge the hypothesis put forward by Finlay & Fenchel and address the question, are there endemic species of microbial eukaryotes? After a brief introduction to the aspects of fungi germane to the debate over microbial species, we will present case histories of species recognition from five familiar genera of free-living fungi. In four cases, phylogenetically recognized species occupy less than global ranges (Neurospora and Saccharomyces from the Ascomycota and Schizophyllum and Lentinula from the Basidiomycota). The fifth example is the only fungus known to have a global distribution, the Ascomycete, Aspergillus fumigatus.
(a) Size of fungi
Finlay & Fenchel (2004) have proposed that the division between micro- and macrobiota lies within the size range of 1–10 mm, and that organisms smaller than 1 mm are likely to have a global distribution (Finlay 2002). In large animals, gene flow requires migration of the entire individual. In sedentary macrobes, bryophyte or ferns, for example, gene flow may be accomplished by microscopic spores. In flowering plants, it may occur via pollen and seeds. In fungi, gene flow is accomplished by microscopic spore dispersal. Some individual thalli of mushrooms species are as large as whales or trees (Smith et al. 1992), but almost all fungi produce spores (meiotic and mitotic propagules), most of which are ca 10 μm in their longest dimension. Even larger spores have been shown to be capable of long-distance dispersal; for example, mitospores of the plant pathogen Puccinia graminis (not smaller than 26×16 μm; Cummins 1971) can travel from South Africa to Australia (Watson & De Sousa 1983), and mitospores of another plant pathogen, Blumeria graminis (not smaller than 24×12 μm; Braun 1995) can cross the North Sea from continental Europe to Britain (Brown et al. 1991). In many fungi, spores function as gametes, so partners in mating need not be in close proximity. The smallest fungi are yeasts (Kurtzman & Fell 1998), e.g. Saccharomyces cerevisiae, and yeasts and their spores are single cells with a largest dimension not greater than 10–15 μm. In addition, on the small side for fungi are the monocentric Chytridiomycota, whose swimming spores are not larger than the yeasts, and whose sporangia can have diameters as large as 100 μm; even these thalli are still an order of magnitude smaller than 1 mm (Sparrow 1960). The remaining fungi, filamentous Chytridiomycota, Zygomycota, Glomeromycota, Ascomycota and Basidiomycota, make vegetative colonies larger than 10 mm in diameter (especially in culture), but they again have spores that are all much smaller than 1 mm. These examples highlight the fact that the small size and high dispersability of propagules allow for the possibility of global distributions of fungi. However, as discussed later, this potential is rarely realized.
(b) Fungal reproduction
Reproductive mode (clonal or recombining) has been thought to be important to speciation by many authors (Fisher 1930; Mayr 1957; Maynard Smith & Szathmary 1995; Coyne & Orr 1998). Recently, Barraclough et al. (2003) considered theoretical aspects of speciation by organisms that can recombine or that are strictly clonal to see if there were any fundamental differences. They conclude that both clonal and recombining organisms will form species, but that recombining organisms may form them faster because they respond more rapidly to selection (Goddard et al. 2005). Barraclough et al. (2003) note that speciation involves not only divergence, but also maintenance, particularly when newly derived species coexist. Here, the most important factor for species maintenance is the adaptation of newly diverged species to different niches; without this adaptation, they cannot coexist whether they are clonal or recombining. For species maintenance in recombining organisms, differential adaptation of coexisting populations must be accompanied by reproductive isolation.
Although species will form in both recombining and clonal organisms, there are aspects unique to each reproductive mode (Barraclough et al. 2003). Sexual reproduction with some outcrossing maintains cohesion within species, whereas reproductive isolation leads to the formation of new species (Coyne & Orr 1998). Recombination has been shown to facilitate more rapid adaptation than clonality (Goddard et al. 2005) and recombining organisms may show larger and more phenotypic differences among species. These effects may increase the chance that newly diverged species will have adapted to different niches and be able to coexist, provided that they are reproductively isolated in sympatry (Felsenstein 1981; Barraclough et al. 2003). On the other hand, clonal organisms can expand their population rapidly and may be better adapted to unpredictably hospitable habitats, e.g. the apparently asexual bdelloid rotifers occupy habitats more prone to unpredictable drought than do their sexual relatives (Ricci 1987; Barraclough et al. 2003). However, clonal organisms should have a higher extinction rate than recombining organisms because they are slower to respond to changing environments (Goddard et al. 2005) and because they are less able to purge their genomes of deleterious mutations (Muller 1964).
For many fungi, it is not a question of whether they reproduce by recombination or clonality, because they can do both. Morphological observation, alone, cannot be used to determine reproductive mode. Recombined progeny result from the sexual processes of mating and meiosis, but progeny of uniparental genetic origin (i.e. functionally clonal) can also result from sexual reproduction by self-fertilization, as well as by clonal reproduction. To make determinations of reproductive mode even more difficult, often fungi that never have been observed to undergo sexual reproduction nevertheless have populations consistent with recombination (Taylor et al. 1999). In fact, only one fungus that lacks the morphology of sexual reproduction has been shown to be exclusively asexual and clonal by population genetic criteria, Penicillium marneffei (Fisher et al. 2005; Vanittanakom et al. 2006). Many fungi that generally self-fertilize (i.e. homothallic and pseudohomothallic species) also retain the ability to outbreed and, therefore, to produce a combination of clonal and recombined progeny. Additionally, in a single species, clonal and recombining reproduction can be temporally or spatially separated. In some fungi causing disease of crop plants, reproduction is clonal in agricultural environments, but elsewhere populations retain the ancestral capacity for sexual recombination (Taylor et al. 1999; Couch et al. 2005). Clonal or inbreeding fungi should be able to invade more distant regions more easily because only one individual is needed to establish a population. Thus, in terms of endemism, one might expect that these fungi would have larger distributions than their obligately outbreeding relatives. Two of the fungi that we will examine can produce both meiotic and mitotic spores (Neurospora and Saccharomyces), two produce only meiotic spores (Schizophyllum and Lentinula), one makes only mitotic spores (A. fumigatus) and two can both self- and cross-fertilize, S. cerevisiae and Neurospora tetrasperma.
(c) Fungal species recognition
Fungal species are recognized by phenotype (MSR), reproductive isolation (BSR) and genetic isolation (PSR; Taylor et al. 2000). MSR is the default means of recognizing fungal species because every properly named fungus is accompanied by a morphological description. The morphological characters have been augmented by other phenotypic characters for fungi of simple morphology (e.g. substrate utilization in yeasts; Kurtzman & Fell 1998) or for those taxa that are industrially important (e.g. water availability of the substrate and growth temperature in Penicillium; Pitt 1979). Reproductive isolation is the basis of BSR, which is assessed by mating tests and has been employed with fungi that can be induced to mate in cultivation. However, BSR has not been widely applied because less than 15% of fungi can be cultivated (P. Crous 2004, personal communication), and of these many cannot be induced to mate. PSR depends on evidence of genetic isolation, which is provided by analysis of the congruence of genealogies based on DNA sequence of appropriately polymorphic loci from a sufficient sampling of individuals (Avise & Wollenberg 1997). PSR has been widely used for fungi, including those that cannot be cultivated, e.g. powdery mildews (Adam et al. 1999) or rusts (Weber et al. 2003).
(d) Fungal case studies
(i) Neurospora
Outcrossing species in the genus Neurospora are associated with recently burned vegetation, or food or food waste that has been heated. Neurospora species have not been shown to prefer particular plant species (Perkins et al. 1976; Perkins & Turner 1988; Turner et al. 2001; Jacobson et al. 2004) and they are considered to be free-living heterotrophs. The early publications on these fungi describe four species, three with eight spores per ascus and one with just four (Shear & Dodge 1927; Tai 1935). Although the pioneering descriptions of the Neurospora species were morphological, it is important to note that both Shear and Dodge, and Tai, conducted mating studies of Neurospora individuals prior to making their morphological descriptions. The clear results of the mating studies likely influenced their ostensibly morphological and taxonomic decisions.
The morphological features recorded by these early authors from small numbers of individuals were challenged by studies of many more individuals by Perkins et al. (1976),
The conidiating species of Neurospora cannot be distinguished from one another by vegetative appearance… Taxonomic diagnosis of the 8-spored heterothallic species [the one 4-spored pseudohomothallic species, N. tetrasperma, is readily recognized] has stressed the size and shape of ascospores, asci and perithecia. Dimensions taken from the literature and from our measurements, however, show that there is a considerable overlap in size between species, and certainly extensive variability within species. In several strains we have noted genotypes that result in completely misleading measurements…
Instead of morphology, Perkins and colleagues, in this and subsequent publications (Perkins & Turner 1988; Turner et al. 2001), advocated the use of mating tests to pairs of tester strains (one for each of the two mating types, designated mat A and mat a) to assign individuals to species. This criterion was used to describe a new outbreeding species, Neurospora discreta (Perkins & Raju 1986). Therefore, the eight-spored, outbreeding Neurospora species would be considered one circumglobal, tropical and subtropical species if recognized solely by morphology. Under BSR using the tester strains, however, each of the four eight-spored species have their own biogeography, which will be discussed later.
