Abstract
The bacterial species definition, despite its eminent practical significance for identification, diagnosis, quarantine and diversity surveys, remains a very difficult issue to advance. Genomics now offers novel insights into intra-species diversity and the potential for emergence of a more soundly based system. Although we share the excitement, we argue that it is premature for a universal change to the definition because current knowledge is based on too few phylogenetic groups and too few samples of natural populations. Our analysis of five important bacterial groups suggests, however, that more stringent standards for species may be justifiable when a solid understanding of gene content and ecological distinctiveness becomes available. Our analysis also reveals what is actually encompassed in a species according to the current standards, in terms of whole-genome sequence and gene-content diversity, and shows that this does not correspond to coherent clusters for the environmental Burkholderia and Shewanella genera examined. In contrast, the obligatory pathogens, which have a very restricted ecological niche, do exhibit clusters. Therefore, the idea of biologically meaningful clusters of diversity that applies to most eukaryotes may not be universally applicable in the microbial world, or if such clusters exist, they may be found at different levels of distinction.
Keywords: bacterial species, species concept, species definition, comparative genomics, genome sequencing
1. Introduction
The total biomass of the smallest organisms on Earth, i.e. the bacteria and the archaea, has been estimated to equal that of terrestrial and marine plants (Whitman et al. 1998), making them the largest unexplored reservoir of biodiversity on Earth. Yet, several important aspects of the biology of these organisms, such as what bacterial (or archaeal) species are, remain poorly understood. Bacterial species are typically demarcated based on a combination of more than one criteria (Vandamme et al. 1996; Stackebrandt et al. 2002), with the criterion proposed by Wayne et al. being by far the most influential one and the reference point for the development of new standards for species. This criterion considers a species to be essentially a collection of strains that are characterized by at least one diagnostic phenotypic trait and whose purified DNA molecules show at least 70% cross-hybridization (DNA–DNA hybridization or DDH; Wayne et al. 1987). Though the 70% DDH standard is pragmatic and universally applicable within the bacterial domain of life (Stackebrandt & Goebel 1994; Brenner et al. 2000; Rossello-Mora & Amann 2001), it has been criticized for being difficult to implement, e.g. cumbersome DDH experiments and inapplicable in environmental surveys, for not being encompassed by any of the eukaryotic species concepts and, most importantly, for resulting in too much phenotypic variation within the named species (Ward 1998; Cohan 2002; Staley 2004; Gevers et al. 2005). The latter is also evident from the fact that only about 4500 bacterial species have been described to date (after removing the 1200 synonyms; Garrity et al. 2004), which contrasts to well over 1 million eukaryotic species, and yet bacteria have been exploring evolutionary adaptations much longer than eukaryotes. Increasingly, the scientific community is finding the species definition based on the 70% DDH standard lacking, which has broader impacts such as for reliable diagnosis of infectious disease agents, intellectual property rights, international regulations for transport and possession of pathogens, bioterrorism agent oversight and reporting, and quarantine. If species designations are not well founded, or phenotype not well circumscribed by the ‘species’, serious confusion and damage may occur.
Owing to the above limitations, the scientific community has also become increasingly ready for a change to a more stringent species definition for bacteria, as is evident in the recent publications by the American Academy for Microbiology (Stahl & Tiedje 2002; Staley 2004). In order for such a definition to emerge, however, a solid understanding of the breadth and importance of the genetic differences among closely related bacteria is required (Konstantinidis & Tiedje 2005). This can only emerge from the extensive characterization of many bacterial groups, and at high resolution. Genome sequencing can reveal, at any resolution level, the genetic differences between two strains and thus can substantially contribute towards this goal. Here, we evaluate the findings emerging from almost a decade of extensive genome sequencing with respect to its correspondence to currently named bacterial species. We argue that, despite the great progress achieved, it is still premature to adopt new standards for species, simply owing to the shortage of adequate data and sampling. Nonetheless, several previously unrecognized (or only partially recognized) trends are becoming evident, such as the importance of ecological, in addition to the evolutionary, relatedness for the biochemical, and hence phenotypic, similarity among bacterial strains.
Genome sequencing has enabled us to start addressing several problematic issues related to the species definition, such as how the 70% DDH gold standard corresponds to sequence and gene-content similarity, as well as to better define the issues that remain to be attacked. The information emerging from genome sequencing, together with information from other approaches such as gene expression studies, should eventually converge to a more soundly based bacterial species definition. Given the present speed in accumulating new data and knowledge, it is conceivable that such a definition may indeed emerge within a few years.
2. What is encompassed by the current species definition?
(a) The extent of intra-species genetic diversity
Our initial analysis of a small number of published DDH values among strains whose genomic sequences have become available revealed that the 70% DDH standard corresponds tightly to approximately 95% average nucleotide identity (ANI; Konstantinidis & Tiedje 2005). (It is important to realize that the DDH method does not measure directly sequence identity but rather the efficiency of hybridization of the DNA molecules. Therefore, the 70% DDH does not correspond to 70% sequence identity; for a recent review of the method see Rosselló-Mora (2006).) ANI is the average nucleotide identity of the total genomic sequence shared between two strains, and our previous studies suggest that ANI is an exceptionally robust and sensitive method for measuring evolutionary relatedness among closely related bacterial strains, i.e. those showing higher than 60% ANI, which, typically, corresponds to greater than 97% 16S rRNA gene sequence identity (Konstantinidis & Tiedje 2005; Konstantinidis et al. in press). The correspondence of 95% ANI with 70% DDH was experimentally verified by the more comprehensive study of Goris et al., which showed, in addition, that ANI is the genome-derived parameter with the greatest correlation with DDH values, among several parameters tested (Goris et al. in press). Therefore, genomes that show higher than 95% ANI should belong to the same species according to the 70% DDH standard.
Using the 95% ANI standard to demarcate genomes into separate species, we have previously shown that species encompass strains that may differ in up to 35% of their gene content, which becomes approximately 20% when the hypothetical and mobile elements are removed from the analysis (Konstantinidis & Tiedje 2005). Hypothetical denotes open reading frames that are not phylogenetically conserved and lack evidence of being protein coding, while mobile denotes plasmid, transposase or prophage genes. There is a growing body of literature which indicates that the hypothetical and mobile elements found in any given genomic sequence are only occasionally responsible for an important phenotype and, in fact, a significant part, if not the majority, of them constitutes essential but not protein-coding sequences (Jackson et al. 2002; Siew & Fischer 2003) or even junk and ephemeral (Lerat & Ochman 2005; Ochman & Davalos 2006) parts of the genome. Collectively, these results demonstrate that the 70% DDH criterion is quite valuable as a first standard approximation for species since it encompasses relatively homogeneous sets of strains which share at least 80% of their genes with a function other than hypothetical or mobile. The question then becomes whether the up to 20% difference in genes with well-recognizable functions is a large enough difference to justify description of further taxonomic subdivisions (more species?) within species.