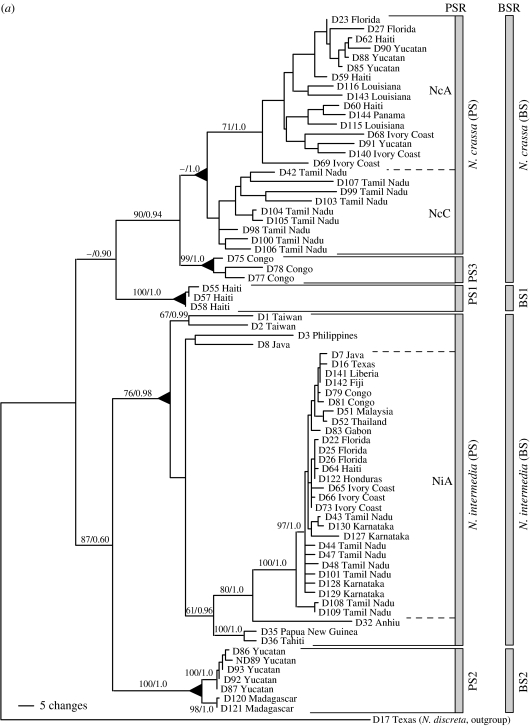
Recently, Dettman et al. (2003a,b) compared methods of fungal species recognition in the five outbreeding Neurospora species recognized by mating to tester strains (Neurospora crassa, Neurospora sitophila, Neurospora intermedia, N. tetrasperma and N. discreta). From a collection of more than 5000 natural isolates, Dettman et al. selected Neurospora individuals to represent all the five outbreeding species with emphases on two species, N. crassa and N. intermedia, and four geographic locations, the Caribbean basin, Africa, India and Asia. For PSR, they compared gene genealogies for four highly polymorphic loci that were sequenced from 147 individuals. For BSR, they compared reproductive success in most of the possible matings for 73 of the individuals used for PSR. PSR found not only the five described species, but also three additional genetically isolated species with few members and narrow endemic ranges, e.g. phylogenetic species 1 (PS1) with three individuals from Haiti, PS2 with seven individuals (five from Yucatan and two from Madagascar), and PS3 with five individuals from Congo. For BSR, Dettman et al. (2003b) focused on just N. crassa and N. intermedia, and the three new phylogenetic species, PS1–PS3. From matings among the 73 individuals representing these species, they found that PSR and BSR identified almost the same species; the only difference was that PSR found PS3, but with BSR, PS3 was indistinguishable from N. crassa (figure 1a). In other words, PS3 was genetically isolated from all other Neurospora species and was reproductively isolated from all species except N. crassa. The comparison among methods of species recognition with Neurospora showed that MSR alone found two species (one with eight ascospores per ascus and one with four), BSR found seven species and PSR found eight (consistent with the observation that genetic isolation precedes reproductive isolation in fungi). The amount of genetic variation seen within the seven species was similar, except for N. discreta, which was the most basal of the outbreeding Neurospora species and which had as much variation as measured by nucleotide polymorphism as the other species combined (Dettman et al. 2003a).
Figure 1.
(a) Phylogenetic species recognition applied to Neurospora and a graphic comparison to biological species recognition applied to the same individuals. Maximum-parsimony (MP) phylogram produced from the combined analysis of DNA sequences from four anonymous nuclear loci (TMI, DMG, TML and QMA loci, a total of 2141 aligned nucleotides). Tree length =916 steps; consistency index =0.651. Labels to the right of the phylogram indicate groups identified by phylogenetic species recognition and biological species recognition. Triangles at nodes indicate that all taxa united by (or distal to) a node belong to the same phylogenetic species. Taxon labels indicate strain number and geographic source. Branch support values for major branches with significant support are indicated by numbers above or below the branches (MP bootstrap proportions/Bayesian posterior probabilities). Figure and legend adapted from Dettman et al. (2003b) with permission of the authors and publisher. (b) Summary of results of phylogenetic species recognition in Neurospora. Neighbour-joining phylogram produced from three loci combined (DMG, TMI and TML) using exemplars of described species, and new phylogenetic species of Neurospora. Figure and legend adapted from Dettman et al. (in press) with permission of the authors and publisher.
The exceptional variation found in N. discreta stimulated studies of species recognition in this taxon (Dettman et al. in press). Neurospora discreta, like the other Neurospora species, had been considered a tropical and subtropical species until its discovery in temperate western North America ranging from New Mexico to 63° N latitude in Alaska (Jacobson et al. 2004). Found beneath the bark, or visible through fissures in the bark, on recently burned woody shrubs and trees, this fungus demonstrates that biologists can fail to perceive even abundant and distinctive microbes. Not only is it common after forest fires in western North America, but also it is almost the only Neurospora species found there; of 500 individuals collected in western North America, 95% were assigned to N. discreta by mating tests (Jacobson et al. 2004). Subsequent to the discovery in North America, N. discreta was found in southern Europe, again associated with recently burned vegetation (Jacobson et al. in press). Rather than being the predominant Neurospora species, N. discreta was found along with similar numbers of individuals of N. crassa, N. sitophila and N. tetrasperma, extending the range of these species well into the temperate zone. However, N. intermedia was not found in these surveys, and although some historic sources place it in Western Europe (see Jacobson et al. in press), it is clearly absent from western North America. PSR was applied to 70 individuals assigned to N. discreta by mating tests (BSR), including individuals from the recently discovered temperate regions. These analyses show that this single biological species contains eight phylogenetic species that have intraspecific and interspecific variation similar to the previously recognized phylogenetic species (Dettman et al. in press; figure 1b).
How has the use of PSR altered our understandings of Neurospora biogeography? Remember that by MSR, the eight-spored Neurospora species would form a single species with a circumglobal tropical and subtropical distribution. Using BSR based on mating to tester strains, Perkins and colleagues recognized four, eight-spored species with the following distributions (Perkins & Turner 1988; Turner et al. 2001). Neurospora intermedia had the broadest, being found in tropical and subtropical regions of Asia, the New World, Africa, India and Australia. Neurospora sitophila had a similar distribution, but was not as commonly encountered in Asia. Neurospora crassa was not found in East Asia or in Australia. Neurospora discreta, while encountered much less frequently than the other species, had a distribution most similar to N. crassa, but with rare occurrences in Australasia.
The biogeography of the two species that received most attention in the Dettman et al. study, N. intermedia and N. crassa, was consistent with the results of Turner et al. (2001) in that both fungi are centred in the tropics and subtropics, but only N. intermedia could be found at higher latitudes in Asia (Turner et al. 2001) and only N. crassa could be found at higher latitudes in North America and Europe (Jacobson et al. 2004, in press). In addition, N. crassa was missing from Asia (Perkins et al. 2001). Although N. intermedia and N. crassa would be one morphological species with a global distribution, by BSR or PSR, they were shown to be two species, each with a different geographic range. While it is true that inferences about geographic distributions of species are dependent on the quality of the sampling, given that sampling in Asia has been thorough enough to find representatives of four outbreeding species, the absence of N. crassa from Asia is difficult to dispute.
Dettman et al. (2003a) also found three new phylogenetic species, PS1, PS2 and PS3, which have narrow geographic distributions: Haiti, Yucatan and Madagascar, and Congo, respectively. When more individuals of PS2 become available, it is likely that distinct Neurospora phylogenetic species will be recognized in Yucatan and Madagascar. Here, again, applying MSR to these fungi would obscure the existence of the three, narrowly endemic, phylogenetic species. Admittedly, ranges inferred from small numbers of individuals are likely to be underestimates. However, given the global scope of individuals in the dataset, the restricted biogeography of the new phylogenetic species is suggestive of narrow endemism.
In the N. discreta clade, examination of the biogeography of phylogenetic species shows that no single phylogenetic species has a global distribution; in general, however, sampling is insufficient to make definitive claims about the complete distributions of the eight phylogenetic species. Nevertheless, sampling in western North America is sufficient to argue that PS4, alone, dominates western North America (Jacobson et al. 2004; Dettman et al. in press) and that none of the seven other species are likely to be found in that region (figure 1b). Therefore, the N. discreta complex represents another group of phylogenetic species with less than global distributions that would be considered to be part of one global species under MSR or BSR. Dettman et al. not only provide additional evidence that Neurospora refutes the ‘everything is everywhere’ hypothesis, but also show that biogeographical inference is very sensitive to methods of species recognition. In sum, what would be two nearly global morphological Neurospora species, one four-spored and one eight-spored, proved to be 15 phylogenetic species, none of them truly global.
(ii) Saccharomyces
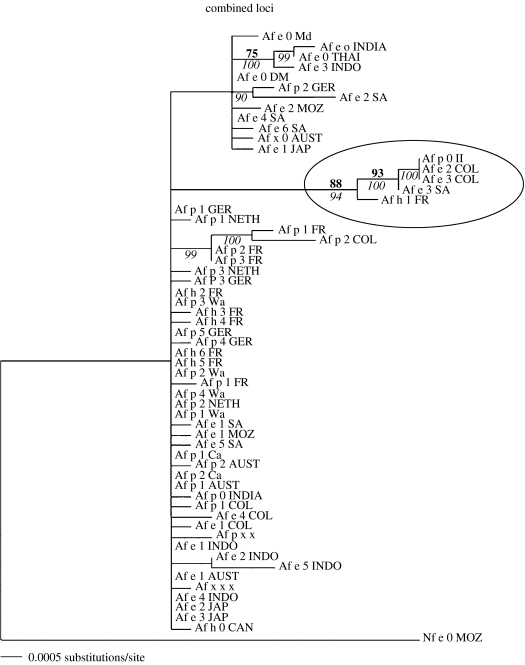
The second group of free-living ascomycetes that we will discuss is the yeast of commerce, S. cerevisiae, and its closest relative, Saccharomyces paradoxus. Unicellular and not larger than 10–15 μm in diameter either as a vegetative cell or a sexual spore (Kurtzman & Fell 1998), these yeasts certainly qualify as microscopic eukaryotes. Morphologically, all species in the group are indistinguishable (Naumova et al. 2003), but were shown by BSR to consist of several reproductively isolated species (Naumov 1987, 1996; Naumov et al. 2000). Although the geographic distributions of these yeast species are not as well studied as those of Neurospora, an examination of the best-known member, S. cerevisiae, and its closest relative, S. paradoxus, challenges the concept of global fungal species. Mating tests between S. cerevisiae and S. paradoxus demonstrate reproductive isolation (Sniegowski et al. 2002). Saccharomyces cerevisiae has been found associated with agriculture, in particular with grapes and wine production, and both S. cerevisiae and S. paradoxus are found associated with oak trees in natural environments (Sniegowski et al. 2002; Johnson et al. 2004).
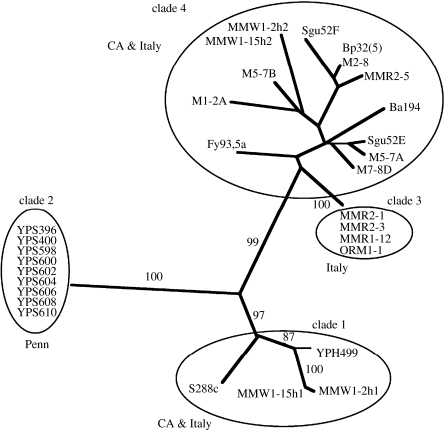
Aa et al. (2006) recently applied PSR to a collection of S. cerevisiae individuals obtained from laboratories, agriculture, wineries and oaks. The laboratory and winery or vineyard isolates were from Italy and California, the oak isolates were from Pennsylvania. The Pennsylvania S. cerevisiae isolates had been shown by BSR to be conspecific with S. cerevisiae individuals from Europe (Sniegowski et al. 2002). Analysis of DNA sequence from four loci (6600 nucleotides) from 27 individuals showed that the Pennsylvania oak individuals were genetically isolated from the California and the Italian isolates, whereas the latter formed three clades, two of which contained isolates from both wine-producing regions (figure 2). A plausible interpretation of the data is that S. cerevisiae isolates involved in commerce have been moved between California and Italy, but that oak individuals have not. The genetic isolation of Pennsylvanian S. cerevisiae from populations in Europe or California shows that this small, free-living eukaryote has genetically isolated populations. Whether or not the population structure in S. cerevisiae correlates with geographic distance, or with ecological factors such as substrate, is not yet known. It is certain, however, that it is not one global, phylogenetic species, and that again, everything is not everywhere.
Figure 2.
Phylogenetic species recognition applied to Saccharomyces cerevisiae. Unrooted distance trees based on four loci combined (CDC 19, FZF1, SSU1 and PHD1). Support of internal branches given as bootstrap percentages from 10 000 resamplings of the data. Construction of trees from the same data using the parsimony optimality criterion yielded trees with essentially the same topology. Figure and legend adapted from Aa et al. (2006) with permission of the authors and publisher.
Saccharomyces paradoxus provides a similar story. First, what was considered to be one biological species was shown by allozyme analysis to consist of two genetically differentiated populations, one in Europe and the other in Far East Asia (Naumov et al. 1997). Second, collections from North America that had been identified as S. paradoxus by mating to tester strains were found to be reproductively isolated from European S. paradoxus individuals (Sniegowski et al. 2002). Recently, Koufopanou et al. (2006) obtained DNA sequence from six genes in 112 individuals from the United Kingdom and Europe, Japan and Canada. The Canadian population showed 5% nucleotide divergence from the European and Asian populations, the latter pair were separated by 1.5% nucleotide divergence (the same as human and chimp). No shared polymorphisms were found among the three populations. For S. paradoxus, what would appear to be a single species with a near global distribution under MSR is revealed to be three species under PSR, each occupying a different continent.
(iii) Schizophyllum
One of the best-studied, free-living Basidiomycota in terms of natural variation and the distribution of alleles conferring mating compatibility is the genetic model organism, S. commune. This decay fungus has been found to decompose more than 150 different plant species throughout the world and it has basidiospores not larger than 8 μm in any dimension (Cooke 1961). Based on BSR, and also on morphology, S. commune has a global distribution (Raper et al. 1958).
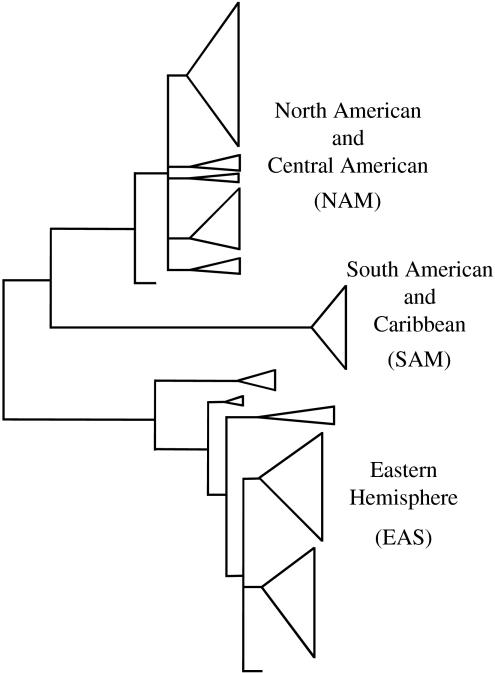
James et al. (1999) characterized 11 polymorphic allozymes for a collection of 136 individuals from North America, Central America, South America, Africa, Australasia and Europe. While alleles at either of the two mating loci showed no subdivision of the global S. commune population (James et al. 1999), allozyme analysis showed genetic differentiation between populations in the Eastern and Western Hemispheres (James et al. 1999). Again, population studies suggest that everything is not everywhere. The most recent examination of S. commune employed nucleic acid variation (James et al. 2001). In this study, the most variable region of the nuclear ribosomal repeat, the intergenic spacer (IGS) was sequenced for 195 individuals from Africa, Asia, Australasia, Europe and in the New World, North, Central and South America. Phylogenetic analysis of IGS variation showed three well-supported clades, one embracing isolates from North and Central America (NAM), a second with isolates from South America (SAM) and a third with the European and Asian/Australasian individuals (EAS; figure 3). James et al. also found evidence of recent migrations, both from South America to the Caribbean, and from Europe to western North America. Based on the fact that S. commune lives on cut logs, the authors speculate that migration and population expansion of S. commune may be tied to human activity (James et al. 2001). Again, what was thought to be one species by MSR or BSR, and perhaps two species by allozyme analysis, proved to be three, geographically distinct species when the rapidly evolving characters underlying nucleic acid polymorphism were used to implement PSR.
Figure 3.
Phylogenetic species recognition applied to Schizophyllum commune based on IGS1 sequence data and heuristic parsimony analysis. Heuristic searches found 9700 equally parsimonious trees. Cartoon of one of 9700 equally parsimonious trees is shown (tree length =285 steps; CI =0.795). Clades are represented by triangles and large clades, NAM, SAM and EAS, represent phylogenetic species. Detailed trees with branch support are to be found in James et al. 2001. Figure and legend modified from those in James et al. (2001) with permission of the authors and publisher.
James & Vilgalys (2001) then investigated S. commune spore dispersal directly by sampling basidiospores in the air at four locations bridging the South American–Caribbean and North American–Central American populations: Guyana, Puerto Rico, Bahamas and Florida. To ‘trap’ basidiospores of S. commune, they exposed, overnight, at each location ca 25 cultures of an unmated, haploid individual from the North American–Central American species. Next, they examined the cultures for evidence of fertilization by spores settling from the air. The mated colonies were then genotyped using four polymorphic loci and, knowing the genotype of the haploid strain proffered as bait, the haploid genotype of the fertilizing spore was deduced for 199 matings. From these data, they estimate that 18 S. commune spores impact each square metre of land per hour. Moreover, the genotype of the spores trapped at each location matched the genotypes of local individuals better than those from distant locations, although the neighbour-joining analysis used to examine this question did not include a test of significance. They did not find evidence of spores traversing the Caribbean Sea from South America to Florida.
These findings dispute the hypothesis that the airborne spore population encompasses all available genotypes across locations and indicate that spores, like individuals on cut logs, show population subdivision (James & Vilgalys 2001). Although more distant genotypes could be present in the airborne spores, perhaps at densities too low to be detected by the spore-trap assay, there is no evidence from resident individuals that distant genotypes become incorporated into local populations.
The taxonomical history of S. commune, initially described as a morphological species of global distribution and then revealed to consist of several biological or phylogenetic species of more narrow distribution, is typical of many mushroom-forming Basidiomycota (Petersen & Hughes 1999). Many of these fungi are symbiotic, either as pathogens (Armillaria, Korhonen 1978; Anderson & Ullrich 1979; Anderson et al. 1980; Heterobasidion, Chase & Ullrich 1990; Johannesson & Stenlid 2003) or as mycorrhizae (Rhizopogon; Kretzer et al. 2003), but others are decay fungi, and provide additional tests for endemism in free-living fungi.