We believe that if the purpose of the species is to be soundly predictive of the phenotypic potential of a strain (as the greater public assumes it to be), then the 70% DDH standard appears too liberal. A 20% difference in functional gene content in a background genome of 5 Mb (like the Escherichia coli genome) translates into a difference of approximately 1000 genes (assuming, on average, one gene per 1 kb of sequence; Konstantinidis & Tiedje 2004). It is reasonable to expect that differences among strains of a species in the presence of 1000 functional genes would be responsible for a range of different phenotypes and for enabling the cell to exploit an array of different ecological niches. In addition, important regulation and expression differences may occur for the genes that are shared between organisms and may have important consequences for the organism's phenotype. Precisely how large a difference in gene content has to be present between two strains before they could be described as separate species, however, would always be an ambiguous question, particularly if the strains were not separated by a substantial genetic distance (see §2d).
Owing to the limited number of available genomic sequences at the time analyses were performed, and in order to gain a large enough dataset, data from different groups of bacteria were pooled together, which results in clear discontinuities in the results reported. Therefore, it remains unclear how the trends described previously apply within a single bacterial species. Towards this direction, we performed a similar analysis to that reported before (Konstantinidis & Tiedje 2005) within five important and contrasting bacterial groups that are presently best represented with genomic sequences. The groups are Streptococcaceae, Staphylococcaceae, Enterobacteriaceae, Shewanellaceae and Burkholderiaceae. These groups sample a variety of genomes sizes, ranging from approximately 1.9 to greater than 8 Mb, and contrasting ecologies, including pathogenic (Staphylococci and Streptococci), opportunistic pathogens (E. coli and Burkholderia) and environmental species (Shewanella and certain species of Burkholderia). Overall, we observed very similar results with these five groups, compared to results of our previous study that pooled together different bacterial groups, in terms of the extent of intra-species gene content and sequence diversity (figure 1a,b).
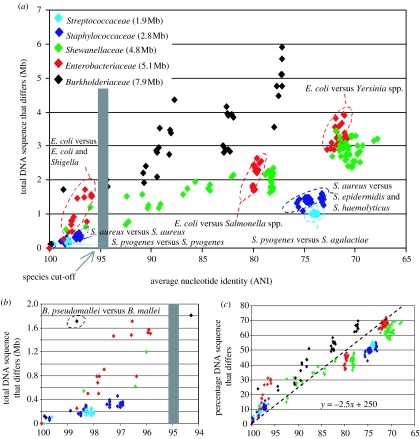
Figure 1.
Genetic diversity within five important bacterial groups. Every data point represents a whole-genome comparison between two genomes and shows the total DNA sequence that differs between the two genomes (y-axes) plotted against the evolutionary distance between the genomes, measured as ANI (x-axes). (a) shows data for all pairs of genomes considered, whereas (b) shows only pairs that share at least 94% ANI. (c) shows the DNA sequence that differs as a percentage of the whole genome of one of the genomes in the pair. Pairs of genomes of the same group are denoted by the same colour. See graph key for group annotation by colour as well as for the average genome size within each group. ANI values were calculated as described previously (Konstantinidis & Tiedje 2005). The grey bars represent approximately 95% ANI that corresponds to the 70% DDH standard for species. Note the formation of ANI-based genetic clusters within the Streptococcaceae, Staphylococcaceae and Enterobacteriaceae groups (denoted by dashed circles), and the lack of clusters (i.e. genetic continuum) within the Burkholderiaceae and Shewanellaceae groups for the same range of ANI values. The clusters within the Streptococcaceae and Staphylococcaceae groups encompass genomes that show small gene-content differences (e.g. S. aureus versus S. aureus circle), and are diagnosable by specific gene-content signatures, e.g. the S. aureus genomes share a total of approximately 1 Mb DNA sequence that is not conserved in the genomes of their closest sequenced relatives, S. agalactiae or S. epidermidis. In contrast, gene-content signatures are less clear in the Enterobacteriaceae group, e.g. the DNA shared by all E. coli genomes, and not found in Salmonella spp. genomes, is less than 200 kb. The clustering seen for the Shewanellaceae group in the range of approximately 70–75% ANI is attributable to a biased selection of the genomes sequenced to represent the most distantly related lineages of the group (i.e. there are seven different species represented by these data points). Sampling within several of these lineages reveals a continuum of genetic diversity for the Shewanellaceae group (represented by data points between 80 and 97% ANI).
(b) The effect of evolution
A strong, linear correlation is observed between the amount of DNA that differs and evolutionary distance over the total evolutionary range covered by the five groups considered (figure 1a), particularly when the DNA differences are plotted as a percentage of the total DNA of the strains (figure 1c), which normalizes for variation in genome size. The equation in the latter case approximates to the relationship: %DNA differ=−2.5× (ANI distance) +250 for all five groups, although strains with larger genomes appear to consistently show slightly higher gene-content differences than strains of medium or small genome size, for comparable values of ANI distance between the strains (e.g. compare data points corresponding to Burkholderia versus those corresponding to Streptococcaceae or Staphylococcaceae in figure 1c). This equation underscores the great interconnection, on a whole-genome scale, between evolutionary relatedness and gene content, which is central for the species definition.
We also observed that strains which show larger than 0.5 Mb genomic differences to any other strain of the same group, also show lower than 98–99% ANI to the latter strain(s). In contrast, all strains that show higher than 98–99% ANI show small genomic differences, i.e. less than 0.2 Mb of their genome differs (figure 1b). Further, these differences tend to be in unstable parts of the genome, i.e. plasmids and prophage genomes (figure 2). The coarse discontinuity observed around 98–99% ANI (see figure 1b) is, however, most probably owing to limited sampling as opposed to clear natural gaps in the genetic continuum. In any case, these findings indicate that considerable evolutionary time must elapse, corresponding, in general, to at least 1–2% genomic nucleotide divergence, before substantial functional gene-content differences start to accumulate in bacterial genomes (for exceptions, see Burkholderia example in §2c).
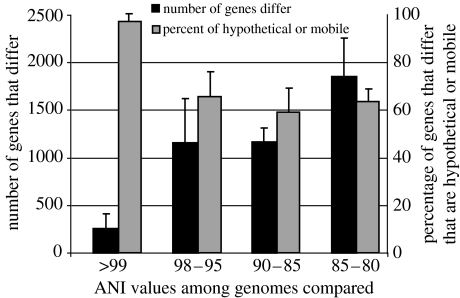
Figure 2.
Gene-content differences among Enterobacteriaceae genomes. Bars represent the average gene-content differences between pairs of Enterobacteriaceae genomes that show a given range of ANI values (x-axis). Black bars represent the total number of genes, on average, that differ (primary y-axis), while grey bars represent what fraction of the former genes is hypothetical, phage, transposase or plasmid genes (secondary y-axis). Error bars represent deviation from the mean, and not standard deviation. Only pairs of genomes (four pairs in total) with considerable gene-content differences were included in the analysis for the higher than 99% ANI category (i.e. pairs of genomes with higher than 99% ANI but very small gene-content differences were not included), while a random selection of six pairs of genomes was included for the remaining three categories of ANI values.