(iv) Lentinula
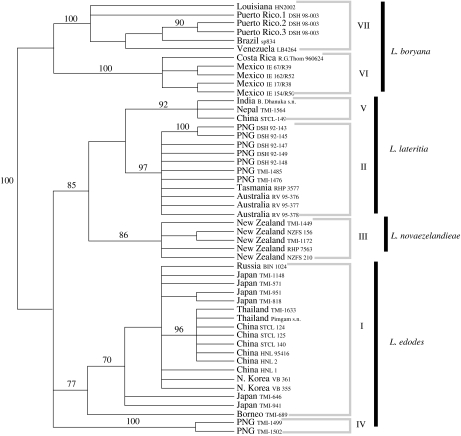
Another carefully studied example is provided by the genus Lentinula, which contains several species of wood decay fungi including the well-known edible mushroom, Lentinula edodes, better known as shiitake. Morphological study of this fungus had resulted in the description of four convincing species of Lentinula, three in the Old World and one in the New World (Pegler 1983). After analysing the sequence for rapidly evolving parts of the rDNA repeat from a worldwide collection including members of all four species, Hibbett (2001) concluded that only one of the four morphological species, Lentinula novaezelandieae, was truly a single, phylogenetic species (figure 4). Each other morphological species contained two phylogenetic species apiece. The two phylogenetic species found in the New World morphological species, Lentinula boryana, were distributed similarly to the phylogenetic species of S. commune, one in South America and the Caribbean, the other in Mexico and Central America (Hibbett 2001).
Figure 4.
Phylogenetic species recognition applied to Lentinula based on ITS sequences. Strict consensus of 3000+ most parsimonious trees (tree length =234 steps; CI=0.861). Bootstrap values greater than 70% are shown above the branches. Numbered groups are phylogenetic species of Lentinula. Morphologically recognized species names are at right. PNG=Papua New Guinea. Figure and legend based on those in Hibbett (2001) with permission of the authors and publisher.
These four examples of fungi that have one broadly distributed morphological species, but two or more biological or phylogenetic species, each with a more narrow distribution, show that small eukaryotes in the fungal kingdom can be endemic like their larger eukaryotic relatives in the plant or animal kingdoms, but that endemism can be perceived only when fungal species are characterized by BSR or PSR. When compared directly in fungi, the fewest species are recognized by MSR, more by BSR and the most by PSR. In this kingdom, genetic isolation precedes reproductive isolation and both of these precede morphological divergence. As mentioned earlier, the key differences between microbes and macrobes that underlie this difference in species recognition involve the number, ease of observation and rate of change of morphological characters.
(e) Maintenance of species
This discussion about the absence or presence of endemism in free-living microbes has focused on the pattern of species and not on the processes of species formation and maintenance. Finlay notes that phylogenetic species may not be reproductively isolated, and therefore might still be considered as one species (Finlay 2002). We would argue that phylogenetic species isolated only by geography (but not by sexual incompatibility) have independent evolutionary trajectories and will inevitably evolve reproductive isolation barriers, if geographic isolation persists (Coyne & Orr 2004). As Kohn (2005) has pointed out, the process of microbial speciation may be best studied experimentally. However, one can make inferences about the roles of geographical and biological isolation barriers in the maintenance of microbial species from studies of natural populations. In the study comparing PSR and BSR in Neurospora (Dettman et al. 2003b), the hybrid matings among N. crassa, N. intermedia, PS1 and PS2, shed light on the barriers to reproduction that maintain species. These barriers can be intrinsic or extrinsic. Intrinsic barriers lead to no progeny or the inviability or infertility of hybrid progeny. Extrinsic barriers are contingent on geographic or environmental parameters and range from the inability to mate due to geographic separation (a very early barrier to mating) to the inability of a viable hybrid progeny to compete favourably with either parent (a very late-acting barrier).
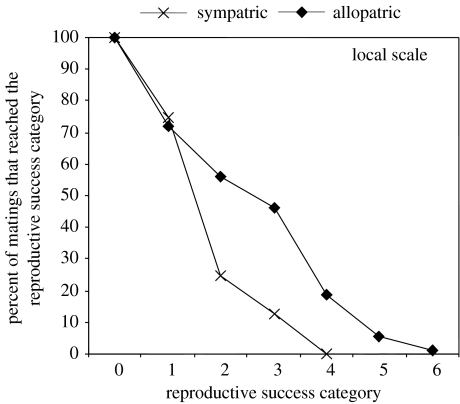
In the course of their study of BSR in Neurospora, Dettman et al. (2003b) made an interesting observation about reproduction and geography. In addition to the obvious result that reproductive success in heterospecific Neurospora matings was significantly lower than in conspecific matings, they found that hybrid matings between geographically separated (allopatric) partners were more successful than those between partners living in the same location (sympatry), and that the allopatric pairings proceeded significantly further in the mating pathway. The appropriate scale of geographic separation at which populations can be considered allopatric is dependent on the biology of the organism; for example, separation sufficient to preclude matings among moles would hardly suffice for gulls. Since the geographic separation needed to achieve allopatry is unclear in fungi, analyses were conducted with sympatry defined at three different scales: the regional scale (i.e. Caribbean, Africa, India or Asia), the sub-regional scale (e.g. Florida, Tamil Nadu or Ivory Coast) and the scale of single collecting sites. At all scales, reproductive barriers were significantly stronger between the sympatric pairs. When the percentages of matings that reached each of the seven stages in the mating pathway were compared for sympatric and allopatric matings (figure 5), it was apparent that the most important barriers were post-fertilization and that far more sympatric pairings than allopatric pairings aborted before significant investment was made in sexual structures. The presence of strengthened barriers to mating between sympatric members of different species provides clear evidence that when extrinsic barriers are absent, stronger intrinsic barriers help to keep Neurospora individuals from forming one global panmictic population.
Figure 5.
Neurospora mating success for sympatric and allopatric heterospecific pairings: the percentage of heterospecific matings between allopatric or sympatric individuals that reached the successive categories of reproductive success. At all geographic scales (regional, sub-regional and local), allopatric matings were significantly more likely than sympatric matings to proceed through the consecutive stages of the sexual cycle. The discrepancy between the allopatric and sympatric curves increases as the scale of sympatry decreases and is most pronounced for local sympatry, shown here. Categories of reproductive success: 0, no response; 1, aborted perithecia; 2, perithecia with no ascospores ejected; 3, perithecia with less than 1% of ejected ascospores having strongly melanized (i.e. black) walls; 4, perithecia with less than 15% black ascospores; 5, perithecia with less than 50% black ascospores; 6, perithecia with more than 50% black ascospores. Figure and legend modified from Dettman et al. (2003b) with permission of the authors and publisher.
The mixture of extrinsic and intrinsic barriers appears to be very effective; no naturally occurring hybrid Neurospora individuals have been found in the collection of ca 3000 N. crassa and N. intermedia individuals. Furthermore, when all matings conducted by Dettman et al. (2003b) were considered, both conspecific and heterospecific, a negative correlation was detected between geographic distance and mating success. Although the presence of conspecific matings in the analysis confounds the issue, the negative correlation is driven by the effect of geography on heterospecific matings. If microbes truly had global ranges, then no amount of geographic separation could suffice to make microbial populations allopatric and one would not see a negative correlation between geography and mating success.
Although hybrid individuals of Neurospora have not been sampled from nature, hybrids are well known from studies of plant pathogenic fungi (Brasier 2000; Schardl & Craven 2003): Fusarium (O'Donnell et al. 2000, 2004), Botrytis (Nielsen & Yohalem 2001), Neotyphodium (Schardl & Craven 2003; Moon et al. 2004), Ophiostoma (Brasier et al. 1998; Konrad et al. 2002), Melampsora (Newcombe et al. 2001) and Heterobasidion (Garbelotto et al. 2004). The difference between these fungi and Neurospora may be explained by the effect of human activity on two key extrinsic factors that separate species, allopatry and ecological inviability in hybrid progeny. Allopatry can be overcome as a barrier to reproduction by the global transport of crop plants and their pathogens, which bring together fungi that would otherwise be geographically separated. Ecological inviability of hybrid progeny due to competition with the parental species can be mitigated by increased access to hosts, as a consequence of farming and forestry or access to new host species by human-facilitated migration. Of course, a fungus does not have to be a symbiont to be affected by human activity. For example, an increase in cut logs associated with forestry may also increase the available habitat for free-living fungi like S. commune. Nevertheless, the contrast between Neurospora and other free-living fungi as compared to plant pathogens highlights the role of extrinsic barriers to reproduction in maintaining species of eukaryotic microbes, and provides indirect evidence for endemism in fungi.
Comparison of free-living fungi such as Neurospora with plant pathogenic fungi can also shed light on mechanisms of speciation (cf. Kohn (2005) for a discussion of the many mechanisms of fungal speciation). Allopatric speciation, where geographic distance provides the early extrinsic mating barrier, is well accepted (at least in microbes). If sibling species are brought together, reproductive barriers developed in allopatry may be strengthened by reinforcement (Noor 1999). Although reinforcement has been controversial, examples have been provided in Drosophila (Ortiz-Barrientos et al. 2004) and other animals (Coyne & Orr 2004; Hoskin et al. 2005) and Neurospora (Dettman et al. 2003b). Sympatric speciation is conceptually more difficult (Coyne & Orr 2004; Kohn 2005). A possible means for parasites to speciate in sympatry postulates that adaptation to different hosts (solving the problem of spatial coexistence of two new species) be coupled with an absence of gene flow between populations on the different hosts (solving the problem of recombination destroying adaptive gene complexes). The coupling could be achieved by one gene that affects both host choice and assortative mating, or by genetically associated pairs of genes that affect each trait separately.