(c) The effect of ecology
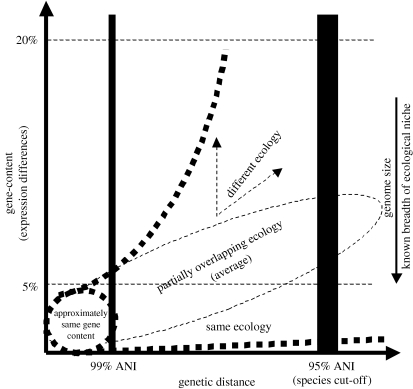
Our comparisons revealed another important trend: the groups that have larger genomes (Burkholderia and E. coli) tend to show larger intra-species genomic differences than groups that have smaller genome sizes (Streptococci and Staphylococci), particularly for strains that show between 95 and approximately 98% ANI (figure 1b). Species with larger genomes are presumably more metabolically versatile and thus able to exploit a larger number of ecological niches. The ecological niche (environment) ultimately determines and shapes the evolution of the gene content of an organism (Moran 2002; Konstantinidis & Tiedje 2004). Therefore, the larger genomic differences observed among strains of the former species could be the outcome of a differential evolution of those strains in response to a series of different ecological niches or habitats. In contrast, the Streptococci and Straphylococci, which are known to have a much narrower ecological niche, i.e. mainly causing specific infections in humans, show substantially smaller genomic differences. In summary, it appears that the phenotypic and ecological potential of strains that show higher than 98–99% ANI, or that are less related at the nucleotide level but share a closely overlapping niche (like the Streptococci), could be more soundly predicted and these may therefore represent more reliable standards for species than the 70% DDH standard. Figure 3 attempts to summarize the conclusions from the analyses of the intra-species genomic differences within the five groups considered.
Figure 3.
Ecological and genetic diversity within bacterial species. Bacterial genomes of the same species according to the 70% DDH criterion, which corresponds to 95% ANI, may show a very broad range of gene-content (and probably gene expression) differences between the two thick, curved, dashed lines. Genomes that, presumably, share a more overlapping ecological niche tend to show smaller gene-content differences than do genomes that are ecologically more differentiated. The former genomes also tend to be smaller in genome size. Accordingly, genomes that show at least 98–99% ANI, or are less identical at the nucleotide level but share a closely overlapping niche, tend to show small gene-content differences.
The coverage with genomic sequences is still too limited (figure 1), however, to allow for more robust conclusions to emerge. Metagenomic studies can be very informative in this respect because they can reveal the importance and extent of gene-content and genetic diversity within a naturally occurring population. Metagenomic approaches also overcome several serious limitations of working with only cultivable strains: most notably, sequenced strains typically originate from very contrasting environments, with little attention to whether or not they share a similar ecological niche or ecological history and whether they are representative of the indigenous community. These issues are less problematic for metagenomics studies, which are typically confined to a specific habitat or to a given niche and can reveal the total heterogeneity of the natural community. In one pioneer study of this kind, Hallam et al. (submitted) employed a metagenomic approach to recover the whole genome of Cenarchaeum symbiosum, an archaeon that lives on the surface of the marine sponge Axinella mexicana (Preston et al. 1996; Schleper et al. 1998). A number of distinct genotypes, which essentially cover the whole gradient of genetic relatedness from 90 to 100% ANI, were found to constitute the dominant natural population of C. symbiosum. Moreover, the genotypes appear to be very similar in terms of gene content, and the limited number of gene-content differences among them was restricted to hypothetical proteins, which is not surprising given that these genotypes probably share an overlapping ecological niche. Although more research is required to consolidate the latter assumption, e.g. it is likely that several distinct ecological niches exist within the same habitat, the C. symbiosum example indicates that a justifiable species may include strains that are much less related to each other than the 95% ANI cut-off in cases where the strains share the same gene content and ecological niche. Some of the clades of important marine groups, such as the Prochlorococcus group, which include considerable sequence but limited gene-content diversity (Coleman et al. 2006), may provide similar examples.
The importance of ecology to gene-content similarity is also evident in the reverse direction, which is nicely exemplified by comparing Burkholderia pseudomallei, an opportunistic pathogen that can be also free-living, with Burkholderia mallei, a specialized, obligatory pathogen derived from a B. pseudomallei-like ancestor. The two genomes available from these species show 98.7% ANI and have 1.7 Mb difference in their genomes, as a result of a substantial genome reduction in the B. mallei lineage, presumably caused by its change to an exclusively pathogenic lifestyle (Holden et al. 2004; Nierman et al. 2004). This example indicates that justifiable species may be found, even among strains that show higher than 98–99% ANI, when severe ecological constraints, which dramatically affect the phenotypic and ecological potential of the organism, have occurred. In summary, evolutionary relatedness coupled to ecological relatedness should eventually make a more predictive species definition than evolutionary relatedness alone (e.g. the 70% DDH standard is, essentially, just a cut-off on evolutionary relatedness).
(d) The pan-genome
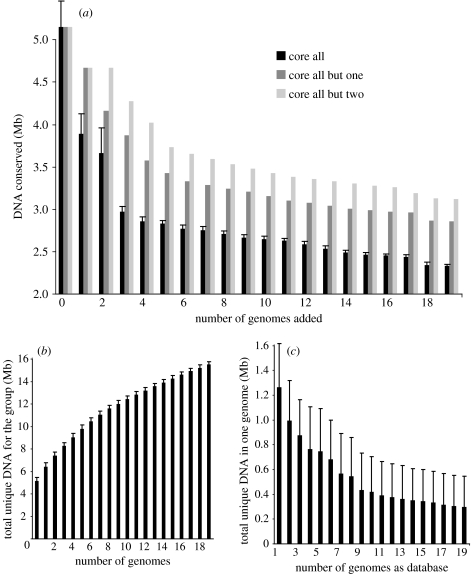
Comparisons among the genomic sequences of the best-sampled species, the E. coli–Shigella spp. (20 genomes, all showing higher than 95% ANI among themselves), revealed that the total amount of unique DNA sequence for the species is more than 15 Mb, which translates to more than 13 000 unique genes, while the total conserved sequence in all genomes is around 2.5 Mb, which is about half of the genome of a typical E. coli strain (5.1 Mb; figure 4a,b). The core genome becomes substantially larger, by approximately 0.5 Mb, when core genes are defined slightly less stringently and are allowed to be missing from one of the 20 genomes. Relaxing the stringency further, e.g. missing in 2 out of the 20 genomes, has a substantially smaller effect on the increase of the core genome (figure 4a). This trend underlines the great importance of gene deletion for intra-species differences and suggests that defining the core genome of a species as the DNA sequence that is conserved in 95% as opposed to all strains of the species (i.e. 19 out of 20 in our example) might be more appropriate. Based on this standard, the conserved core for E. coli–Shigella spp. genomes is approximately 3 Mb, which corresponds to approximately 2800 genes. Further, the total gene reservoir for this species, i.e. its pan-genome (Tettelin et al. 2005), appears unsaturated by the available 20 genomes since our calculations indicate that any new strain sequenced is expected to have a significant number of novel genes, particularly when the strain shows lower than 98–99% ANI (or different ecology?) to any other already sequenced strain (figure 4c). These findings are consistent with our previous study that used a smaller set of genomes (Konstantinidis & Tiedje 2005). Subsequent analysis of other bacterial groups, such as Streptococcus agalactiae (Tettelin et al. 2005), also revealed extensive intra-species gene diversity. These results highlight the remarkable promiscuity of bacteria in taking up DNA from outside sources and the great diversity and plasticity of bacterial species.