Phytophagous insects provide the most applicable model for speciation of pathogenic fungi (Dres & Mallet 2002), but insects can use behaviour to achieve host preference and assortative mating. With plant pathogenic fungi, there may not be animal-like behaviours to support assortative mating or host preferences, although the activity of insects to facilitate mating in rusts (Craigie 1927) might provide an isolation barrier akin to plant pollination. Giraud et al. (2006) have considered the problem of sympatric speciation of pathogenic fungi that broadcast spores and gametes with no means of selecting hosts or mating partners. Using simulations, they find that sympatric speciation can occur in an ancestral population, provided it is polymorphic for host preference and if its gametes do not disperse so that mating is favoured for partners selected for growth on the same host. Conversely, for fungi whose gametes can disperse long distances, e.g. among the many fungi showing host preference, Armillaria (Basidiomycota; Anderson & Ullrich 1979), Heterobasidion (Basidiomycota; Chase & Ullrich 1990), Sclerotinia (Ascomycota; Carbone & Kohn 2001) or Magnaporthe (Ascomycota; Couch et al. 2005), allopatry would seem to be the most effective premating reproductive barrier. If everything were everywhere, it is difficult to see how new species could form in these fungi, or in fungi without host preference, like Schizophyllum or Neurospora.
Given the four examples of free-living fungi with less than global distributions noted earlier, and the many other examples of symbiotic pathogens or mutualists with even narrower distributions, one might think that there could be no truly global fungal species. However, just as there are macrofauna with global distributions, e.g. the Arctic tern or Norway rat, there is at least one example of a carefully studied fungus with a global population, A. fumigatus.
(f) Aspergillus fumigatus
This ascomycete causes disease in humans, most often in those undergoing immune system suppression in preparation for bone marrow or organ transplants (Latgé 1999). It can be found in almost any soil as well as composting vegetation, owing in part to its ability to grow at 37°C (Raper & Fennell 1965). When medical mycologists first investigated large collections of clinical and environmental isolates of A. fumigatus using polymorphisms caused by a mobile repeated element (Debeaupuis et al. 1997), they found no correlation between DNA fingerprint and either geography, environment or disease. PSR and population structure were then investigated by two different research groups, each working with different worldwide collections of individuals and each employing different polymorphic DNA regions as population genetic markers (Pringle et al. 2005; Rydholm et al. 2006). Pringle et al. (2005) assembled a collection of 63 A. fumigatus individuals from every continent except Antarctica and sequenced five polymorphic loci for each individual. They found two phylogenetic species, one comprising just five of the individuals. Both species had global distributions with no indication of endemism (figure 6). Rydholm et al. (2006) analysed DNA sequence from three sequenced intergenic regions for 70 isolates, again from every continent except Antarctica. This study found one global species with no population structure. They also analysed sequence from the three loci for another 33 isolates collected from five sites in North America and Europe to search for any correlation between genetic and geographic distances; none was found. Aspergillus fumigatus, therefore, has a global distribution.
Figure 6.
Phylogenetic species recognition applied to Aspergillus fumigatus. Bayesian analysis of five loci combined with separate substitution models for each partition. Parsimony bootstrap support above 70% is given in bold. Bayesian posterior probability above 90 is italicized. The phylogeny is rooted using Neosartorya fischeri as an outgroup. Figure and legend modified from Pringle et al. (2005) with permission of the authors and publisher.
These two research groups also investigated reproductive mode in A. fumigatus by population genetic means and neither of them were able to reject the null hypothesis of recombination (Pringle et al. 2005; Rydholm et al. 2006). In addition, Pöggeler (2002), Dyer et al. (2003) and Paoletti et al. (2005) found that the genes known to be required for mating in fungi that can reproduce sexually in cultivation are also found in A. fumigatus. Furthermore, Paoletti et al. (2005) showed that A. fumigatus individuals of each of the two mating types were found in equal proportion throughout the range of the species. Therefore, not only is A. fumigatus a single global species, but it also appears to reproduce by recombination as well as clonal means.
How does A. fumigatus maintain a global population when other fungi do not? One hypothesis postulates that A. fumigatus can maintain a global reach due to the small size of its mitospores, 2–3 μm in their largest dimension (Latgé 1999). However, as noted at the outset, far larger fungal spores can move from South Africa to Australia (Watson & De Sousa 1983), weakening the argument that the unique distribution of A. fumigatus is a consequence of its very small spores. The other hypothesis is that the environment favoured by A. fumigatus, composting vegetation, has been greatly expanded by human activity (Pringle et al. 2005). If this hypothesis is correct, then A. fumigatus should show evidence of a recent population expansion. The analysis of Rydholm et al. (2006) supports an expansion by showing low genetic diversity among, and a recent common ancestor for, A. fumigatus individuals as compared to the closely related species, Neosartorya fischeri. A completely satisfying explanation for the global distribution of A. fumigatus awaits further study. However, the expansion of suitable environment, aided by human activity, appears to provide a part of the answer.
These few examples of free-living fungi show that small eukaryotes exhibit species-level geographic distributions similar to those of their large relatives. Some fungi have narrowly endemic ranges, some have continental ranges, and at least one has a global range. In all the cases, species recognition based on morphology is more inclusive than that based on reproductive or genetic isolation. Therefore, hypotheses of global ranges for small eukaryotes based on studies of morphospecies must be viewed with caution. Until these hypotheses have been challenged with studies employing BSR and PSR, any conclusions about microbial community structure are premature.
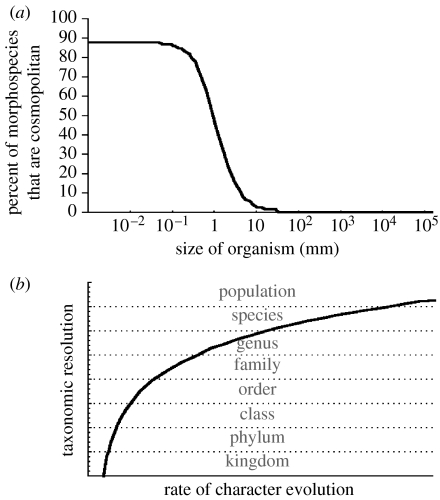
(g) Reconciling Finlay & Fenchel's MSR observations with fungal endemism
How then, may we reconcile the results of fungal studies using PSR or BSR with Finlay & Fenchel's demonstration of a dramatic decrease in the percentage of endemic protist morphospecies when organismal size drops below 1–10 mm (figure 7a)? We believe that there are two reasons that explain why MSR, BSR and PSR find the same species (or nearly the same species) in macrobes but not in microbes. The number of morphological characters is the most potentially obvious reason. At one extreme, no matter how many morphological characters one might find in a single-celled organism, a two-celled organism might have twice as many. As the number of cells increases to the point that macroscopic morphology emerges, the number of characters would increase even faster, for the possible permutations of cell organization are enormous. A corollary of the relationship of organismal size and the number of morphological characters is that such characters are easier to observe in larger organisms. Doubtless, there are microscopic characters that could be used to distinguish between, for instance, black and grizzly bears, but they are not needed. There are more than enough macroscopic characters to accomplish the task.
Figure 7.
A graphical depiction of a plausible explanation for the differences observed between studies of microbial endemism using PSR, BSR and MSR: morphological characters evolve more slowly than those used for PSR or BSA, thereby recognizing taxonomic groups broader than species and masking endemism. (a) The observation of Finlay & Fenchel that the percent of morphospecies that are cosmopolitan is much higher in smaller organisms, with a sharp point of inflexion ca 1–10 mm. Data from fungi suggest that morphological characters evolve more slowly in small organisms, thereby recognizing coarser taxonomic groups and obscuring endemism. (b) Relationship between rate of character evolution and taxonomic resolution. Faster evolving characters yield greater resolution for more refined taxonomic distinctions. Slower evolving characters yield greater resolution for coarser taxonomic distinctions. PSR is usually performed on characters that are polymorphic among isolates, and thus it is appropriate for resolution of species. BSR is implicitly performed on reproductive isolating factors, which generally evolve rapidly and are naturally appropriate for the resolution of biological species. MSR, like PSR, is ideally performed on characters that are polymorphic among isolates. However, if no or few morphological characters are polymorphic among isolates, morphospecies will map to coarser taxonomic groups than the species observed by BSR and PSR.
The second reason for the discrepancy between microbe and macrobe in the level of species recognition among MSR, BSR and PSR is evident from the case histories of fungi. Endemism may be demonstrated only when the characters used to make taxonomic assessment evolve at a rate that is fast enough to yield evolved differences among species (figure 7a). Such characters may be molecular evidence of genetic isolation, reproductive isolating barriers or morphological characters. Evidence from the fungi discussed earlier shows that genetic isolation precedes reproductive isolation and that morphological differentiation comes last. The rate of nucleotide substitution in fungi (10−8–10−9 nucleotide substitutions per nucleotide position per year), a key factor in phylogenetics, has been shown to be similar to that in bacteria and macroscopic eukaryotes (Kasuga et al. 2002). If the rate of phylogenetic character evolution is similar in microbes and macrobes, it must be that the rate of morphological character evolution is higher in larger organisms, where canalized but modular and highly evolvable systems of development permit rapid morphological innovation (Kirshner & Gerhart 1998). From the perspective of biological function, the cells of unicellular organisms must be able to perform every function required for life, and this requirement imposes a constraint on the evolution of those cells.
We would not expect the remarkable morphological diversity found among the specialized cells composing the various organs of macrobes to occur in unicellular species. From the perspective of biological form, if we try to imagine all of the unique ways to arrange very small numbers of identical spherical cells, we see that the number of possible arrangements increases much faster than the number of cells. Considering only those arrangements where the imaginary edges connecting the cell vertices are collinear or form a right angle, we find that two cells have only one arrangement (linear) and three cells have two possible arrangements (one linear and one planar), but four cells have at least nine (one linear, four planar and four three-dimensional). Simply, organismal shape can be changed faster by moving cells than by changing cell shape. If there is a correlation between the rate of morphological character evolution and the size of the organism, then the observation in figure 7a is a natural consequence of the decreasing resolution of morphological characters for species-level taxonomy, due to their diminished rates of evolutionary change in smaller organisms.
We submit that the fungal comparisons of species recognition based on morphology, genetic isolation and reproductive isolation show that these microbes can be narrowly endemic or demonstrate a global population, and everything in between. Finlay & Fenchel and others who claim that morphology, alone, can be used to recognize species in other microbial eukaryotes must challenge their hypotheses with studies of genetic and reproductive isolations. This type of research already has been initiated, but, as yet, with too few loci that may evolve too slowly (Katz et al. 2005; Foissner 2006). When PSR benefits from multiple genealogies of genes evolving at appropriate rates, we predict that all types of eukaryotic microbes will be found to have the diversity of geographic ranges found in fungi as well as large plants and animals.
Acknowledgments
We thank M. Donohue for discussion about fungal species recognition. Writing of this manuscript was supported by US National Science Foundation grants DEB-0316710 and DEB 0516511 to J.W.T. D.J. is also supported, in part, by US National Science Foundation grant MCB-0417282, awarded to David D. Perkins, Stanford University.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Species and speciation in micro-organisms’.
References
- Aa, E., Townsend, J. P., Adams, R. I., Nielsen, K. M. & Taylor, J. W. 2006. Population structure and gene evolution in Saccharomyces cerevisiae FEMS Yeast Res.6, 702–715. (doi:10.1111/j.1567-1364.2006.00059.x) [DOI] [PubMed]
- Adam L, Ellwood S, Wilson I, Saenz G, Xiao S, Oliver R.P, Turner J.G, Somerville S. Comparison of Erysiphe cichoracearum and E. cruciferarum and a survey of 360 Arabidopsis thaliana accessions for resistance to these two powdery mildew pathogens. Mol. Plant Microbe Interact. 1999;12:1031–1043. doi: 10.1094/MPMI.1999.12.12.1031. [DOI] [PubMed] [Google Scholar]
- Anderson J.B, Ullrich R.C. Biological species of Armillaria mellea in North America. Mycologia. 1979;71:402–414. [Google Scholar]
- Anderson J.B, Korhonen K, Ullrich R.C. Relationships between European and North American biological species of Armillaria mellea. Exp. Mycol. 1980;4:87–95. doi:10.1016/0147-5975(80)90053-5 [Google Scholar]
- Avise J.C, Wollenberg K. Phylogenetics and the origin of species. Proc. Natl Acad. Sci. USA. 1997;94:7748–7755. doi: 10.1073/pnas.94.15.7748. doi:10.1073/pnas.94.15.7748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas-Becking L.G.M. W.P. van Stockum and Zoon; The Hague, The Netherlands: 1934. Geobiologie of inleiding tot de milieukunde. [Google Scholar]
- Barraclough T.G, Birky C.W, Jr, Burt A. Diversification in sexual and asexual organisms. Evolution. 2003;57:2166–2172. doi: 10.1111/j.0014-3820.2003.tb00394.x. doi:10.1554/02-339 [DOI] [PubMed] [Google Scholar]
- Brasier C.M. The rise of hybrid fungi. Nature. 2000;405:134–135. doi: 10.1038/35012193. doi:10.1038/35012193 [DOI] [PubMed] [Google Scholar]
- Brasier C.M, Kirk S.A, Pipe N.D, Buck K.W. Rare interspecific hybrids in natural populations of the Dutch elm disease pathogens Ophiostoma ulmi and Ophiostoma novo-ulmi. Mycol. Res. 1998;102:45–57. doi:10.1017/S0953756297004541 [Google Scholar]
- Braun U. Gustav Fischer; Jena, Germany: 1995. The powdery mildews (Erysiphales) of Europe. [Google Scholar]
- Brown J.K.M, Jessop A.C, Navideh Rezanoor H. Genetic uniformity in barley and its powdery mildew pathogen. Proc. R. Soc. B. 1991;246:83–90. [Google Scholar]
- Carbone I, Kohn L.M. A microbial population-species interface: nested cladistic and coalescent inference with multilocus data. Mol. Ecol. 2001;10:947–964. doi: 10.1046/j.1365-294x.2001.01244.x. doi:10.1046/j.1365-294X.2001.01244.x [DOI] [PubMed] [Google Scholar]
- Chase T.E, Ullrich R.C. Genetic basis of biological species in Heterobasidion annosum: Mendelian determinants. Mycologia. 1990;82:67–72. [Google Scholar]
- Coleman W, Suarez A, Goff L.J. Molecular delineation of species and syngens in volvocacean green algae (Chlorophyta) J. Phycol. 1994;30:80–90. doi:10.1111/j.0022-3646.1994.00080.x [Google Scholar]
- Cooke W.B. The genus Schizophyllum. Mycologia. 1961;53:575–599. [Google Scholar]
- Couch B.C, Fudal I, Lebrun M.H, Tharreau D, Valent B, van Kim P, Nottéghem J.-L, Kohn L.M. Origins of host-specific populations of the blast pathogen Magnaporthe oryzae in crop domestication with subsequent expansion of pandemic clones on rice and weeds of rice. Genetics. 2005;170:613–630. doi: 10.1534/genetics.105.041780. doi:10.1534/genetics.105.041780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J.A, Orr H.A. The evolutionary genetics of speciation. Phil. Trans. R. Soc. B. 1998;353:2878–2305. doi: 10.1098/rstb.1998.0210. doi:10.1098/rstb.1998.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J.A, Orr H.A. Sinauer Associates, Inc; Sunderland, MA: 2004. Speciation. [Google Scholar]
- Craigie J.H. Discovery of the function of pyncnia of the rust fungi. Nature. 1927;120:765–767. [Google Scholar]
- Cummins G.B. Springer; New York, NY: 1971. The rust fungi of cereals, grasses and bamboos. [Google Scholar]
- de Vargas C, Norris R, Zaninetti L, Gibb S.W, Pawlowski J. Molecular evidence of cryptic speciation in planktonic foraminifers and their relation to oceanic provinces. Proc. Natl Acad. Sci. USA. 1999;96:2864–2868. doi: 10.1073/pnas.96.6.2864. doi:10.1073/pnas.96.6.2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaupuis J.P, Sarfati J, Chazalet V, Latgé J.P. Genetic diversity among clinical and environmental isolates of Aspergillus fumigatus. Infect. Immun. 1997;65:3080–3085. doi: 10.1128/iai.65.8.3080-3085.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettman J.R, Jacobson D.J, Taylor J.W. A multilocus genealogical approach to phylogenetic species recognition in the model eukaryote Neurospora. Evolution. 2003a;57:2703–2720. doi: 10.1111/j.0014-3820.2003.tb01514.x. doi:10.1554/03-073 [DOI] [PubMed] [Google Scholar]
- Dettman J.R, Jacobson D.J, Turner E, Pringle A, Taylor J.W. Reproductive isolation and phylogenetic divergence in Neurospora: comparing methods of species recognition in a model eukaryote. Evolution. 2003b;57:2721–2741. doi: 10.1111/j.0014-3820.2003.tb01515.x. doi:10.1554/03-074 [DOI] [PubMed] [Google Scholar]
- Dettman, J. R., Jacobson, D. J. & Taylor, J. W. In press. Multilocus sequence data reveal extensive phylogenetic species diversity within the Neurospora discreta complex. Mycologia [DOI] [PubMed]
- Dres M, Mallet J. Host races in plant-feeding insects and their importance in sympatric speciation. Phil. Trans. R. Soc. B. 2002;357:471–492. doi: 10.1098/rstb.2002.1059. doi:10.1098/rstb.2002.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunigan D.D, Fitzgerald L.A, Van Etten J.L. Phycodnaviruses: a peek at genetic diversity. Virus Res. 2006;117:119–132. doi: 10.1016/j.virusres.2006.01.024. doi:10.1016/j.virusres.2006.01.024 [DOI] [PubMed] [Google Scholar]