Figure 4.
(a) Genetic diversity within the 20 sequenced E. coli–Shigella spp. genomes. Bars show the total DNA sequence (y-axis) that remains conserved in all genomes (black bars), in all but one (dark grey), and in all but two (light grey), with the inclusion of more genomes in the analysis (x-axis). (b) The total unique DNA sequence in all genomes, (c) as well as the amount of DNA sequence that remains specific to one genome as the number of sequenced genomes increases is also shown. Therefore, (a) represents the total conserved core sequence for the group, (b) shows the total gene diversity for the group and (c) shows the amount of strain-specific (novel) DNA with increased coverage of the group with genomic sequences. Bars represent the mean of 20 different random combinations of genomes and error bars represent one standard deviation from the mean. The bars are therefore representative of the ‘average’ strain for the group, which has a genome size of approximately 5.1 Mb and shows maximum 97–98% ANI to any other strain of the group. The relatively large error bars in (c) are attributable to strains with either a very small number of novel genes (because there is another very identical (e.g. higher than 99% ANI) genome in the dataset, such as for genomes of the E. coli O157 and S. flexneri lineages), or a large number of novel genes (usually for genomes showing lower than 97% ANI to all other genomes in the dataset). The analysis was based on reciprocally best-matched conserved 1 kb fragments, using the Blast algorithm (Altschul et al. 1997), and a minimum cut-off for calling a fragment conserved of 50% identity (nucleotide level) over an alignable region of at least 700 out of the 1000 bases of the fragment. Comparable trends were observed when the analysis was restricted to complete genomes and/or coding sequences only. Therefore, the impact of the draft status of several of the genomic sequences used in the analysis on the trends shown is expected to be minor.
It is important to realize, however, that the majority of these novel genes, particularly when strains show higher than 98% ANI to already sequenced strains (e.g. the E. coli case; figure 2) or species with narrow ecological niches (Streptococci), involve, by and large, hypothetical and mobile genes. Therefore, in the cases described here, the majority (but probably not all, because some genes could be ecologically important; Boyd & Brussow 2002) of the intra-species gene-variable pool may consist of ephemeral intruders of the genome (since they are not evolutionarily conserved), that are probably continuously recycled and, presumably, have minor, if any, ecological importance. Therefore, the total gene reservoir of species may appear mathematically infinite (Tettelin et al. 2005) but, in practical terms, it may approach saturation with a few genomic sequences for ecologically uniform species. Consistent with this interpretation, Tettelin et al. (2005) predicted that there would be less than 50 novel genes in the next (eighth) genome of S. agalactiae, which appears to be an ecologically and phenotypically uniform species (2.1–2.2 Mb is the average genome size for this species). However, these gene differences may simply be the presence of a transient prophage genome. On the other hand, the number of novel genes in the next (21st) E. coli genome is expected to be approximately 300 when this genome shows lower than 98% ANI to any other E. coli genome (figure 4c). This constitutes a large genetic difference and indicates that the 20 genomes of E. coli–Shigella spp. are probably too heterogeneous to be considered the same species.
In conclusion, some, even substantial, gene-content differences may be observable within currently named species and this might continue to be the case even if more stringent standards for species are adopted. The importance of these genes for splitting a species, however, should be determined by evaluating their ecological and phenotypic potential for the organism as well as potential genetic discontinuity between strains of the species (see also clusters in §3).
(e) What is an ecotype and what is a ‘species’?
Although the ecotype is not recognized as an official rank of bacterial taxonomy (Brenner et al. 2000), the concept of ecotype is important for microevolutionary studies because it describes a collection of strains that shows some level of ecological distinctiveness within its species (Schloter et al. 2000; Cohan 2001). In other words, ecotypes preserve the full, or nearly full, phenotypic and ecological potential that characterizes the species and are able to exploit only a slightly different ecological niche compared with its species, such as growth (or loss of growth) on a new carbon substrate or the ability to cause increased/decreased disease symptoms for a pathogenic species. An ecotype would therefore be expected to have only a small gene-content (or expression) difference compared with other ecotypes of the same species, or if larger differences exist, they would be carried by unstable parts of the genome such as plasmids. Previous attempts to elucidate the genetic basis of the ecological distinctiveness of ecotypes were hampered by the fact that ecotypes were discovered by the sequence analysis of a single or a few genes in the genome (Cohan 2001), which were probably irrelevant to the ecological distinctiveness, or by an observable phenotypic property without elucidating the underlying genetic elements responsible for the property (Schloter et al. 2000). Therefore, advancing the concept of ecotype would require a solid understanding of the genetic basis of the ecological distinctiveness of the ecotype and distinguishing the latter from the ecological distinctiveness of the species. Genome sequencing may be particularly helpful in this context because it reveals, at a very fine scale, the full genetic potential of an organism.
Our analysis reveals that justifiable ecotypes are observable among homogeneous groups of strains (e.g. Streptococci), even among almost identical strains of the E. coli and Salmonella groups. For example, several of the sequenced Salmonella strains that show higher than 98.5% ANI among themselves are characterized by different levels of pathogenicity, e.g. typhi versus typhimurium pathovars, and our genomic comparisons reveal that there are small gene-content and plasmid DNA differences among these strains which could account for their pathogenic differentiation (figures 1b and 2). The almost identical genomes of Bacillus anthracis, Bacillus cereus (Hoffmaster et al. 2004) or Yersinia pestis (Chain et al. 2004) provide similar examples as well. In contrast, the E. coli genomes that show large gene-content differences (and typically lower than 98% ANI to any other E. coli genome), or the B. mallei in the previous example, cannot be considered as ecotypes of the same species or B. pseudomallei, respectively. Considering such genomes as different species appears more appropriate given their extensive functional and, presumably, ecological differentiation. Ecotypes may also be observable among strains that may have identical gene content but show a slight ecological differentiation as a result of gene-expression differences caused by environmental adaptations or sequence divergence, like in the Streptococci or Staphylococci groups.
3. Are there clusters or a continuum of diversity?
An important issue that remains unresolved is whether bacteria exhibit a genetic continuum in nature, which is not supportive of a transparent species definition, or whether there are coherent sequence/genomic clusters, as are evident for most eukaryotic organisms. Clusters are typically defined as collections of strains that are more related, e.g. in terms of sequence identity and gene content, among themselves than to strains outside the cluster. It is possible that some environments support clusters whereas others do not. For instance, bacteria that evolve in close association with a host, e.g. obligate pathogens or symbionts, and exhibit limited dispersal hence mixing outside the host, are likely to exhibit well-defined genetic clusters as an effect of their coevolution with their host (allopatric speciation). This scenario may be generally applicable to bacteria that have a very narrow ecological niche and their dispersal between inhabitable niches is restricted by survival and/or distance factors. Bacterial (or archaeal) species living in isolated, hyperthermal lakes such as Sulfolobus spp. (Papke et al. 2003; Whitaker et al. 2003) may be examples of this kind. In contrast, environments like soil, with great complexity, mixing and slow growth rates (Grey & Williams 1971), which would facilitate mixing of populations before the divergence of populations takes place, may support a continuum of diversity (the importance of selection is discussed below). Accordingly, bacterial groups that are able to exploit a variety of ecological niches and can disperse more freely between their habitable niches (like many soil bacteria) are more likely to exhibit a continuum of diversity than coherent sequence clusters (for a more extensive discussion of bacterial species biogeography see Martiny et al. (2006) and Ramette & Tiedje (in press)).
The data from the five groups evaluated here appear to support this model. For example, the obligatory pathogenic species of Staphylococci and Streptococci with a (presumably) narrow ecological niche, which is also evident by their small genome size, appear to exhibit clear genomic clusters in terms of ANI relatedness (see circles, figure 1a). Unless sampling biases (see §3a) have obscured our results, the clusters observed for these groups are unequivocal owing to the high precision of the ANI measurement. On the contrary, the environmental Shewanella and Burkholderia species appear to form a continuum of genetic diversity in the region that corresponds to distinct clusters in the Staphylococci and Streptococci groups or to the 70% DDH standard (figure 1a). The Enterobacteriaceae group also appears to show distinct clusters as evidenced by the comparison between E. coli and its closest sequenced relative, the Salmonella spp. (figure 1a). The recent description of Escherichia albertii (Huys et al. 2003), a new species that probably spans the genetic gap between E. coli and Salmonella (Hyma et al. 2005), as well as analysis of environmental E. coli isolates (Byappanahalli et al. 2006; Ishii et al. 2006) indicate, however, that the genomic clusters recovered in this group may be the effect of sampling biases rather than naturally occurring clusters. The latter is also more consistent with our prediction that E. coli, owing to its opportunistic lifestyle—namely the possibility to thrive in humid soils and freshwater systems (Ishii et al. 2006) and in a variety of animals—as well as its average-to-large genome size, is more likely to show a continuum of diversity as opposed to genomic clusters. Definitely, a better understanding of the breadth of the ecological niche and diversity of even the best-studied bacterium is still required.
(a) Clusters as an effect of biased sampling
The E. coli example demonstrates, however, the need for extensive and unbiased sampling. Sampling biases may have indeed confounded several studies that claimed to have recovered distinct genomic clusters from surveys of a large number of strains, including the E. coli group. First, most of these surveys involved pathogenic organisms where there is a strong bias towards characterizing isolates that cause certain symptoms in patients. This pool of strains, however, is unlikely to be representative of the naturally occurring genetic diversity, particularly in cases where pathogenicity factors are carried in mobile genetic elements, have only recently evolved in the lineage, or involve only a few of the genes in the genome. In all these cases, it is very likely to observe a population burst of one, or a few, genotype(s), which may obscure the presence of other genotypes. Even, in cases where asymptomatic patients and/or healthy individuals are sampled, there is usually a biased prescreening of the isolates, either owing to the selectivity of the isolation method or the requirement to conform to specific ‘species standards’ in order for an isolate to be further processed. Atypical or rare isolates, which are likely to span the continuum of diversity between groups with distinct symptoms or phenotypes, are usually discarded before further analysis. Lastly but very importantly, even if sequenced-based clusters are identifiable, this would not guarantee that those clusters encompass phenotypically or ecologically uniform strains, as this is evident in the E. coli group (figures 1b and 4). In fact, when comparing strains that are well separated by evolutionary time (e.g. E. coli versus Salmonella; figure 1b), clear clusters will be always observable but are not necessarily meaningful for the species definition. Clusters that encompass highly uniform strains and are associated with a distinct phenotype or ecological behaviour are the desirable units. Such clusters may be observable within the Streptococci and Staphylococci groups (figure 1b).
4. Implications for the species concept
The species concept is the theoretical framework that attempts to describe what bacterial species are and explain how they are formed; the species definition is how the concept is exercised in practice. Therefore, advancing the species concept is essential for a better species definition. The present working hypothesis for the bacterial species concept is that bacteria form coherent genomic clusters that are characterized by distinctive phenotypic properties (for theories and reviews on the species concept, see the following excellent references: Templeton (1989), Dykhuizen & Green (1991), Rossello-Mora & Amann (2001), Cohan (2002) and Gevers et al. (2005)). The clusters are thought to be created as the effect of two major forces, selection and recombination (geographical isolation, which has also been advocated as cohesive force for speciation was previously mentioned). The most favourable species concepts for bacteria presently in practice, the biological (Mayr 1997) and the phylogenetic (Hull 1997) concepts (the recently proposed ecotype concept (Cohan 2002) can be seen as a variation of the phylogenetic concept), are essentially based on the differential weighting of the importance of these processes.
The amount of genomic evidence that can be actually used to quantify the importance of selection and recombination for the species concept is currently too limited to allow for robust conclusions. Several trends have started to emerge, however, and are summarized later. Although selection is probably strong enough to purge diversity in well-mixed, stable and nutrient-rich environments, like inside host cells or under laboratory conditions, this may not be the case in environments with great heterogeneity and slow growth. Consistent with this interpretation, Hallam et al. (submitted) recovered a range of genotypes of C. symbiosum living in the complex surface of its sponge host, and some of the genotypes may have diverged several million years ago based on their sequence difference. In contrast, the metagenomic analysis of the natural assemblage of the acid mining drainage (AMD) biofilm, a less complex and higher cell biomass environment, recovered more homogeneous populations (Tyson et al. 2004). The genetic continuum seen in the Burkholderia and Shewanella groups (figure 1), which typically live in more complex environments, is also in agreement with the previous interpretation. Further technological improvements on detecting selection and assembling single genotypes from the environment as well as sampling the natural populations over time will be required to further advance understanding on how selection is actually acting on and shaping bacterial populations.
Recombination on the other hand is thought to occur more frequently within species (whatever is the actual species unit for recombination) than between species, and this difference in the rate of recombination is considered to be a cohesive force responsible for the creation and maintenance of species. This assumption is essentially based on a limited number of laboratory experiments that have shown a gradual decrease in the efficiency of recombination with larger sequence difference between the recombining parts (Vulic et al. 1997; Majewski et al. 2000). If recombination rate is falling gradually with increasing sequence divergence and there are no clear cut-offs in terms of sequence identity to this process, then recombination cannot be informative for the species definition. Furthermore and despite efforts to infer the natural rate and magnitude of recombination based on the sequencing of a limited number of genes in the genome (Feil et al. 2001; Spratt et al. 2001), the recombination rate, at the whole-genome level, for most bacterial groups is essentially unknown. The findings from a few recent metagenomic surveys are not conclusive either. For instance, Tyson et al. (2004) and Nesbo et al. (2006) observed high and genome-wide recombination between genotypes of the dominating archaeon population in the AMD biofilm and the hyperthermophilic bacteria of genus Thermotoga, respectively, but Hallam et al. (submitted) did not observe similar rates in a different archaeon living on sponges. The latter study, as well as several comparative studies of available whole-genome sequences of Staphylococcus aureus species (Hughes & Friedman 2005), suggests that the observable recombination is too functionally and spatially restricted in the genome to represent a major cohesive force for species. The lesson from these pilot studies is that studying recombination within the same environment or ecological niche will be particularly fruitful in the future. In all cases, however, it appears that there is presently no easy way to incorporate selection and recombination in the species concept without first advancing our understanding of their rate and importance for natural bacterial populations. For these reasons, the current species definition appears more pragmatic and operational than a definition that attempts to materialize the current, limited knowledge about selection and recombination.