- Dyer P.S, Paoletti M, Archer D.B. Genomics reveals sexual secrets of Aspergillus. Microbiology. 2003;149:2301–2303. doi: 10.1099/mic.0.C0119-0. doi:10.1099/mic.0.C0119-0 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Skepticism towards Santa Rosalia, or why are there so few kinds of animals? Evolution. 1981;35:124–138. doi: 10.1111/j.1558-5646.1981.tb04864.x. doi:10.2307/2407946 [DOI] [PubMed] [Google Scholar]
- Fenchel T, Finlay B.J. The ubiquity of small species: patterns of local and global diversity. Bioscience. 2004;54:777–784. doi:10.1641/0006-3568(2004)054[0777:TUOSSP]2.0.CO;2 [Google Scholar]
- Fenchel T, Finlay B.J. The diversity of microbes: resurgence of the phenotype. Phil. Trans. R. Soc. B. 2006;361:1965–1973. doi: 10.1098/rstb.2006.1924. doi:10.1098/rstb.2006.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B.J. Global dispersal of free-living microbial eukaryotic species. Science. 2002;296:1061–1063. doi: 10.1126/science.1070710. doi:10.1126/science.1070710 [DOI] [PubMed] [Google Scholar]
- Finlay B.J, Fenchel T. Cosmopolitan metapopulations of free-living microbial eukaryotes. Protist. 2004;155:237–244. doi: 10.1078/143446104774199619. doi:10.1078/143446104774199619 [DOI] [PubMed] [Google Scholar]
- Fisher R.A. Oxford University Press; Oxford, UK: 1930. The genetical theory of natural selection. [Google Scholar]
- Fisher M.C, De Hoog S, Vanittanakom N. Low effective dispersal of asexual genotypes in heterogeneous landscapes by the endemic pathogen Penicillium marneffei. PLoS Pathogens. 2005;2:159–165. doi: 10.1371/journal.ppat.0010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner W. Biogeography and dispersal of micro-organisms: a review emphasizing protists. Acta Protozoologica. 2006;45:111–136. [Google Scholar]
- Garbelotto M, Gonthier P, Linzer R, Nicolotti G, Otrosina W. A shift in nuclear state as the result of natural interspecific hybridization between two North American taxa of the basidiomycete complex Heterobasidion. Fungal Genet. Biol. 2004;41:1046–1051. doi: 10.1016/j.fgb.2004.08.003. doi:10.1016/j.fgb.2004.08.003 [DOI] [PubMed] [Google Scholar]
- Geiser D.M, Pitt J.I, Taylor J.W. Cryptic speciation and recombination in the aflatoxin producing fungus Aspergillus flavus. Proc. Natl Acad. Sci. USA. 1998;95:388–393. doi: 10.1073/pnas.95.1.388. doi:10.1073/pnas.95.1.388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser D.M, Dorner J.W, Horn B.W, Taylor J.W. The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genet. Biol. 2000;31:169–179. doi: 10.1006/fgbi.2000.1215. doi:10.1006/fgbi.2000.1215 [DOI] [PubMed] [Google Scholar]
- Giraud T, Villareal L, Austerlitz F, Le Gac M, Lavigne C. Importance of the life cycle in sympatric host race formation and speciation of pathogens. Phytopathology. 2006;96:280–287. doi: 10.1094/PHYTO-96-0280. [DOI] [PubMed] [Google Scholar]
- Goddard M.R, Godfray H.C, Burt A. Sex increases the efficacy of natural selection in experimental yeast populations. Nature. 2005;434:636–640. doi: 10.1038/nature03405. doi:10.1038/nature03405 [DOI] [PubMed] [Google Scholar]
- Hibbett D.S. Shiitake mushrooms and molecular clocks: historical biogeography of Lentinula. J. Biogeogr. 2001;28:231–241. doi:10.1046/j.1365-2699.2001.00528.x [Google Scholar]
- Hoskin C.J, Higgie M, McDonald K.R, Moritz C. Reinforcement drives rapid allopatric speciation. Nature. 2005;437:1353–1356. doi: 10.1038/nature04004. doi:10.1038/nature04004 [DOI] [PubMed] [Google Scholar]
- Jacobson D.J, et al. Neurospora in temperate forests of western North America. Mycologia. 2004;96:66–74. [PubMed] [Google Scholar]
- Jacobson, D. J., et al in press. New findings of Neurospora in Europe and comparisons of diversity in temperate climates on continental scales. Mycologia [DOI] [PubMed]
- James T.Y, Vilgalys R. Abundance and diversity of Schizophyllum commune spore clouds in the Caribbean detected by selective sampling. Mol. Ecol. 2001;10:471–479. doi: 10.1046/j.1365-294x.2001.01224.x. doi:10.1046/j.1365-294x.2001.01224.x [DOI] [PubMed] [Google Scholar]
- James T.Y, Porter D, Hamrick J.L, Vilgalys R. Evidence for limited intercontinental gene flow in the cosmopolitan mushroom, Schizophyllum commune. Evolution. 1999;53:1665–1677. doi: 10.1111/j.1558-5646.1999.tb04552.x. doi:10.2307/2640430 [DOI] [PubMed] [Google Scholar]
- James T.Y, Moncalvo J.M, Li S, Vilgalys R. Polymorphism at the ribosomal DNA spacers and its relation to breeding structure of the widespread mushroom Schizophyllum commune. Genetics. 2001;157:149–161. doi: 10.1093/genetics/157.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson H, Stenlid J. Molecular markers reveal genetic isolation and phylogeography of the S and F intersterility groups of the wood-decay fungus Heterobasidion annosum. Mol. Phylogenet. Evol. 2003;29:94–101. doi: 10.1016/s1055-7903(03)00087-3. doi:10.1016/S1055-7903(03)00087-3 [DOI] [PubMed] [Google Scholar]
- Johnson L.J, Koufopanou V, Goddard M.R, Hetherington R, Schafer S.M, Burt A. Population genetics of the wild yeast Saccharomyces paradoxus. Genetics. 2004;166:43–52. doi: 10.1534/genetics.166.1.43. doi:10.1534/genetics.166.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasuga T, White T.J, Taylor J.W. Estimation of nucleotide substitution rates in eurotiomycete fungi. Mol. Biol. Evol. 2002;19:2318–2324. doi: 10.1093/oxfordjournals.molbev.a004056. [DOI] [PubMed] [Google Scholar]
- Katz L.A, McManus G.B, Snoeyenbos-West O.L.O, Griffin A, Pirog K, Costas B, Foissner W. Reframing the ‘Everything is everywhere’ debate: evidence for high gene flow and diversity in ciliate morphospecies. Aquat. Microb. Ecol. 2005;41:55–65. [Google Scholar]
- Kirshner M, Gerhart J. Evolvability (perspective) Proc. Natl Acad. Sci. USA. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. doi:10.1073/pnas.95.15.8420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn L.M. Mechanisms of fungal speciation. Annu. Rev. Phytopathol. 2005;43:279–308. doi: 10.1146/annurev.phyto.43.040204.135958. doi:10.1146/annurev.phyto.43.040204.135958 [DOI] [PubMed] [Google Scholar]
- Konrad H, Kirisits T, Riegler M, Halmschlager E, Stauffer C. Genetic evidence for natural hybridization between the Dutch elm disease pathogens Ophiostoma novo-ulmi ssp novo-ulmi and O. novo-ulmi ssp americana. Plant Pathol. 2002;51:78–84. doi:10.1046/j.0032-0862.2001.00653.x [Google Scholar]
- Korhonen K. Interferility and clonal size in the Armillaria mellea complex. Karstenia. 1978;18:31–42. [Google Scholar]
- Koufopanou V, Hughes J, Bell G, Burt A. The spatial scale of genetic differentiation in a model organism: the wild yeast Saccharomyces paradoxus. Phil. Trans. R. Soc. B. 2006;361:1941–1946. doi: 10.1098/rstb.2006.1922. doi:10.1098/rstb.2006.1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzer A.M, Luoma D.L, Molina R, Spatafora J.W. Taxonomy of the Rhizopogon vinicolor species complex based on analysis of ITS sequences and microsatellite loci. Mycologia. 2003;95:480–487. [PubMed] [Google Scholar]
- Kurtzman C.P, Fell J.W. Elsevier; Amsterdam, The Netherlands; New York, NY: 1998. The yeasts: a taxonomic study. [Google Scholar]
- LaChance M.-A. Here and there or everywhere? Bioscience. 2004;54:884. doi:10.1641/0006-3568(2004)054[0884:HATOE]2.0.CO;2 [Google Scholar]
- Latgé J.P. Aspergillus fumigatus and Aspergillosis. Clin. Microbiol. Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Szathmary E. W.H. Freeman; Oxford, UK: 1995. The major transitions in evolution. [Google Scholar]
- Mayr E. American Association for the Advancement of Science; Washington, DC: 1957. The species problem. [Google Scholar]
- McCauley E, Murdoch W.W. Predator–prey dynamics in environments rich and poor in nutrients. Nature. 1990;343:455–457. doi:10.1038/343455a0 [Google Scholar]
- Moon C.D, Craven K.D, Leuchtmann A, Clement S.L, Schardl C.L. Prevalence of interspecific hybrids amongst asexual fungal endophytes of grasses. Mol. Ecol. 2004;13:1455–1467. doi: 10.1111/j.1365-294X.2004.02138.x. doi:10.1111/j.1365-294X.2004.02138.x [DOI] [PubMed] [Google Scholar]