5. Gene expression and lessons from eukaryotic organisms
In addition to gene presence, gene expression plays an important role for the phenotypic or ecological potential of an organism. Different gene regulation or expression levels for genes that are shared between organisms may constitute another level of differentiation (and thus speciation) between organisms. Current knowledge on the importance of these processes is too limited to be summarized here, while studying these processes will require further technological developments. The recent findings from genomic projects of higher eukaryotic organisms may be particularly relevant or at least stimulating for bacteria as well. Most prominently, the sequencing project of our closest relative, the chimpanzee, revealed that we share, on average, approximately 98.7% nucleotide identity in the common parts of the genome while only 3% of the genome differs (Chimpanzee Consortium 2005). Accordingly, it appears that differential gene expression during development, as opposed to gene-content differences, is more important for the morphological difference between human and the chimpanzee (Enard et al. 2002). Although the organization of the cells of the higher eukaryotes is very complex compared with that of bacterial cells, this finding draws attention to the possibility that expression differences may be very important among strains of the same bacterial species. In any case, it is worth noting that strains, which show higher than 98% ANI, are expected to show minimum gene expression and regulation differences because there is, presumably, a limited number of non-neutral point mutations, e.g. our analyses show that, typically, 60–70% of the nucleotide changes are synonymous at this level. Accordingly, the gene presence should determine, by and large, the phenotypic differences among strains related at this level. Therefore, relatively more attention should probably be given to the genome as opposed to the transcriptome for such strains.
6. Advancing methods for studying species diversity
Another important issue related to the species definition is the robustness of the methods employed to characterize bacterial strains and their genetic diversity. Although there is usually sufficient understanding of the relative limitations and advantages of the available methods, their absolute robustness as well as how to contrast and compare datasets derived by different methods remain more obscure. Genomic sequences can assist towards this direction because they offer resolution at any level and can be used as the unequivocal reference standard upon which methods can be evaluated and calibrated. Our work with the DDH method as well as recent studies with single nucleotide polymorphism (SNP; Alland et al. 2003; Pearson et al. 2004; Filliol et al. 2006) are examples of this kind. SNP analysis, however, cannot be generically applied in different groups of bacteria without a substantial amount of prior research to identify the informative SNP sites, and therefore its importance as a standard method for species definition may be limited.
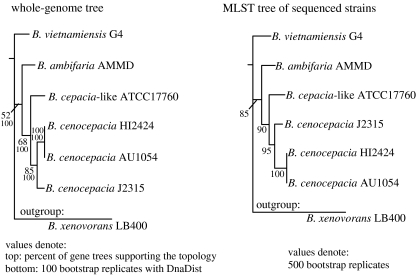
One of the methods that has gained increasing popularity for performing intra-species diversity studies but was never compared to whole-genome-level relatedness is multilocus sequence typing (MLST; Maiden et al. 1998). We performed a pilot evaluation of the MLST method by comparing the whole-genome-based phylogeny of seven available genomes of Burkholderia strains with the MLST-based phylogeny, which employed the full sequences of seven genes that are commonly used for MLST applications in the Burkholderiaceae group (Baldwin et al. 2005). Our results show that the MLST-based phylogeny is very congruent with the whole-genome-based phylogeny (figure 5) and this appears to be independent of the genes used, i.e. different combinations of seven genes give equally robust phylogenies. The genomes compared, however, typically show lower than 95% ANI among themselves; therefore, these results are relevant for inter-species comparisons. When we performed a similar analysis with genomes showing higher than 95% identity, the congruence between the derived phylogenies was substantially lower, albeit still statistically significant. It appears that the selection of the genes is more important for studies that target intra-species diversity, since many genes show weak phylogenetic signal at this level (Konstantinidis et al. submitted). However, this level of diversity is more interesting to study with respect to the species definition. With the increasing availability of genomic sequence, it is now possible to identify the best-performing genes (or methods) for phylogenetic purposes via comparisons with the whole-genome-derived phylogenies, and hence advance the methods and their accuracy.
Figure 5.
Whole-genome-based evaluation of the MLST method for the Burkholderiaceae group. The whole-genome tree was constructed based on the concatenated alignment of 2589 genes that are shared in all seven Burkholderia genomes, using the dnaML version of the Phylip package (Felsenstein 2004). The MSLT tree is based on the concatenated alignments of the seven full-length genes that are frequently used in MLST applications in the Burkholderia group (i.e. recA, gyrB, lepA, phaC, trpB, gltB and atpD; Baldwin et al. 2005) and was built with the same method as the whole-genome tree.
7. Summary and recommendations
Despite the very attractive attempts to reconcile the eukaryotic and bacterial species under the same biological species concept or its variations (Templeton 1989; Hull 1997), we believe that bacteria exhibit biology which is too different compared with eukaryotic organisms, even the asexual ones, for this to be fruitful. Not only that, but our understanding of bacterial biology is only now starting to advance satisfactorily towards this direction. For example, the great promiscuity of bacteria to take up genetic material from outside sources, as evident in the 20 genomic sequences of E. coli available, as well as the initial evidence that suggest that bacteria may frequently show a continuum of genetic diversity in nature, are poles apart from the norms in eukaryotic biology.
More and higher resolution sampling is required to allow for more definitive conclusions to be drawn. For instance, although our preliminary results with other bacterial groups indicate that the results for the E. coli group represented here (figures 2 and 4) may be more universally applicable for free-living bacteria, this remains to be more fully assessed. Especially needed are data on whether coherent clusters exist in nature for bacteria and what part of the gene/genetic diversity within the clusters may constitute junk or neutral DNA. Alternative methodologies to whole-genome sequencing, like DNA–DNA microarrays or improved MLST approaches (Konstantinidis et al. submitted), or increased pace in genome sequencing, offer promise that the needed datasets will soon be available.
Another major conclusion from the results presented here (e.g. figure 1) is that, even with the whole genomic sequences available, the decision when to split species to produce more species, or what are the boundaries between a species and its ecotypes, may not always be unequivocal. In fact, our discussion of some of the results, e.g. the differences between small and medium size genomes, was intentionally stretched for demonstration purposes. The reality is that given the overwhelming diversity of bacterial species revealed, there will be always ‘grey areas’ of decision related to the species definition, where practical versus completely standardized criteria would be more useful. Besides, classifying strains into species is probably very artificial compared to what is ongoing in nature.
Currently, it appears more pragmatic and efficient to preserve the current species definition than to replace it, because it is serviceable as a first level of screening and current phylogenetic knowledge is too limited for a universal and sound change in the definition. More stringent standards could, however, be adopted when there is sufficient understanding of important ecological differences among closely related organisms, e.g. those showing higher than 95% ANI. Related to this, a measure of standardized or ‘absolute’ relatedness to the type strain(s) of a species, measured by ANI or ANI-predicting methods like an optimized MLST application (Konstantinidis et al. submitted), as well as the place of isolation, are very important information and should accompany any newly described isolate. This strategy would be also very helpful in the ‘grey areas’ of relatedness because, while it does not require a decision about species designation to be made, it gives the full perspective of the genetic (and potentially ecological) relatedness of the isolate to what is already known. The fact that ANI is, first, a simple, robust and pragmatic measure for all bacteria at the species level and probably up to the family level, and second, that it overcomes many of the limitations of traditional methods greatly magnifies its importance and potential for such finer-scale systematic studies.