- Muller H.J. The relevance of mutation to mutational advance. Mutation Res. 1964;1:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- Naumov G.I. Genetic-basis for classification and identification of the ascomycetous yeasts. Stud. Mycol. 1987:469–475. [Google Scholar]
- Naumov G.I. Genetic identification of biological species in the Saccharomyces sensu stricto complex. J. Ind. Microbiol. 1996;17:295–302. doi:10.1007/BF01574704 [Google Scholar]
- Naumov G.I, Naumova E.S, Sniegowski P.D. Differentiation of European and Far East Asian populations of Saccharomyces paradoxus by allozyme analysis. Int. J. Syst. Bacteriol. 1997;47:341–344. doi: 10.1099/00207713-47-2-341. [DOI] [PubMed] [Google Scholar]
- Naumov G.I, James S.A, Naumova E.S, Louis E.J, Roberts I.N. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Syst. Evol. Microbiol. 2000;50:1931–1942. doi: 10.1099/00207713-50-5-1931. [DOI] [PubMed] [Google Scholar]
- Naumova E.S, Bulat S.A, Mironenko N.V, Naumov G.I. Differentiation of six sibling species in the Saccharomyces sensu stricto complex by multilocus enzyme electrophoresis and UP-PCR analysis. Antonie Van Leeuwenhoek Int. J. Gen. Mol. Microbiol. 2003;83:155–166. doi: 10.1023/a:1023328320228. doi:10.1023/A:1023328320228 [DOI] [PubMed] [Google Scholar]
- Newcombe G, Stirling B, Bradshaw H.D. Abundant pathogenic variation in the new hybrid rust Melampsora×columbiana on hybrid poplar. Phytopathology. 2001;91:981–985. doi: 10.1094/PHYTO.2001.91.10.981. [DOI] [PubMed] [Google Scholar]
- Nielsen K, Yohalem D.S. Origin of a polyploid Botrytis pathogen through interspecific hybridization between Botrytis aclada and B. byssoidea. Mycologia. 2001;93:1064–1071. [Google Scholar]
- Noor M.A.F. Reinforcement and other consequences of sympatry. Heredity. 1999;83:503–508. doi: 10.1038/sj.hdy.6886320. doi:10.1038/sj.hdy.6886320 [DOI] [PubMed] [Google Scholar]
- O'Donnell K, Kistler H.C, Tacke B.K, Casper H.H. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl Acad. Sci. USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. doi:10.1073/pnas.130193297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K, Ward T.J, Geiser D.M, Kistler H.C, Aoki T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 2004;41:600–623. doi: 10.1016/j.fgb.2004.03.003. doi:10.1016/j.fgb.2004.03.003 [DOI] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Counterman B.A, Noor M.A.F. The genetics of speciation by reinforcement. PLoS Biol. 2004;2:2256–2263. doi: 10.1371/journal.pbio.0020416. doi:10.1371/journal.pbio.0020416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti M, et al. Evidence for sexuality in the opportunistic fungal pathogen Aspergillus fumigatus. Curr. Biol. 2005;15:1242–1248. doi: 10.1016/j.cub.2005.05.045. doi:10.1016/j.cub.2005.05.045 [DOI] [PubMed] [Google Scholar]
- Pegler D.N. The genus Lentinula (Tricholomataceae tribe Collybieae) Sydowia. 1983;36:227–239. [Google Scholar]
- Perkins D.D, Raju N.B. Neurospora discreta, a new heterothallic species defined by its crossing behavior. Exp. Mycol. 1986;10:323–338. doi:10.1016/0147-5975(86)90019-8 [Google Scholar]
- Perkins D.D, Turner B.C. Neurospora from natural populations—toward the population biology of a haploid eukaryote. Exp. Mycol. 1988;12:91–131. doi:10.1016/0147-5975(88)90001-1 [Google Scholar]
- Perkins D.D, Turner B.C, Barry E.G. Strains of Neurospora collected from nature. Evolution. 1976;30:281–313. doi: 10.1111/j.1558-5646.1976.tb00910.x. doi:10.2307/2407702 [DOI] [PubMed] [Google Scholar]
- Petersen R.H, Hughes K.W. Species and speciation in mushrooms. Bioscience. 1999;49:440–452. doi:10.2307/1313552 [Google Scholar]
- Pitt J.I. Academic Press; London, UK: 1979. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. [Google Scholar]
- Pöggeler S. Genomic evidence for mating abilities in the asexual pathogen Aspergillus fumigatus. Curr. Genet. 2002;42:153–160. doi: 10.1007/s00294-002-0338-3. doi:10.1007/s00294-002-0338-3 [DOI] [PubMed] [Google Scholar]
- Pringle A, Baker D.M, Platt J.L, Wares J.P, Latge J.P, Taylor J.W. Cryptic speciation in the cosmopolitan and clonal human pathogenic fungus Aspergillus fumigatus. Evolution. 2005;59:1886–1899. doi:10.1554/04-241.1 [PubMed] [Google Scholar]
- Raper J.R, Fennell D.I. Williams and Wilkins; Baltimore, MD: 1965. The genus Aspergillus. [Google Scholar]
- Raper J.R, Krongelb G.S, Baxter M.G. The number and distribution of incompatibility factors in Schizophyllum commune. Am. Nat. 1958;92:221–232. doi:10.1086/282030 [Google Scholar]
- Ricci C. Ecology of bdelloids—how to be successful. Hydrobiologica. 1987;147:117–127. doi:10.1007/BF00025734 [Google Scholar]
- Rydholm C, Szakacs G, Lutzoni F. Low genetic variation and no detectable population structure in Aspergillus fumigatus compared to closely related Neosartorya species. Eukaryotic Cell. 2006;5:650–657. doi: 10.1128/EC.5.4.650-657.2006. doi:10.1128/EC.5.4.650-657.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schardl C.L, Craven K.D. Interspecific hybridization in plant-associated fungi and oomycetes: a review. Mol. Ecol. 2003;12:2861–2873. doi: 10.1046/j.1365-294x.2003.01965.x. doi:10.1046/j.1365-294X.2003.01965.x [DOI] [PubMed] [Google Scholar]
- Shear C.L, Dodge B.O. Life histories and heterothallism of the red bread-mold fungi of the Monila sitophila group. J. Agric. Res. 1927;34:1019–1042. [Google Scholar]
- Smith M.L, Bruhn J.N, Anderson J.B. The fungus Armillaria bulbosa is among the largest and oldest living organisms. Nature. 1992;356:428–431. doi:10.1038/356428a0 [Google Scholar]
- Sniegowski P.D, Dombrowski P.G, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEMS Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Sparrow F.K., Jr . University of Michigan; Ann Arbor, MI: 1960. Aquatic phycomycetes. [Google Scholar]
- Stoeck T, Przybos E, Schmidt H.J. Eur. J. Protistol. 1998;34:348. [Google Scholar]
- Tai F.L. Two new species of Neurospora. Mycologia. 1935;27:328–330. [Google Scholar]
- Taylor J.W, Jacobson D.J, Fisher M.C. The evolution of asexual fungi: reproduction, speciation and classification. Annu. Rev. Phytopathol. 1999;37:197–246. doi: 10.1146/annurev.phyto.37.1.197. doi:10.1146/annurev.phyto.37.1.197 [DOI] [PubMed] [Google Scholar]
- Taylor J.W, Jacobson D.J, Kroken S, Kasuga T, Geiser D.M, Hibbett D.S, Fisher M.C. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. doi:10.1006/fgbi.2000.1228 [DOI] [PubMed] [Google Scholar]
- Taylor, J. W., Turner, E., Pringle, A., Dettman, J. R. & Johannesson, H. 2006. Fungal species: thoughts on their recognition, maintenance and selection. In Fungi in the Environment (British Mycological Society Symposia No. 25) (ed. G. M. Gadd, S. C. Watkinson, P. S. Dyer), ch. 15. Cambridge, UK: Cambridge University Press.
- Telford R.J, Vandvik V, Birks H.J.B. Dispersal limitations matter for microbial morphospecies. Science. 2006;312:1015. doi: 10.1126/science.1125669. doi:10.1126/science.1125669 [DOI] [PubMed] [Google Scholar]
- Turner B.C, Perkins D.D, Fairfield A. Neurospora from natural populations: a global study. Fungal Genet. Biol. 2001;32:67–92. doi: 10.1006/fgbi.2001.1247. doi:10.1006/fgbi.2001.1247 [DOI] [PubMed] [Google Scholar]
- Vanittanakom N, Cooper C.R, Fisher M.C, Sirisanthana T. Penicillium marneffei infection and recent advances in the epidemiology and molecular biology aspects. Clin. Microbiol. Rev. 2006;19:95–110. doi: 10.1128/CMR.19.1.95-110.2006. doi:10.1128/CMR.19.1.95-110.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson I.A, De Sousa C.N.A. Long distance transport of spores of Puccinia graminis tritici in the southern hemisphere. Proc. Linn. Soc. NSW. 1983;106:311–321. [Google Scholar]
- Weber R.W, Webster J, Engel G. Phylogenetic analysis of Puccinia distincta and P. lagenophorae, two closely related rust fungi causing epidemics on Asteraceae in Europe. Mycol. Res. 2003;107:15–24. doi: 10.1017/s0953756202006937. doi:10.1017/S0953756202006937 [DOI] [PubMed] [Google Scholar]
- Whitaker R.J, Grogan D.W, Taylor J.W. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. doi:10.1126/science.1086909 [DOI] [PubMed] [Google Scholar]