Acknowledgments
We thank the Institute for Genomic Research (TIGR) and the Sanger Center for permission to use preliminary sequence data. This work was supported by the National Science Foundation (awards DEB0516252 and DEB-00755564), the DOE's Genomics:GTL Program and the Center for Microbial Ecology. K.T.K. is grateful to the Bouyoukos Fellowship Program for supporting his Ph.D. studies in the laboratory of J.M.T.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Species and speciation in micro-organisms’.
Present address: Max Planck Institute for Marine Microbiology, Celsiusstrasse 1, Bremen 28359, Germany.
References
- Alland D, et al. Modeling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J. Bacteriol. 2003;185:3392–3399. doi: 10.1128/JB.185.11.3392-3399.2003. doi:10.1128/JB.185.11.3392-3399.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F, Madden T.L, Schaffer A.A, Zhang J, Zhang Z, Miller W, Lipman D.J. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. doi:10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin A, et al. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 2005;43:4665–4673. doi: 10.1128/JCM.43.9.4665-4673.2005. doi:10.1128/JCM.43.9.4665-4673.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd E.F, Brussow H. Common themes among bacteriophage-encoded virulence factors and diversity among the bacteriophages involved. Trends Microbiol. 2002;10:521–529. doi: 10.1016/s0966-842x(02)02459-9. doi:10.1016/S0966-842X(02)02459-9 [DOI] [PubMed] [Google Scholar]
- Brenner D, Staley J, Krieg N. Classification of prokaryotic organisms and the concept of Bacterial speciation. Springer; New York, NY: 2000. Bergey's manual of systematic bacteriology. [Google Scholar]
- Byappanahalli M.N, Whitman R.L, Shively D.A, Sadowsky M.J, Ishii S. Population structure, persistence, and seasonality of autochthonous Escherichia coli in temperate, coastal forest soil from a Great Lakes watershed. Environ. Microbiol. 2006;8:504–513. doi: 10.1111/j.1462-2920.2005.00916.x. doi:10.1111/j.1462-2920.2005.00916.x [DOI] [PubMed] [Google Scholar]
- Chain P.S, et al. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl Acad. Sci. USA. 2004;101:13 826–13 831. doi: 10.1073/pnas.0404012101. doi:10.1073/pnas.0404012101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Consortium. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. doi:10.1038/nature04072 [DOI] [PubMed] [Google Scholar]
- Cohan F.M. Bacterial species and speciation. Syst. Biol. 2001;50:513–524. doi: 10.1080/10635150118398. [DOI] [PubMed] [Google Scholar]
- Cohan F.M. What are bacterial species? Annu. Rev. Microbiol. 2002;56:457–487. doi: 10.1146/annurev.micro.56.012302.160634. doi:10.1146/annurev.micro.56.012302.160634 [DOI] [PubMed] [Google Scholar]
- Coleman M.L, Sullivan M.B, Martiny A.C, Steglich C, Barry K, Delong E.F, Chisholm S.W. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. doi:10.1126/science.1122050 [DOI] [PubMed] [Google Scholar]
- Dykhuizen D.E, Green L. Recombination in Escherichia coli and the definition of biological species. J. Bacteriol. 1991;173:7257–7268. doi: 10.1128/jb.173.22.7257-7268.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard W, et al. Intra- and interspecific variation in primate gene expression patterns. Science. 2002;296:340–343. doi: 10.1126/science.1068996. doi:10.1126/science.1068996 [DOI] [PubMed] [Google Scholar]
- Feil E.J, et al. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl Acad. Sci. USA. 2001;98:182–187. doi: 10.1073/pnas.98.1.182. doi:10.1073/pnas.98.1.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein, J. 2004 Phylip (Phylogeny Inference Package) version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle
- Filliol I, et al. Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of other DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J. Bacteriol. 2006;188:759–772. doi: 10.1128/JB.188.2.759-772.2006. doi:10.1128/JB.188.2.759-772.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity G, Bell J, Lilburn T. Springer; New York, NY: 2004. Bergey's manual of systematic bacteriology. [Google Scholar]
- Gevers D, et al. Opinion: re-evaluating prokaryotic species. Nat. Rev. Microbiol. 2005;3:733–739. doi: 10.1038/nrmicro1236. doi:10.1038/nrmicro1236 [DOI] [PubMed] [Google Scholar]
- Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P. & Tiedje, J. M. In press. DNA–DNA hybridization values and their relation to whole genome sequence similarities, Int. J. Syst. Evol. Microbiol [DOI] [PubMed]
- Grey T, Williams S. Microbial productivity in soil. Symp. Soc. Gen. Microbiol. 1971;21:255–286. [Google Scholar]
- Hallam, S. J., Konstantinidis, K. T., Brochier, C., Putnam, N., Schleper, C., Preston, C. M., de la Torre, J., Richardson, P. M. & DeLong, E. F. Submitted. Genomic analysis of a symbiotic marine crenarchaeon, Cenarchaeum symbiosum [DOI] [PMC free article] [PubMed]
- Hoffmaster A.R, et al. Identification of anthrax toxin genes in a Bacillus cereus associated with an illness resembling inhalation anthrax. Proc. Natl Acad. Sci. USA. 2004;101:8449–8454. doi: 10.1073/pnas.0402414101. doi:10.1073/pnas.0402414101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holden M.T, et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl Acad. Sci. USA. 2004;101:14 240–14 245. doi: 10.1073/pnas.0403302101. doi:10.1073/pnas.0403302101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes A.L, Friedman R. Nucleotide substitution and recombination at orthologous loci in Staphylococcus aureus. J. Bacteriol. 2005;187:2698–2704. doi: 10.1128/JB.187.8.2698-2704.2005. doi:10.1128/JB.187.8.2698-2704.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull D.L. The ideal species concept—and why we can't get it. Chapman and Hall; London, UK: 1997. Species: the units of biodiversity. [Google Scholar]
- Huys G, Cnockaert M, Janda J.M, Swings J. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int. J. Syst. Evol. Microbiol. 2003;53:807–810. doi: 10.1099/ijs.0.02475-0. doi:10.1099/ijs.0.02475-0 [DOI] [PubMed] [Google Scholar]
- Hyma K.E, Lacher D.W, Nelson A.M, Bumbaugh A.C, Janda J.M, Strockbine N.A, Young V.B, Whittam T.S. Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J. Bacteriol. 2005;187:619–628. doi: 10.1128/JB.187.2.619-628.2005. doi:10.1128/JB.187.2.619-628.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S, Ksoll W.B, Hicks R.E, Sadowsky M.J. Presence and growth of naturalized Escherichia coli in temperate soils from Lake Superior watersheds. Appl. Environ. Microbiol. 2006;72:612–621. doi: 10.1128/AEM.72.1.612-621.2006. doi:10.1128/AEM.72.1.612-621.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.H, Harrison S.H, Herring P.A. A theoretical limit to coding space in chromosomes of bacteria. Omics. 2002;6:115–121. doi: 10.1089/15362310252780861. doi:10.1089/15362310252780861 [DOI] [PubMed] [Google Scholar]
- Konstantinidis K.T, Tiedje J.M. Trends between gene content and genome size in prokaryotic species with larger genomes. Proc. Natl Acad. Sci. USA. 2004;101:3160–3165. doi: 10.1073/pnas.0308653100. doi:10.1073/pnas.0308653100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis K.T, Tiedje J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl Acad. Sci. USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. doi:10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis, K. T., Ramette, A. & Tiedje, J. M. In press. Genomic evaluations and improvements on single and multi locus sequence typing methods for studying intra-species diversity. J. Appl. Environ. Microbiol
- Lerat E, Ochman H. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res. 2005;33:3125–3132. doi: 10.1093/nar/gki631. doi:10.1093/nar/gki631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden M.C, et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl Acad. Sci. USA. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. doi:10.1073/pnas.95.6.3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majewski J, Zawadzki P, Pickerill P, Cohan F.M, Dowson C.G. Barriers to genetic exchange between bacterial species: Streptococcus pneumoniae transformation. J. Bacteriol. 2000;182:1016–1023. doi: 10.1128/jb.182.4.1016-1023.2000. doi:10.1128/JB.182.4.1016-1023.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiny J.B, et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol. 2006;4:102–112. doi: 10.1038/nrmicro1341. doi:10.1038/nrmicro1341 [DOI] [PubMed] [Google Scholar]
- Mayr E. Columbia University Press; New York, NY: 1997. Systematics and the origin of species from the viewpoint of a zoologist. [Google Scholar]
- Moran N.A. Microbial minimalism: genome reduction in bacterial pathogens. Cell. 2002;108:583–586. doi: 10.1016/s0092-8674(02)00665-7. doi:10.1016/S0092-8674(02)00665-7 [DOI] [PubMed] [Google Scholar]
- Nesbo C.L, Dlutek M, Doolittle W.F. Recombination in Thermotoga: implications for species concepts and biogeography. Genetics. 2006;172:759–769. doi: 10.1534/genetics.105.049312. doi:10.1534/genetics.105.049312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman W.C, et al. Structural flexibility in the Burkholderia mallei genome. Proc. Natl Acad. Sci. USA. 2004;101:14 246–14 251. doi: 10.1073/pnas.0403306101. doi:10.1073/pnas.0403306101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Davalos L.M. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. doi:10.1126/science.1119966 [DOI] [PubMed] [Google Scholar]
- Papke R.T, Ramsing N.B, Bateson M.M, Ward D.M. Geographical isolation in hot spring cyanobacteria. Environ. Microbiol. 2003;5:650–659. doi: 10.1046/j.1462-2920.2003.00460.x. doi:10.1046/j.1462-2920.2003.00460.x [DOI] [PubMed] [Google Scholar]
- Pearson T, et al. Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc. Natl Acad. Sci. USA. 2004;101:13 536–13 541. doi: 10.1073/pnas.0403844101. doi:10.1073/pnas.0403844101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston C.M, Wu K.Y, Molinski T.F, DeLong E.F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc. Natl Acad. Sci. USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. doi:10.1073/pnas.93.13.6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramette, A. N. & Tiedje, J. M. In press. Biogeography: an emerging cornerstone for understanding prokaryotic diversity, ecology, and evolution. Microb. Ecol [DOI] [PubMed]
- Rosselló-Mora R. DNA–DNA reassociation methods applied to microbial taxonomy and their critical evaluation. Springer; Berlin, Germany: 2006. Molecular identification, systematics, and population structure of prokaryotes. [Google Scholar]
- Rossello-Mora R, Amann R. The species concept for prokaryotes. FEMS Microbiol. Rev. 2001;25:39–67. doi: 10.1111/j.1574-6976.2001.tb00571.x. doi:10.1111/j.1574-6976.2001.tb00571.x [DOI] [PubMed] [Google Scholar]
- Schleper C, DeLong E.F, Preston C.M, Feldman R.A, Wu K.Y, Swanson R.V. Genomic analysis reveals chromosomal variation in natural populations of the uncultured psychrophilic archaeon Cenarchaeum symbiosum. J. Bacteriol. 1998;180:5003–5009. doi: 10.1128/jb.180.19.5003-5009.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloter M, Lebuhn M, Heulin T, Hartmann A. Ecology and evolution of bacterial microdiversity. FEMS Microbiol. Rev. 2000;24:647–660. doi: 10.1111/j.1574-6976.2000.tb00564.x. doi:10.1111/j.1574-6976.2000.tb00564.x [DOI] [PubMed] [Google Scholar]
- Siew N, Fischer D. Analysis of singleton ORFans in fully sequenced microbial genomes. Proteins. 2003;53:241–251. doi: 10.1002/prot.10423. doi:10.1002/prot.10423 [DOI] [PubMed] [Google Scholar]
- Spratt B.G, Hanage W.P, Feil E.J. The relative contributions of recombination and point mutation to the diversification of bacterial clones. Curr. Opin. Microbiol. 2001;4:602–606. doi: 10.1016/s1369-5274(00)00257-5. doi:10.1016/S1369-5274(00)00257-5 [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel B.M. Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 1994;44:846–849. [Google Scholar]
- Stackebrandt E, et al. Report of the ad hoc committee for the re-evaluation of the species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. doi:10.1099/ijs.0.02360-0 [DOI] [PubMed] [Google Scholar]
- Stahl, D. A. & Tiedje, J. M. 2002 Microbial ecology and genomics: a crossroads of opportunity. American Society for Microbiology, Colloquia reports. Available at: http://www.asm.org/Academy/index.asp?bid=2124 [PubMed]
- Staley J.T. Speciation and bacterial phylospecies. ASM Press; Washington, DC: 2004. Micobial diversity and bioprospecting. [Google Scholar]
- Templeton A.R. The meaning of species and speciation: a genetic prespective. Sinauer Associates; Sunderland, MA: 1989. Speciation and its consequences. [Google Scholar]
- Tettelin H, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc. Natl Acad. Sci. USA. 2005;102:13 950–13 955. doi: 10.1073/pnas.0506758102. doi:10.1073/pnas.0506758102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson G.W, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. doi:10.1038/nature02340 [DOI] [PubMed] [Google Scholar]
- Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vulic M, Dionisio F, Taddei F, Radman M. Molecular keys to speciation: DNA polymorphism and the control of genetic exchange in enterobacteria. Proc. Natl Acad. Sci. USA. 1997;94:9763–9767. doi: 10.1073/pnas.94.18.9763. doi:10.1073/pnas.94.18.9763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward D.M. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1998;1:271–277. doi: 10.1016/s1369-5274(98)80029-5. doi:10.1016/S1369-5274(98)80029-5 [DOI] [PubMed] [Google Scholar]
- Wayne L.G, et al. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 1987;37:463–464. [Google Scholar]
- Whitaker R.J, Grogan D.W, Taylor J.W. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. doi:10.1126/science.1086909 [DOI] [PubMed] [Google Scholar]
- Whitman W.B, Coleman D.C, Wiebe W.J. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. doi:10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]